ABSTRACT
It has been indicated that the Golgi apparatus contributes to autophagy, but how it is involved in autophagosome formation and maturation is not well understood. Here we show that amino acid starvation causes trans-Golgi derived membrane fragments to colocalize with autophagosomes. Depletion of the Golgi stacking protein GORASP2/GRASP55, but not GORASP1/GRASP65, increases both MAP1LC3 (LC3)-II and SQSTM1/p62 levels. We demonstrate that GORASP2 facilitates autophagosome-lysosome fusion by physically linking autophagosomes and lysosomes through the interactions with LC3 on autophagosomes and LAMP2 on late endosomes/lysosomes. Furthermore, we provide evidence that GORASP2 interacts with BECN1 to facilitate the assembly and membrane association of the phosphatidylinositol 3-kinase (PtdIns3K) UVRAG complex. These findings indicate that GORASP2 plays an important role in autophagosome maturation during amino acid starvation.
Abbreviations
ATG14: autophagy related 14; BafA1: bafilomycin A1; BSA: bovine serum albumin; CQ: chloroquine; EBSS: earle’s balanced salt solution; EM: electron microscopy; EEA1: early endosome antigen 1; GFP: green fluorescent protein; GORASP1/GRASP65: golgi reassembly stacking protein 1; GORASP2/GRASP55: golgi reassembly stacking protein 2; LAMP1: lysosomal-associated membrane protein 1; LAMP2: lysosomal-associated membrane protein 2; MAP1LC3: microtubule associated protein 1 light chain 3; MTOR: mechanistic target of rapamycin kinase; PBS: phosphate-buffered saline; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol 3-phosphate; PK: protease K; PNS: post-nuclear supernatant; RFP: red fluorescent protein; SD: standard deviation; TGN: trans-Golgi network; UVRAG: UV radiation resistance associated.
Introduction
Autophagy is an important cellular response to nutrient starvation [Citation1–Citation3], which can be induced by different conditions. For example, amino acid depletion inactivates MTOR (mechanistic target of rapamycin kinase), whereas glucose depletion activates AMP-activated protein kinase (AMPK) [Citation1], both of which subsequently activate ULK1/2 (unc-51 like autophagy activating kinase 1/2), ATG13, ATG101 and RB1CC1 [Citation1]. These autophagy regulators activate class III phosphatidylinositol 3-kinase (PtdIns3K) complex, which involves PIK3C3/VPS34, BECN1/Beclin 1, ATG14/ATG14L (autophagy related 14), PIK3R4/VPS15 and AMBRA1 [Citation1]. The phosphatidylinositol 3-phosphate (PtdIns3P)-enriched phagophore recruits the ATG12-ATG5-ATG16L1 protein complex and MAP1LC3B/LC3 (microtubule associated protein 1 light chain 3) to initiate 2 ubiquitin-like reactions that help phagophore to elongate and recruit membranes to form autophagosomes [Citation4]. Upon fusion of the double membrane, fully formed autophagosomes fuse with lysosomes to generate autolysosomes for degradation of the sequestrated materials [Citation1]. RAB7A GTPase and its effectors [Citation5], including the SNAP [Soluble NSF (N-ethylmaleimide-sensitive factor) Attachment Protein] Receptor (SNARE) complex consisting of STX17, SNAP29, and VAMP7/8 [Citation6], TECPR1 (tectonin beta-propeller repeat containing 1) [Citation7], the HOPS complex [Citation8], and the PtdIns3K UVRAG complex formed by BECN1, PIK3C3, PIK3R4, UVRAG and SH3GLB1/BIF-1 [Citation9,Citation10], are involved in autophagosome-lysosome fusion.
Several membrane organelles have been implicated as the membrane source for autophagosomes. The endoplasmic reticulum (ER) [Citation11], mitochondria [Citation12] and plasma membrane [Citation13] provide lipid and some key proteins for autophagosome formation. The Golgi apparatus also contributes to autophagosome formation. For example, ATG9A, a transmembrane protein cycling between the trans-Golgi network (TGN) and endosomes, relocates to autophagosomes upon amino acid starvation in mammalian cells, which implies that the Golgi may deliver membranes to autophagosomes [Citation14]. Furthermore, adaptor protein complex 1 (AP1)-mediated clathrin vesicles deliver membranes from the TGN to autophagosomes [Citation15]. Additionally, BECN1 and the PtdIns3K complex are concentrated on the TGN [Citation16], and ATG14 promotes the translocation of BECN1 from the TGN to autophagosomes [Citation17]. Thus, the Golgi plays a fundamental role in the regulation of autophagy.
The Golgi apparatus, an essential organelle for protein and lipid modifications, trafficking, and sorting, exists as stacks of flattened cisternal membranes in most eukaryotic cells. Formation of this stacked structure depends on 2 Golgi reassembly stacking proteins (GORASPs), GORASP1/GRASP65 (golgi reassembly stacking protein 1) and GORASP2/GRASP55, which are localized to the cis and medial-trans cisternae, respectively. These peripheral membrane proteins form trans-oligomers from adjacent cisternae to hold Golgi cisternae together into stacks [Citation18,Citation19]. During mitosis, GORASP proteins are phosphorylated, which inhibits their oligomerization, resulting in Golgi cisternal unstacking; while in telophase, dephosphorylation allows GORASPs to re-oligomerize and Golgi cisternae to restack [Citation20].
How the Golgi responds to nutrient deprivation and how it contributes to autophagy remain elusive. In this study, we investigated the role of the Golgi in autophagy and made an unexpected finding that GORASP2 contributes to autophagosome maturation upon amino acid starvation by physically linking autophagosomes and lysosomes and by facilitating the assembly of the class III PtdIns3K UVRAG complex.
Results
Amino acid starvation induces Golgi derived vesicles to colocalize with autophagosomes
To determine how the Golgi responds to amino acid starvation, HeLa cells were treated with Earle’s Balanced Salt Solution (EBSS) and bafilomycin A1 (BafA1) for 1–8 h as indicated (), and stained for LC3, GOLGA2/GM130 and TGOLN2/TGN46 to reveal the autophagosomes, cis-Golgi, and trans-Golgi, respectively. Consistent with previous reports [Citation21], this treatment resulted in autophagosome accumulation, as indicated by the increasing number of LC3 puncta over time (). Interestingly, the Golgi, especially trans-Golgi, generated vesicular structures that colocalized with autophagosomes (). These results demonstrate that amino acid starvation triggers mild Golgi fragmentation. We then determined the effect of EBSS plus BafA1 treatment on the level of major Golgi structural proteins. While most proteins decreased or did not change, the GORASP2 protein level significantly increased upon starvation (), indicating that GORASP2 might play a role in amino acid starvation-induced autophagy.
