ABSTRACT
The membrane origins of autophagosomes have been a key unresolved question in the field. The earliest morphologically recognizable structure in the macroautophagy/autophagy itinerary is the double-membraned cup-shaped phagophore. Newly formed phosphatidylinositol 3-phosphate (PtdIns3P) on the membranes destined to become phagophores recruits WIPI2, which, in turn, binds ATG16L1 to define the sites of autophagosome formation. Here we review our recent study showing that membrane recruitment of WIPI2 requires coincident detection of PtdIns3P and RAB11A, a protein that marks recycling endosomes. We found that multiple core autophagy proteins are more tightly associated with the recycling endosome compartment than with endoplasmic reticulum (ER)-mitochondrial contact sites. Furthermore, biochemical isolation of the recycling endosomes confirmed that they recruit autophagy proteins. Finally, fixed and live-cell imaging data revealed that recycling endosomes engulf autophagic substrates. Indeed, the sequestration of mitochondria after mitophagy stimulation depends on early autophagy regulators. These data suggest that autophagosomes evolve from the RAB11A compartment.
Autophagy involves the sequestration of cytoplasmic contents into phagophores that mature into double-membraned autophagosomes, which ultimately fuse with lysosomes to enable degradation of the autophagic contents. Autophagosomes derive from cup-shaped, double-membraned precursors called phagophores, whose membrane origins have been a key unresolved issue in the field.
Conceptually, phagophores could either be formed de novo by membranes from multiple sources, or derive predominantly from one donor platform compartment, possibly with inputs from additional diverse sources. Most of the literature considering this question has focused on the identification of intracellular compartments that have membrane contacts with phagophores and have reported such interactions with the ER and ER-mitochondrial contract sites. This has led to the prevailing view that phagophores derive from such ER-associated compartments. However, phagophores have contacts with many other compartments, including endosomes. Importantly, if phagophores evolved from a donor platform distinct from the ER or ER-mitochondrial contact sites, then one may still see ER-phagophore contacts if such events were occurring at a later stage in the autophagosome biogenesis itinerary. As a crude analogy, if one were studying the origins of late endosomes, then one would be wary of claiming that they originated from compartments with which they shared membrane contacts, such as the ER – and indeed, late endosomes evolve from early endosomes with which they do not have frequent contacts.
In 2013, we reported that vesicles carrying different autophagy proteins, ATG9 and ATG16L1, travel via different endocytic routes from distinct sites on the plasma membrane and meet in recycling endosomes. We, and others, assumed that these ATG proteins then trafficked from the recycling endosomes to the site of phagophore formation. Now, our data suggest that the RAB11A-positive compartments (recycling endosomes) are indeed the membranes from which phagophores evolve [Citation1].
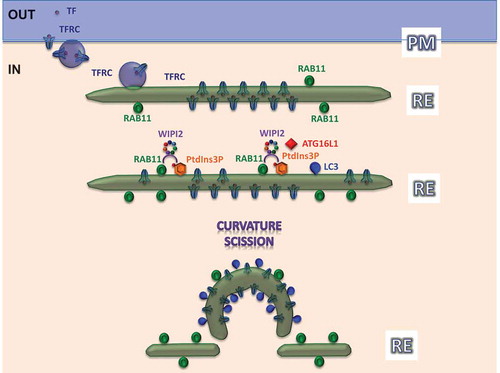
We reached this conclusion based on several observations. The recruitment of WIPI2 to phagophore precursor membranes is a key step in autophagosome biogenesis, as WIPI2 binds ATG16L1 and the latter protein dictates where autophagosomes form. WIPI2 binds membranes enriched in PtdIns3P. Because this lipid is also associated with membranes that are not primary platforms for autophagosome biogenesis (i.e., early/late endosomes), we hypothesized that WIPI2 membrane binding was mediated both by PtdIns3P as well as a protein-protein interaction. Such so-called coincident detection is a theme for many protein-phosphoinositide interactions. We found that WIPI2 membrane association requires an interaction with RAB11A, in addition to PtdIns3P. This WIPI2-RAB11A interaction is critical for phagophore formation. RAB11A is a marker for recycling endosomes, and WIPI2 also colocalizes with other markers for this compartment, such as RAB10 or TFRC (transferrin receptor). The importance of this interaction was underlined when we artificially localized RAB11A to early endosomes, a PtdIns3P-enriched compartment where phagophores do not normally form. This experiment caused WIPI2 recruitment to early endosomes.