Figure 1. Amino acid starvation induces Golgi derived vesicles to colocalize with autophagosomes. (a) Golgi-derived fragments colocalize with autophagosomes upon amino acid starvation. HeLa cells were incubated with growth medium (ctr) or with EBSS and 400 nM BafA1 (E + B) for the indicated times, stained for LC3, TGOLN2, GOLGA2, and DNA. The four columns on the right are the enlarged areas indicated in the first column on the left. Scale bar: 10 µm on the left, 1 µm on the right. (b) Quantification of (a) for the average number of LC3 puncta per cell. Values are shown as mean ± SD from three independent experiments; statistical significance of the results was assessed by Student’s t-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (c) Quantification of (a) for the percentage of TGOLN2 and GOLGA2 signals that colocalized with LC3. (d) Western blots of LC3 and major Golgi proteins in HeLa cells treated as in (a). (e) Quantification of (d) for the GORASP2 protein level. (f) Electron microscopy pictures of HeLa cells treated with growth medium (ctr), BafA1, EBSS, or EBSS with 400 nM BafA1 (E + B) for 4 h. Scale bar: 500 nm. Cis- and trans-sides of the Golgi are indicated.
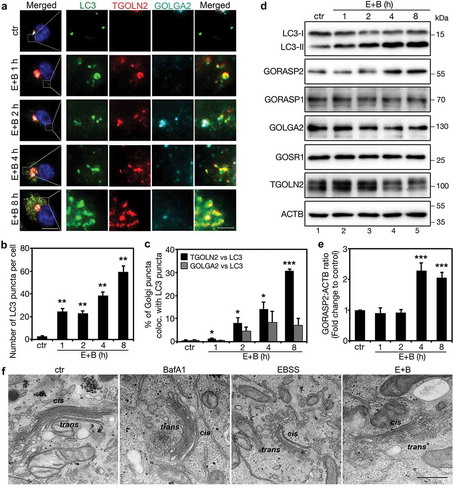
Similarly, EBSS alone or together with other lysosome inhibitors, NH4Cl or chloroquine (CQ), or with the MTOR inhibitor rapamycin, also induced the trans-Golgi to form and release vesicular structures that colocalized with autophagosomes. The same treatments also increased LC3 and GORASP2 protein levels (Figures S1 and S2(A-D)). As long-term amino acid starvation resulted in cell death, we used 4 h treatment in our following experiments. Since EBSS contains relatively low glucose level than growth medium, we treated cells with low glucose with or without BafA1 for 4 h, and the results showed that glucose starvation did not affect the level of tested Golgi structural proteins including GORASP2 (Figure S2E), suggesting that the effect of EBSS on GORASP2 was caused by amino acid starvation.
To further investigate how amino acid starvation affects Golgi morphology, we performed electron microscopy (EM). Compared to growth medium (control condition) or growth medium with BafA1, EBSS treatment or EBSS with BafA1 significantly reduced the cisternal length and caused fenestration of the Golgi cisternae in particular in the medial-trans cisternae ( and S3). Notably, many autophagosomes were observed in the close proximity of the Golgi stacks ( and S3), consistent with a role of the Golgi in autophagy.
GORASP2 depletion inhibits autophagosome maturation
As the protein level of GORASP2 was increased by amino acid starvation ( and S1D), we investigated its role in autophagy. Knockdown of GORASP2, but not GORASP1, significantly increased the number of autophagosomes under both control and amino acid starvation conditions (). GORASP2 depletion also increased both LC3-II and SQSTM1/p62 protein levels (), which was not observed in control or GORASP1 RNAi treated cells. These results show that GORASP2 is required for autophagy mediated protein degradation, indicating that GORASP2 may be involved in autophagosome-lysosome fusion.
Figure 2. GORASP2 depletion results in autophagosome accumulation. (a) GORASP2 depletion increases the number of autophagosomes. HeLa cells were transfected with control (ctri), GORASP2 (GORASP2i or G2i) or GORASP1 (GORASP1i or G1i) RNAi as indicated for 72 h, treated with growth medium (ctr), 50 µM chloroquine (CQ), EBSS or EBSS with 50 µM CQ (E+ CQ) for 4 h, and stained for LC3, TGOLN2, GOLGA2, and DNA. Scale bar: 5 µm. (b) Quantification of (a) for the average number of LC3 puncta per cell. (c) Western blots of indicated proteins in HeLa cells treated with growth medium (ctr), CQ, EBSS, or EBSS with CQ (E+ CQ) for 4 h. Asterisk indicates an unspecific band.
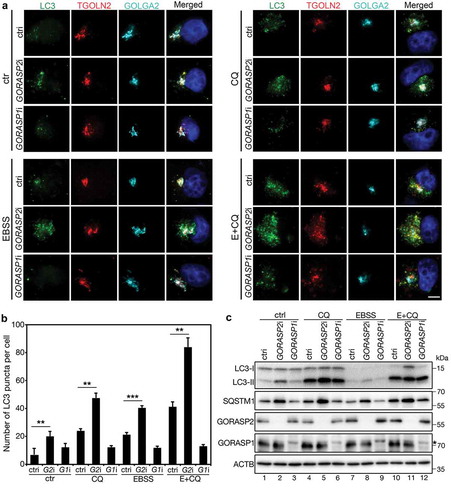
To further clarify the role of GORASP2 in autophagosome maturation, we determined the effect of GORASP2 depletion on autophagic flux in HeLa cells that express the RFP-GFP-LC3 reporter [LC3 tagged by both the red (RFP) and green (GFP) fluorescent proteins] [Citation22]. This dual tagged LC3 construct emits both red and green fluorescence lights upon excitation at neutral pH as in autophagosomes. Upon autophagosome-lysosome fusion, GFP is quenched by the acidic pH in the autolysosomes, and thus only red fluorescence is seen. This reporter allows us to distinguish autolysosomes from autophagosomes [Citation23]. Under control condition, depletion of GORASP2, but not GORASP1, not only increased the number of RFP-GFP-LC3 puncta, but also increased the percentage of LC3 positive puncta that are in yellow (Figure S4). Similar results were obtained after amino acid starvation, whereas addition of BafA1, which neutralizes lysosome pH and inhibits autophagosome-lysosome fusion [Citation24,Citation25], increased the number of the dual-color autophagosomes under all experimental conditions (Figure S4). These results support our hypothesis that GORASP2 facilitates autophagosome-lysosome fusion.
GORASP2 is targeted to autophagosomes and lysosomes upon amino acid starvation
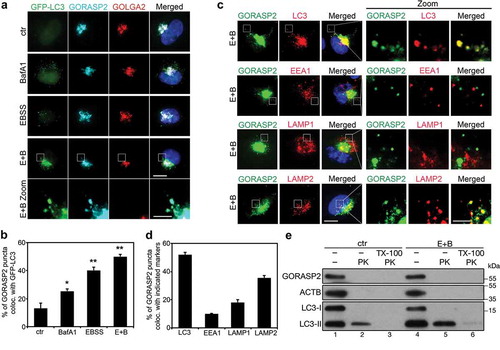
To further investigate how GORASP2 facilitates autophagosome maturation, we determined its subcellular localization under starvation conditions. Upon amino acid starvation, GORASP2 formed puncta outside of the Golgi region, which colocalized well with autophagosomes indicated by GFP-LC3 () or endogenous LC3 (). In addition, GORASP2 in puncta also colocalized with LAMP2 (late endosomes and lysosomes), but not with EEA1 (early endosomes) or LAMP1 (late endosomes) (). To determine whether GORASP2 is located on the surface or is engulfed into autophagosomes, we performed a protease K (PK) protection assay. In this experiment, we used LC3 as a marker. The membrane bound form of LC3, LC3-II, is localized both in the lumen and on the surface of autophagosomes, and thus was partially protected from protease digestion. Its soluble form, LC3-I, as well as the cytoplasmic protein, ACTB, were both degraded. Like LC3-I, GORASP2 was also completely degraded (), suggesting that GORASP2 localized to the surface of the autophagosomes upon amino acid starvation. These results indicate that GORASP2 functions at the surface of autophagosomes rather than a substrate for autophagy mediated degradation.
Figure 3. GORASP2 is localized to autophagosomes and lysosomes upon starvation. (a) GORASP2 colocalizes with GFP-LC3 upon starvation. GFP-LC3 HeLa cells were treated with growth medium (ctr), BafA1, EBSS or EBSS and 400 nM BafA1 (E + B) for 4 h, stained for GORASP2, GOLGA2 and DNA. The bottom row shows higher magnifications of the indicated area in the above row. Scale bar: 10 µm in the upper four rows, 3 µm in the bottom row. (b) Quantification of (a) for the percentage of GORASP2 puncta that colocalized with GFP-LC3. (c) GORASP2 colocalizes with LC3 and LAMP2 but not EEA1 or LAMP1 upon starvation. HeLa cells were treated with EBSS and 400 nM BafA1 (E + B) for 4 h, stained for GORASP2, LC3, EEA1, LAMP1, or LAMP2 and DNA as indicated. The three rows on the right are higher magnifications of the boxed area in the three rows on the left. Scale bars: 10 µm in the left rows, 3 µm in the right rows. (d) Quantification of (c) for the percentage of GORASP2 puncta that colocalized with LC3, EEA1, LAMP1 or LAMP2. (e) Western blot of the proteinase K protection assay. HeLa cells were treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h, then the collected PNS were equally divided into three tubes, one left untreated, one was incubated with 2.5 μg/ml protease K (PK) only, and one was treated with both PK and 1% TritonX-100 (TX-100) for 10 min.
GORASP2 binds LC3 and LAMP2 to facilitate autophagosome-lysosome fusion
Since GORASP2 forms oligomers and functions as a membrane tether in Golgi stacking [Citation19], we determined the possibility that it may function in a similar way to link autophagosomes and lysosomes and facilitate their fusion. In support of this hypothesis, knockout of GORASP2 by CRISPR/Cas9 [Citation26] significantly reduced LC3-LAMP2 colocalization (). To determine how GORASP2 is targeted to autophagosomes, we tested the possibility that GORASP2 might interact with LC3, as they colocalized upon amino acid starvation (). Indeed, GFP-LC3, but not GFP alone, co-immunoprecipitated with GORASP2 under amino acid starvation (). The interaction between GORASP2 and LC3 was significantly increased by BafA1 or EBSS treatment, while a combination of both yielded a much stronger effect (). These results indicate that there is a basal level of autophagy and GORASP2-LC3 interaction under growth condition, whereas amino acid starvation increases autophagy level and enhances GORASP2-LC3 interaction. Given that GORASP2 is required for autophagosome-lysosome fusion () and that GORASP2 puncta colocalize with LAMP2 (), we also determined and confirmed GORASP2-LAMP2 interaction using a similar approach (), although this interaction was not regulated by the nutrient level ().
Figure 4. GORASP2 binds LC3 and LAMP2 to facilitate autophagosome-lysosome fusion. (a-d) GORASP2 knockout reduces LC3-LAMP2 colocalization. (a) Wild type (WT) or GORASP2 knockout (KO) HeLa cells were treated with EBSS and 400 nM BafA1 (E + B) for 4 h, and stained for LC3, LAMP2, and DNA. The bottom row (Zoom) shows a higher magnification of the boxed area in the upper row. Scale bars: 10 µm in the upper row, 1 µm in the lower row. (b) Quantification of (a) for the percentage of LAMP2 puncta that colocalized with LC3 in WT or GORASP2 knockout cells. (c-d) GORPAS2 binds LC3. (c) GFP-LC3 or GFP expressing HeLa cells were transfected with GORASP2-MYC for 16 h, treated with EBSS and 400 nM BafA1 (E + B) for 4 h, and immunoprecipitated with a GFP antibody followed by western blotting. (d) GFP-LC3 HeLa cells were transfected with GORASP2-MYC, then treated with growth medium (ctr), BafA1, EBSS or EBSS and 400 nM BafA1 (E + B) for 4 h, and immunoprecipitated with a GFP antibody followed by western blotting. (e-f) GORASP2 binds LAMP2. (e) GFP or GORASP2-GFP expressing HeLa cells were treated with EBSS and 400 nM BafA1 (E + B) for 4 h, and immunoprecipitated with a GFP antibody followed by western blotting. (f) HeLa cells were transfected with GORASP2-GFP, treated with indicated medium for 4 h, and immunoprecipitated with a GFP antibody. (g) GORASP2 facilitates LC3-LAMP2 interaction. mRFP-GFP-LC3 cells treated with EBSS and BafA1 were immunoprecipitated with a GFP antibody in the presence of nothing (lane 1), or increasing amount of BSA (as control, lanes 2 and 3) or His-tagged GORASP2 (lanes 4 and 5), followed by western blot for LAMP2, His and GFP. Ponceau stain shows the input and arrows indicate the position of indicated proteins. .
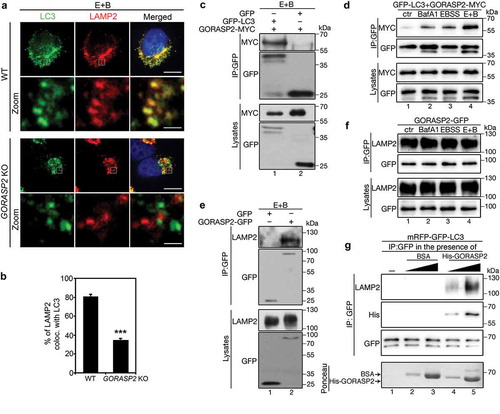
The finding that GORASP2 bound to LC3 on autophagosomes and LAMP2 on lysosomes triggered us to consider GORASP2 as a linker between autophagosomes and lysosomes through the interactions with LC3 and LAMP2. To test this hypothesis, we determined the effect of GORASP2 on the interaction between LC3 and LAMP2. While the LC3-LAMP2 interaction was very weak and barely detectable, adding purified recombinant GORASP2 into the reaction significantly increased LC3-LAMP2 interaction in a dose-dependent manner (, lanes 4 and 5 vs. 1). In contrast, adding BSA (bovine serum albumin) had no effect. In this assay, GORASP2, but not BSA, co-immuoprecipitated with LC3 and LAMP2 (). Thus, GORASP2 facilitates autophagosome-lysosome fusion through enhancing LC3-LAMP2 interaction after amino acid starvation, which is similar to the role of GORASP2 in autophagosome maturation after glucose starvation [Citation27].
GORASP2 binds the PtdIns3K UVRAG complex and facilitates its assembly
Autophagosome-lysosome fusion involves multiple proteins, including BECN1 [Citation9]. Therefore, we determined the interaction between GORASP2 and BECN1 by co-immunoprecipitation. GORASP2 co-precipitated with BECN1 after amino acid starvation, while GORASP1 co-purified with GOLGA2, a known binding protein for GORASP1, but not BECN1 (). Reciprocal co-immunoprecipitations confirmed the GORASP2-BECN1 interaction (). In comparison with control condition, amino acid starvation or BafA1 treatment significantly enhanced GORASP2-BECN1 interaction, and a combination of both exhibited an even stronger effect (), suggesting that starvation increases GORASP2-BECN1 association.