Compatible with normal WIPI2 localization to recycling endosomes, we observed that key proteins involved in the early (WIPI2, ZFYVE1/DFCP1, ATG14 and BECN1/Beclin 1), and later (Atg8-family members) steps of autophagosome formation localize to the same compartment, and these associations are resistant to cytoskeletal disruption. While such proteins also show apparent colocalization with ER-mitochondrial contact sites, these associations are largely lost when the cytoskeleton is disrupted. This suggests that the autophagy proteins are associated with the recycling endosomes while the apparent colocalizations with ER-mitochondrial contact sites (which have been the dogma in the field) are either weaker, or are simply the consequence of the ER being essentially everywhere in the cell. Our experiments suggest that WIPI2 association with the recycling endosomes occurs at the sites that are subsequently conjugated with LC3 and that WIPI2 leaves the membranes after LC3 attaches.
When autophagy is induced (serum and amino acid starvation or SMER28 stimulation), TFRC accumulates on recycling endosomes on LC3-positive structures. Using different electron microscopy techniques, we demonstrated that these structures are double-membraned autophagosomes. The extracellular domain of TFRC (as a transmembrane protein) and its endocytosed ligand, TF (transferrin), which traffic to recycling endosomes particularly during starvation, are concentrated between the inner and outer autophagosome membranes, thus providing additional support and clues as to how recycling endosomes may evolve into phagophores (). The functional nature of these structures is supported by the autophagy-dependent degradation of TFRC. We used these observations to develop a new approach to biochemically isolate recycling endosomes using TF conjugated with iron beads (TF-Ferrofluid assay). This technique confirmed that TF is recruited in starvation conditions predominantly to recycling endosomes, which also contain autophagic proteins, including lipidated LC3, but not early endosomes, endoplasmic reticulum or Golgi markers.
Figure 1. The figure shows how TFRC is endocytosed together with its ligand, TF (purple dot), from the plasma membrane (PM) to recycling endosomes (RE) during starvation. The extracellular domain of this receptor is concentrated between the inner and outer evolving phagophore membranes, labeled by RAB11A and PtdIns3P. RAB11A and PtdIns3P constitute a ‘coincident-detection’ signature for WIPI2 membrane recruitment. ‘Curvature’ and ‘Scission’ are important events for the evolution of phagophores from the recycling endosome, which need further mechanistic exploration.
We showed that autophagic substrates, such as SQSTM1/p62 and mutant HTT (huntingtin), are engulfed by a RAB11A-positive compartment. Importantly, our live cell imaging experiments confirmed that RAB11A-positive membranes engulf mitochondria in cells exposed to a range of stimuli inducing mitochondrial autophagy (mitophagy), which shares its core machinery with canonical autophagy. This process requires proteins regulating early steps in autophagy.
Consistent with the data described above, the RAB11A-derived membrane that enwraps damage mitochondria contains TFRC, whereas depletion of RAB11A and WIPI2 delay degradation of depolarized mitochondria and TFRCs.
In summary, we think that our data provide strong support for the model that recycling endosomes are membrane platforms that recruit key machinery that enables phagophore formation. Indeed, phagophores appear to evolve from domains of the RAB11A-positive compartment, compatible with the localization of the extracellular domain of TFRC and its ligand between the inner and outer autophagosome membranes. While our data do not support the view that the endoplasmic reticulum or ER-mitochondrial contact sites are the primary platforms from which phagophores evolve, we do observe frequent contacts between these compartments and nascent autophagosomes. Compatible with previous suggestions arguing that the lipidation enzymes for LC3 are ER derived, we observed a tighter association between ATG3 and the ER than recycling endosomes or autophagosomes (the exception among the ATG proteins we studied). Thus, we think that the ER likely plays roles in phagophore formation distinct from serving as the primary membrane platform.
Acknowledgments
We are grateful to the UK Dementia Research Institute (funded by the MRC, Alzheimer’s Research UK and the Alzheimer’s Society) (DCR), and the Wellcome Trust (Principal Research Fellowship to DCR (095317/Z/11/Z)), a Strategic Grant to Cambridge Institute for Medical Research (100140/Z/12/Z) for funding.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Puri C, Vicinanza M, Ashkenazi A, et al. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev Cell. 2018 Apr 9;45(1):114–131. PMID: 29634932.