Figure 5. GORASP2 interacts with BECN1 upon starvation. (a) GORASP2 but not GORASP1 binds BECN1. GFP, GORASP2-GFP or GORASP1-GFP expressing HeLa cells were treated with EBSS and 50 µM CQ for 4 h and immunoprecipitated with a GFP antibody followed by western blotting for BECN1, GOLGA2 and GFP. *, IgG heavy chain. (b-d) Starvation enhances GORASP2-BECN1 interaction. HeLa cells were co-transfected GFP vector as control or GORASP2-GFP with FLAG-BECN1, then treated with EBSS and 400 nM BafA1 (E + B) for 4 h and immunoprecipitated with a GFP antibody (b) or FLAG antibody (c) followed by western blotting. (d) GORASP2-GFP transfected HeLa cells were treated with growth medium (ctr), BafA1, EBSS, or EBSS and 400 nM BafA1 (E + B) for 4 h and immunoprecipitated with a GFP antibody followed by western blotting for BECN1 and GFP. (e) GORASP2 schematic domain structure. Indicated are the myristic acid (myr) for membrane association, the N-terminal GRASP domain for dimerization and oligomerization, and the C-terminal Serine/Proline-Rich (SPR) domain for regulation. (f-g) C-terminus of GORASP2 binds BECN1. HeLa cells were transfected with GFP, or GFP-tagged the N-terminus (aa1-212, NT), C-terminus (aa213-452, CT), or full length (FL) GORASP2, or indicated truncation mutants for 16 h, treated with EBSS and 400 nM BafA1 (E + B) for 4 h and immunoprecipitated with a GFP antibody followed by western blotting of GFP and BECN1.
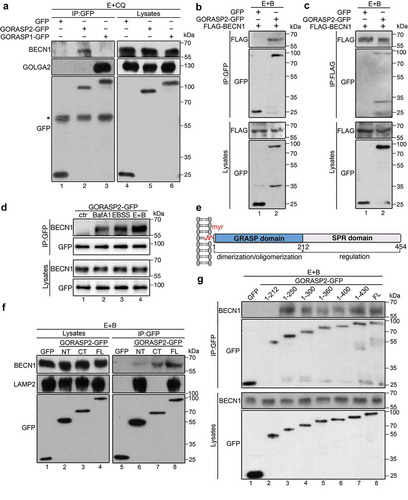
We then mapped the domain structure of GORASP2 that interacts with BECN1 using our available GORASP2 truncation mutants [Citation19,Citation27]. We found that the C-terminus of GORASP2 interacted with BECN1, likely via the aa212-250 region (). This is consistent with the domain structure and function of GORASP2 in Golgi stacking. While its N-terminal GRASP domain forms dimers and trans-oligomers, its C-terminal Serine/Proline Rich (SPR) domain is heavily phosphorylated during mitosis and regulates the oligomerization property of the N-terminal GRASP domain () [Citation19,Citation20].
BECN1 is a major component of the PtdIns3K complex, and class III PtdIns3K complex is known to mediate autophagosome-lysosome fusion by enriching PtdIns3P at the interface [Citation28]. There are three different PtdIns3K complexes that play distinct roles in autophagy [Citation28]. The ATG14 complex generates PtdIns3P on phagophore to recruit certain ATG components to expand the phagophore [Citation29,Citation30]; the UVRAG complex generates PtdIns3P at the interface between autophagosome and lysosome to facilitate autophagosome and lysosome fusion [Citation9]; whereas the RUBCN/Rubicon complex [Citation30] inhibits the role of the UVRAG complex in autophagosome maturation (). In addition to BECN1, GORASP2 co-immunoprecipitated with PIK3C3 (), the core protein of all three PtdIns3K complexes, indicating that GORASP2 interacts with the PtdIns3K complexes rather than BECN1 alone.
Figure 6. GORASP2 interacts with the PtdIns3K UVRAG complex and facilitates its assembly. (a) Diagram showing the components in the three distinct PtdIns3K complexes. (b-f) GORASP2 binds the PtdIns3K UVRAG complex. HeLa cells were transfected with indicated proteins, treated with indicated medium for 4 h, and analyzed by immunoprecipitation and western blotting. (g) Endogenous GORASP2 interacts with the PtdIns3K UVRAG complex. NRK cells were treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h and immunoprecipitated with a GORASP2 antibody followed by western blotting for BECN1, VPS34, UVRAG and GORPAS2. (h) GORPAS2 facilitates the assembly of the PtdIns3K UVRAG complex. Wild type (WT) or GORASP2 knockout (KO) HeLa cells were transfected with FLAG-BECN1, treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h and immunoprecipitated with a FLAG antibody followed by western blotting for UVRAG, SH3GLB1, VPS34, GORASP2 and FLAG. ACTB was used as loading control. Asterisk indicates the IgG heavy chain. (i-j) GORASP2 directly interacts with BECN1 and UVRAG. (i) GST and GST-tagged BECN1 and SH3GLB1 proteins were incubated with recombinant His-tagged GORASP2, pulled down with glutathione beads, and blotted for GORAP2. BSA was included as a negative control. (j) MBP and MBP-tagged UVRAG were incubated with recombinant His-tagged GORASP2, pulled down with amylose beads, and blotted for GORASP2. (k) GORASP2 facilitates BECN1 and UVRAG interaction. MBP and MBP-tagged UVRAG were incubated with recombinant GST-tagged BECN1 in the presence of BSA (as negative control) or His-GORASP2, pulled down with amylose beads, and blotted for BECN1. Ponceau stain shows the input and arrows indicate the position of indicated proteins.
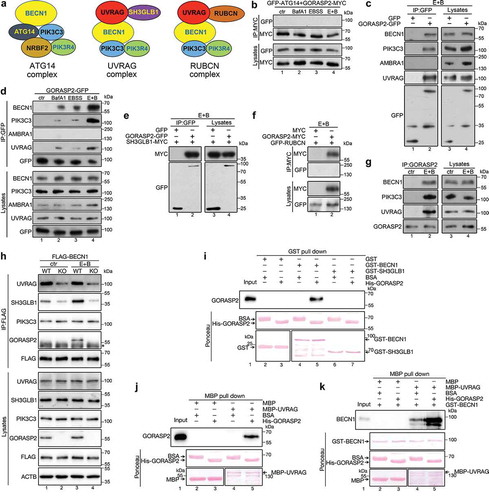
To specify the PtdIns3K complex that GORASP2 interacts with, we performed co-immunoprecipitation of GORASP2 with some core components of the three complexes. We excluded the interaction between GORASP2 and the ATG14 complex, since GORASP2 did not bind to ATG14 () or AMBRA1 (). Further experiments also failed to detect an interaction between GORASP2 and RUBCN, a protein in the RUBCN complex (). Positive interactions were detected between GORASP2 and the UVRAG complex, as GORASP2 copurified with UVRAG () and SH3GLB1 (), two components of the UVRAG complex. Moreover, the interactions between GORASP2, BECN1, PIK3C3 and UVRAG were confirmed by co-immunoprecipitation of endogenous proteins (). These results demonstrate that GORASP2 interacts with the UVRAG complex, but not with the ATG14 or RUBCN complex. As UVRAG complex functions in autophagosome-lysosome fusion [Citation9], these results support the function of GORASP2 in autophagosome maturation.
Since GORASP2 interacts with the PtdIns3K UVRAG complex, it was of our interest to investigate whether GORASP2 regulates its assembly. To do this, we assessed the interactions of BECN1 with UVRAG, SH3GLB1 and PIK3C3 in wild type and GORASP2 knockout cells. While GORASP2 knockout had no effect on BECN1-PIK3C3 interaction, it significantly reduced the interaction of BECN1 with UVRAG and SH3GLB1 (), suggesting a role of GORASP2 in regulating the PtdIns3K UVRAG complex assembly. To confirm this result, we determined the interaction of GORASP2 with the UVRAG complex in vitro using purified GORASP2, BECN1, UVRAG, and SH3GLB1. Pull down results showed that GORASP2 directly interacted with BECN1 and UVRAG, but not SH3GLB1 (). Furthermore, the interaction between BECN1 and UVRAG was significantly enhanced by the addition of GORASP2 (), demonstrating that GORASP2 facilitates the PtdIns3K UVRAG complex assembly.
GORASP2 enhances BECN1 membrane association upon starvation
It has been previously shown that BECN1 is concentrated on the Golgi under control conditions [Citation16,Citation17], which was confirmed in our study by fluorescence microscopy (). Upon amino acid starvation, BECN1 was redistributed to puncta structures (autophagosomes) (), which fits the idea that BECN1 shuttles between the Golgi and autophagosomes [Citation16]. In GORASP2 knockout cells, BECN1 displayed a diffused cytosolic pattern, suggesting that GORASP2 deletion reduced BECN1 membrane association (). To verify this observation, we performed subcellular fractionation of wild type and GORASP2 knockout HeLa cells cultured under control and amino acid starvation conditions. We separated membranes from cytosol by ultracentrifugation of the post-nuclear supernatant (PNS) and analyzed BECN1 level in each fraction by western blot. Under control condition, a large fraction of BECN1 was found in the membrane-bound fraction, while GORASP2 knockout significantly reduced the amount of BECN1 in the membrane fraction. Under amino acid starvation, more BECN1 was associated with membranes, while GORASP2 deletion reduced this amount (). To confirm the specificity, we exogenously expressed GORAPS2 in GORASP2 knockout cells, which increased BECN1 membrane association under control condition (). These results demonstrate that GORASP2 facilitates BECN1 membrane association.
Figure 7. GORASP2 facilitates BECN1 membrane association upon starvation. (a) BECN1 partially relocates to autophagosomes upon amino acid starvation. HeLa cells were treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h, stained for BECN1, TGOLN2, GORASP2 and DNA. Scale bars: 2 µm. (b) GORASP2 knockout reduces BECN1 membrane association. Wild type (WT) or GORASP2 knockout (KO) cells were stained for BECN1, TGOLN2, GORASP2 and DNA. Scale bar: 20 µm. (c) Wild type (WT) or GORASP2 knockout (KO) HeLa cells treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h, collected to generate PNS, which was separated into cytosol (cyt) and membranes (mem) by ultracentrifugation. Equal volume proportions of samples were analyzed by western blotting for BECN1, TUBA and GOSR1. (d) Quantitation of (c) for the percentage of BECN1 in the membrane fraction. (e) The reduced membrane association of BECN1 in GORASP2 knockout HeLa cells can be rescued by exogenously expressed GORASP2. GORASP2 KO HeLa cells were transfected with MYC or GORASP2-MYC and analyzed as in (c). (f) Quantification of (e) for the percentage of BECN1 in membrane fractions. (g-h) GORASP2 competes with GLIPR2 for BECN1 interaction. (g) HeLa cells were co-transfected with GORASP2-GFP, GLIPR2-MYC and FLAG-BECN1, treated with indicated medium for 4 h, immunoprecipitated with a FLAG antibody, and blotted for MYC, GFP and FLAG. (h) HeLa cells were co-transfected with GORASP2-GFP and GLIPR2-MYC, treated with growth medium (ctr) or EBSS and 400 nM BafA1 (E + B) for 4 h, immunoprecipitated with a GFP or MYC antibody, and blotted for GFP, MYC and BECN1. (i) Working model. Upon amino acid starvation, trans-Golgi derived vesicles translocate to autophagosomes, whereas GORASP2 binds LC3 and LAMP2 to mediate autophagosome-lysosome fusion. Additionally, it facilitates assembly of the PtdIns3K UVRAG complex at the interface of autophagosome and lysosome to facilitate autophagosome maturation.
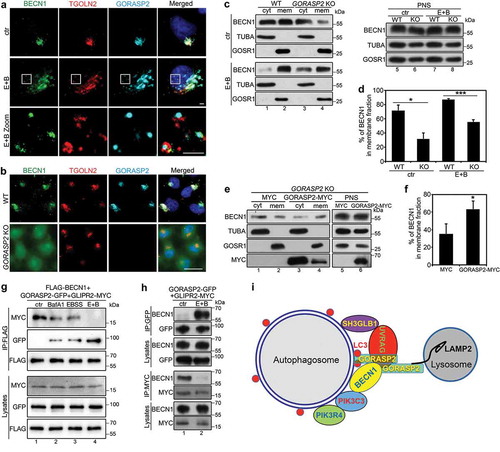
It has been previously shown that GLIPR2/GAPR1 interacts with BECN1 on the Golgi to inhibit autophagy [Citation16], while our results suggested that GORASP2 bound to BECN1 under starvation conditions to facilitate the PtdIns3K UVRAG complex formation and autophagosome maturation. To determine how GORASP2 and GLIPR2 interplay to regulate BECN1 function, we determined the interactions between GORASP2, GLIPR2, and BECN1 under different conditions. Interestingly, our results showed that BECN1 preferentially bound to GLIPR2 under growth condition, while it interacted more with GORASP2 under amino acid starvation condition (). These results indicate that both GORASP2 and GLIPR2 are important regulators for BECN1, and a balance between these two proteins controls BECN1 localization and function at the Golgi or autophagosomes.
Discussion
In this study, we made an unexpected finding that the Golgi stacking protein GORASP2/GRASP55 contributes to autophagosome maturation by linking autophagosomes and late endosomes/lysosomes through the interaction with LC3 and LAMP2 and by facilitating the PtdIns3K UVRAG complex assembly (). This is the first study that links GORASP2, autophagosome/lysosome membrane tethering, and PtdIns3K complex assembly.
Upon amino acid starvation, the Golgi underwent partial fragmentation in the order from trans to cis, and Golgi-derived fragments colocalized with autophagosomes (), consistent with previous reports suggesting a role of the Golgi in autophagosome formation [Citation14,Citation15]. Given that GORASP2 level increased upon amino acid starvation () and that only a relatively small proportion of GORASP2 was targeted to autophagosomes (), the amount of GORASP2 in the Golgi should not be significantly reduced. Therefore, GORASP2 retargeting from the Golgi to autophagosomes was unlikely the main cause of Golgi fragmentation. In a parallel study, we found that, unlike amino acid starvation, glucose starvation does not significantly affect Golgi morphology, but also causes GORASP2 retargeting to autophagosome-lysosome interface [Citation27], suggesting that GORASP2 plays an essential role in autophagosome maturation under different autophagy-inducing conditions. Amino acid depletion directly inactivates MTOR, whereas glucose starvation activatesMAPK8/c-Jun amino-terminal kinase and IKBKB/IκB kinase, and indirectly inhibits MTOR through AMPK [Citation1]. Thus, activation of different signaling pathways might have distinct consequences for downstream effectors on the Golgi and lead to specialized functional consequences. Alternatively, amino acid starvation directs TFEB (transcription factor EB) translocation to nucleus to initiate transcription of a large amount of autophagy and lysosome related proteins [Citation31], and the increased protein synthesis triggers increased Golgi transport to autophagosomes, which is comparably milder in glucose starvation. The exact mechanism of how GORASP2 is targeted to autophagy upon amino acid starvation is not well understood. GORASP2-containing fragmented vesicles derived from the Golgi could traffic to autophagosomes to help their expansion, although it is also possible that GORASP2 is recruited to autophagosomes directly from the cytosol by LC3.
GORASP2 and GORASP1 are originally discovered as homologs in mammalian cells with similar molecular features and functions in Golgi stacking [Citation32]. Both GORASPs regulate Golgi stacking, ribbon linking, cell cycle progression, and unconventional secretion [Citation32–Citation35]. GORASP1 is also involved in spindle dynamics during mitosis [Citation36] and apoptosis [Citation37]; while GORASP2 facilitates autophagy [Citation27,Citation38]. Consistent with the role of GORASP2 in autophagy, we found that depletion of GORASP2 resulted in autophagosome accumulation ( and S4). It is possible that depletion of GORASP2 causes Golgi defects, such as missorting of lysosomal enzymes, which contribute to autophagosome accumulation. However, our results indicate that this is an invalid explanation. For example, CQ treatment increased the number of LC3 puncta () and the level of LC3-II protein () in GORASP2 knockdown cells, suggesting that the lysosome is still functional in GORASP2 depleted cells. Additionally, GORASP1 knockdown also results in Cathepsin D missorting [Citation39] but not autophagosome accumulation, suggesting a specific role of GORASP2 in autophagy.
Our study revealed a novel role for GORASP2 as a membrane linker in autophagy. Under either glucose starvation [Citation27,Citation40,Citation41] or amino acid starvation (), GORASP2 interacts with LC3 on autophagosomes and LAMP2 on lysosomes and functions as a membrane linker to facilitate autophagosome-lysosome fusion (). Interestingly, the HOPS tethering complex [Citation8] is also involved in autophagosome-lysosome tethering. While the relationship between GORASP2 and the HOPS complex is a major future effort of our study, the significant reduction of LC3-LAMP2 colocalization in GORASP2 knockout cells indicates an important role for GORASP2 in autophagosome-lysosome linking ().
In addition to directly linking autophagosomes and lysosomes, we provided strong evidence that GORASP2 facilitates the assembly of the PtdIns3K-UVRAG complex. First, under amino acid starvation condition, GORASP2 interacted with multiple components of the PtdIns3K UVRAG complex, including BECN1, PIK3C3, UVRAG, and SH3GLB1, but not with ATG14 or RUBCN, proteins specific for the PtdIns3K ATG14 and RUBCN complexes, respectively (). Second, GORASP2 facilitated BECN1’s membrane association. Under growth conditions, BECN1 was concentrated on the Golgi; upon amino acid starvation, it colocalized with GORASP2 on autophagosomes (). Depletion of GORASP2 significantly reduced BECN1 membrane association under both control and amino acid starvation conditions, which was rescued by exogenously expressed GORASP2 (). In addition, GORASP2 depletion reduced BECN1 interaction with UVRAG and SH3GLB1 in cells (), while adding purified GORASP2 enhanced the BECN1-UVRAG interaction in vitro (), demonstrating that GORASP2 facilitates the PtdIns3K UVRAG complex formation. In regard to how GORASP2 facilitates UVRAG complex assembly, we speculate that GORASP2 first helps UVRAG to bind BECN1 to form a complex, and then dissociates from these proteins once a stable UVRAG complex forms. This may explain why GORASP2 was not co-purified with the UVRAG complex in the previous reports, although the exact mechanisms await future investigation. Given the important role of this complex in autophagosome-lysosome fusion, these results confirmed that GORASP2 plays a critical role in autophagosome maturation.
The alternative interaction of BECN1 with GLIPR2 and GORASP2 is striking, as BECN1 almost exclusively interacted with GLIPR2 under growth condition and with GORASP2 after amino acid starvation (). Both GORASP2 and BECN1 largely remained on the Golgi even after amino acid starvation (); thus, the mechanism that regulates BECN1 interaction with GLIPR2 and GORASP2 remains as a mystery. GLIPR2 is known as a lipid raft-associated protein on the Golgi [Citation42]. Through binding to BECN1, GLIPR2 inhibits BECN1 redistribution to non-Golgi vesicles that contribute to autophagosome formation [Citation16]. Conversely, BECN1 preferably interacted with GORASP2 upon amino acid starvation, presumably on autophagosomes (), indicating that GORASP2 functions as a positive regulator for BECN1 autophagosome localization and function in autophagosome maturation. In conclusion, our study uncovered a novel role for the Golgi stacking protein GORASP2 in autophagosome maturation.
Materials and methods
Cell lines
HeLa and normal rat kidney (NRK) cells were cultured in Dubelcco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, 11965092) supplemented with 10% super calf serum (Gemini, 100–510) and 100 units/ml penicillin-streptomycin (Thermo Fisher Scientific, 15140122) at 37°C with 5% CO2. HeLa cells stably expressing GFP, GORASP2-GFP, or GORASP1-GFP were previously generated by Dr. Yi Xiang [Citation19]. GORASP2 knockout HeLa cells were established previously [Citation26], mRFP-GFP-LC3 expressing HeLa cells [Citation43] were kindly provided by Dr. David Rubinsztein (University of Cambridge). GFP-LC3 HeLa cells were generated by transfection of GFP-LC3 plasmid into HeLa cells followed by 1 µg/ml puromycin (Thermo Fisher Scientific, A1113803) selection and fluorescence activated cell sorting (FACS) of GFP positive cells.
Reagents, plasmids, and antibodies
Constructs for GFP-tagged full length GORASP1 and GORASP2 were previously described [Citation19]. GFP-LC3 was purchased from Addgene (22405, deposited by Dr. Jayanta Debnath) [Citation44] and was used to generate the GFP-LC3-expressing HeLa cells. pEGFP-RUBCN was purchased from Addgene (21636, deposited by Dr. Tamotsu Yoshimori) [Citation45]. pBICEP-CMV2 3xFLAG-BECN1 and GLIPR2-MYC were kindly provided by Dr. Beth Levine (UT Southwestern) [Citation16]. MYC-SH3GLB1 was kindly provided by Dr. Yoshinori Takahashi (Penn State University) [Citation46]. GST-SH3GLB1 was kindly provided by Dr. Hong-Gang Wang (Penn State College of Medicine) [Citation47]. GFP-ATG14 and GST-BECN1 were kindly provided by Dr. Kun-Liang Guan (University of California, San Diego) [Citation48]. GORASP2 was constructed in the pcDNA3.1/myc-His (-) A vector or pET-30a(+) vector as GORASP2-MYC or His-GORASP2, respectively. UVRAG was constructed in the pMAL-c2X vector as MBP-UVRAG.
Antibodies used in this study include monoclonal antibodies against LAMP1 [Developmental Studies Hybridoma Bank (DSHB), H4A3], LAMP2 (DSHB, H4B4), GFP (Proteintech Group, 66002–1-lg), GOLGA2 (BD Transduction Laboratories, 610823), TUBA (DSHB, 4A1), GOSR1 (BD Transduction Laboratories, 611184), PIK3C3 (Santa Cruz Biotechnology, sc-365404), AMBRA1 (Santa Cruz Biotechnology, sc-398204), SH3GLB1 (Santa Cruz Biotechnology, sc-374146), UVRAG (Cell Signaling Technology, 5320), FLAG (Sigma-Aldrich, M1804) and ACTB (Sigma-Aldrich, AC-15); polyclonal antibodies against GORASP2 (Proteintech Group, 10598–1-AP), GORASP1 (UT465, Joachim Seemann, UT Southwestern), GOLGA2 (N73, Joachim Seemann), TGOLN2 (Bio-Rad, AHP500), LC3 (MBL International, PM036), SQSTM1 (Sigma-Aldrich, P0067), BECN1 (Santa Cruz Biotechnology, 48341), ATG9A (GeneTex, GTX128427), EEA1 (Santa Cruz Biotechnology, sc-6415), MAN1A1 (Abcam, ab140613), MTOR (Cell Signaling Technology, 2983), p-RPS6KB1 (Cell Signaling Technology, 9234) and RPS6KB1 (Cell Signaling Technology, 9202). MYC antibody was kindly provided by Dr. David Sheff (University of Iowa).
Cell transfection
Polyethylenimine was used for transient transfection of plasmids according to manufacturer’s instructions (Polysciences, Inc., 23966–1). Cells were used 16 h after transfection. For knockdown experiments, HeLa cells or mRFP-GFP-LC3 expressing cells were transfected with Lipofectamine RNAiMAX according to manufacturer’s instructions (Thermo Fisher Scientific, 13778150) and cells were used 72 h after transfection. Control non-specific RNAi (AM4636) was purchased from Applied Biosystems. Human GORASP2 and GORASP1 targeting sequences were previously described [Citation19,Citation39].
Immunofluorescence microscopy
Immunofluorescence microscopy was performed as described previously [Citation26,Citation49]. Briefly, HeLa cells were fixed in 3% paraformaldehyde (Fisher Scientific, AC416780030) in Phosphate-buffered saline PBS (Invitrogen, 21600069) for 15 min followed by quenching with 50 mM NH4Cl in PBS for 10 min and permeabilized with 0.3% Triton-X-100 in PBS for 10 min. Permeabilized cells were blocked with 0.2% gelatin and 0.04% NaN3 in PBS for 10 min followed by incubation with primary antibodies for 1 h at room temperature, washed with PBS, and incubated with secondary antibodies for 1 h at room temperature. Cells were stained for DNA with Hoechst 33342 (Tocris Bioscience, 5117) (1 μg/ml) and coverslips were mounted with Moviol. Wide-field fluorescence microscopy was performed on a Zeiss Observer Z1 using a 63x/1.4 oil objective at a Z-step of 0.5 µm; shown are max projections. Axiovision software was used for image acquisition and analysis. For quantification, ImageJ was used to count the number of LC3 puncta and for the colocalization analysis following the user’s manual.
Electron microscopy
EM was performed as previously described [Citation49]. Briefly, cells were plated in 6-well dishes. After 24 hours of culture, cells were fixed with 2.5% glutaraldehyde (Sigma-Aldrich, G5882) and then processed for Epon embedding (Electron Microscopy Sciences, 14,120). Sections of 60 nm were mounted onto Formvar-coated nickel grids (Electron Microscopy Sciences, FF200-Ni) and double contrasted with 2% uranyl acetate (Electron Microscopy Sciences, 22400) for 5 min and 3% lead citrate (Electron Microscopy Sciences, 17810) for 5 min. Grids were imaged using a JEM 1400plus TEM imaging system, utilizing an AMT XR81M-B camera. EM images were taken from the perinuclear region of the cell where Golgi membranes were normally concentrated. Golgi stacks and Golgi clusters were identified using morphological criteria and quantified using standard stereological techniques. They had to contain a nuclear profile with an intact nuclear envelope. A cisterna was defined as a membrane-bound structure in the Golgi cluster whose length is at least 4x its width, normally 20–30 nm in width and longer than 150 nm and a stack is the set of flattened, disk-shaped cisternae resembling a stack of plates [Citation50].
Subcellular fractionation
Subcellular fractionation was performed as previously described [Citation51]. Briefly, cells were washed with homogenization buffer (0.25 M sucrose, 1 mM EDTA, 1 mM magnesium acetate, 10 mM HEPES-KOH, pH 7.2, and protease inhibitors) and resuspended in 800 μl of homogenization buffer by pipetting. Cells were cracked with a ball bearing homogenizer as monitored under a microscope by trypan blue (Thermo Fisher Scientific, 15250016) exclusion to a breakage of 75–80%. The homogenate was centrifuged for 10 min at 1000 x g, 4°C. The PNS was isolated and subjected to ultracentrifugation in a TLA55 rotor at 120,000 x g for 1 h. The supernatant (cytosol) was collected, and the membranes in the pellet were resuspended in homogenization buffer. Equal volume fractions of the PNS, cytosol, and membrane fractions were analyzed by western blotting.
Modulation of autophagy
To induce autophagy and check Golgi phenotype under amino acid starvation condition, HeLa or NRK cells cultured in control medium were extensively washed with PBS and then treated with the following medium for 4 h unless stated: control medium (DMEM medium, 10% super calf serum and 100 units/ml penicillin-streptomycin); control medium with 400 nM BafA1 (LC Laboratories, B-1080), 50 μM CQ (Sigma-Aldrich, C6628), or 5 μM rapamycin (LC Laboratories, R-5000); EBSS (prepared following Sigma-Aldrich E6132 recipe); EBSS supplemented with 400 nM BafA1, 50 μM CQ, or 20 mM NH4Cl; glucose starvation medium [DMEM without glucose (Thermo Fisher Scientific, 11966025), 10% super calf serum and 100 units/ml penicillin-streptomycin]; or glucose starvation medium with 400 nM BafA1.
Immunoprecipitation
To determine the interaction between GORASP2 and other proteins, HeLa cells transfected with indicated plasmids, or GFP, GORASP2-GFP, and GORASP1-GFP stable HeLa cells, were lysed in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP40 and EDTA-free protease inhibitors (Roche, 11873580,001). Protein A Resin (GenScript, L00210) or Protein G Resin (GenScript, L00209) were pre-incubated with indicated antibodies for 2 h at 4ºC. Lysate was cleared by centrifugation, and incubated with the pretreated beads overnight at 4ºC, subsequently washed and analyzed by western blotting.
To determine the interaction between LC3 and LAMP2, BSA (EMD Millipore, 2930-100GM) or purified His-tagged GORASP2 was introduced into the co-immunoprecipitation assay. Briefly, mRFP-GFP-LC3 cells were lysed and protein concentration was measured, BSA or His-GORASP2 was added into the cell lysates at a 1:2500 (lower level) or 1:500 (higher level) ratio, mixed with GFP antibodies for co-immunoprecipitation.
In vitro pull down assay
GST and GST-tagged BECN1 and SH3GLB1 were expressed in BL21 (DE3) bacteria and purified with glutathione Sepharose 4B beads (GE Healthcare, 17075605); MBP and MBP-tagged UVRAG were expressed in BL21 (DE3) bacteria and purified with Amylose beads (New England Biolabs, E8021L); His-GORASP2 were expressed in BL21 (DE3) bacteria and purified with Ni-NTA beads (Thermo Fisher Scientific, 88222).
To determine GORASP2 interaction with BECN1 and SH3GLB1, 5 μg of GST, GST-tagged BECN1 or SH3GLB1 proteins were incubated with 5 μg of His-tagged GORASP2 or BSA (as negative control) in reaction buffer (20 mM Tris-HCl, pH8.0, 150 mM NaCl, 1% Triton X-100) at 4ºC overnight, glutathione Sepharose 4B beads were then added into the mixture for another 30 min. After extensive washing, bound proteins were analyzed by western blot with an anti-GORASP2 antibody. The same procedure was applied to the interaction between UVRAG and GORASP2. In Brief, 5 μg of MBP or MBP-UVRAG were incubated with 5 μg of His-tagged GORASP2 proteins in the same reaction buffer, and amylose beads were used to pull down the protein complex.
To determine UVRAG and BECN1 interaction, 5 μg MBP or MBP-tagged UVRAG proteins were incubated with 5 μg of GST-tagged BECN1 proteins in the presence of 5 μg of BSA or His-GORASP2 in the reaction buffer described above at 4ºC overnight, and amylose beads were used to pull down the protein complex.
Protease K protection assay
Protease K protection assay was performed as previously described [Citation52]. Briefly, HeLa cells were split onto 10 cm dishes and cultured in control medium. On the second day, one dish was treated with control medium and the other with EBSS and 400 nM BafA1 for 4 h. Cells were then collected and resuspended in HBS buffer (20 mM HEPES pH 7.4, 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, protease inhibitor), homogenized with a ball-baring homogenizer and centrifuged to generate PNS. Each PNS was equally divided into three tubes, one was left untreated, one was incubated with 2.5 μg/ml Protease K (Thermo Fisher Scientific, AM2542), and the other was treated with both protease K and 1% Triton X-100 (from 20% stock) for 10 min on ice. Protease K was then inhibited by adding 1 mM PMSF (from 100 mM stock in isopropanol) and incubated on ice for 10 min. Proteins in each sample were precipitated by methanol/chloroform. The pellets were dissolved in sample buffer (50 mM Tris-HCl, pH 6.8, and 2% SDS) and analyzed by western blotting.
Quantification and statistical analysis
All data represent the mean ± SD (standard deviation) of at least three independent experiments. At least 20 cells were counted for colocalization analysis or 300 cells for puncta number measurement. A statistical analysis was conducted with two-tailed Student’s t-test. Differences in means were considered statistically significant at p < 0.05. Significance levels are: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Analyses were performed using ImageJ. Figures were assembled with Photoshop CS5.
Supplemental Material
Download Zip (26.5 MB)Acknowledgments
We thank Dr. David Rubinsztein (University of Cambridge) for the mRFP-GFP-LC3 cell line, Dr. Beth Levine (UT Southwestern) for pBICEP-CMV2 3xFLAG-BECN1 and GLIPR2-MYC constructs, Dr. Yoshinori Takahashi (Penn State University) for MYC-SH3GLB1 construct, Dr. Kun-Liang Guan (University of California, San Diego) for GFP-ATG14 and GST-BECN1 constructs, Dr. Hong-Gang Wang (Penn State College of Medicine) for GST-SH3GLB1 construct, Dr. Joachim Seemann for GORASP1 antibodies, Dr. David Sheff (University of Iowa) for MYC antibodies, Dr. John Schiefelbein (University of Michigan) for mannitol, and members of the Wang lab for helpful and constructive discussions and comments on the project.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental materials
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156.
- Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3:588–596.
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42.
- Walczak M, Martens S. Dissecting the role of the Atg12–atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424–425.
- McEwan David G, Popovic D, Gubas A, et al. PLEKHM1 Regulates Autophagosome-Lysosome Fusion through HOPS Complex and LC3/GABARAP Proteins. Mol Cell. 2015;57:39–54.
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269.
- Chen D, Fan W, Lu Y, et al. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012;45:629–641.
- Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25::1327–1337.
- Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8::688–699.
- Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151.
- Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521.
- Hailey DW, Kim PK, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667.
- Ravikumar B, Moreau K, Jahreiss L, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757.
- Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900.
- Guo Y, Chang C, Huang R, et al. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci. 2012;125:1706–1715.
- Shoji-Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206.
- Sun Q, Fan W, Chen K, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216.
- Wang Y, Seemann J, Pypaert M, et al. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. Embo J. 2003;22:3279–3290.
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251.
- Zhang X, Wang Y. GRASPs in golgi structure and function. Front Cell Dev Biol. 2015;3:84.
- Pilli M, Arko-Mensah J, Ponpuak M, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234.
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460.
- Vicinanza M, Korolchuk VI, Ashkenazi A, et al. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015;57:219–234.
- Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438.
- Marino ML, Pellegrini P, Di Lernia G, et al. Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem. 2012;287:30664–30676.
- Bekier ME 2nd, Wang L, Li J, et al. Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol Biol Cell. 2017;28:2833–2842.
- Zhang X, Wang L, Lak B, et al. GRASP55 senses glucose deprivation through O-GlcNAcylation to promote autophagosome-lysosome fusion. Dev Cell. 2018;45:245–261,e246.
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362.
- Lu J, He L, Behrends C, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920.
- Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476.
- Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42–ra42.
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330.
- Duran JM, Kinseth M, Bossard C, et al. The role of GRASP55 in golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587.
- Rabouille C, Linstedt AD. GRASP: a multitasking tether. Front Cell Dev Biol. 2016;4:1.
- Gee HY, Noh SH, Tang BL, et al. Rescue of deltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146:746–760.
- Sutterlin C, Polishchuk R, Pecot M, et al. The golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222.
- Cheng JP, Betin VM, Weir H, et al. Caspase cleavage of the golgi stacking factor GRASP65 is required for Fas/CD95-mediated apoptosis. Cell Death Dis. 2010;1:e82.
- Dupont N, Jiang S, Pilli M, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. Embo J. 2011;30:4701–4711.
- Xiang Y, Zhang X, Nix DB, et al. Regulation of protein glycosylation and sorting by the golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659.
- Zhang X, Wang Y. GRASP55 facilitates autophagosome maturation under glucose deprivation. Mol Cell Oncol. 2018;5:e1494948.
- Zhang X, Wang Y. The golgi stacking protein GORASP2/GRASP55 serves as an energy sensor to promote autophagosome maturation under glucose starvation. Autophagy. 2018;14:1649–1651.
- Eberle HB, Serrano RL, Füllekrug J, et al. Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J Cell Sci. 2002;115:827–838.
- Moreau K, Ravikumar B, Renna M, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317.
- Fung C, Lock R, Gao S, et al. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806.
- Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396.
- Takahashi Y, Tsotakos N, Liu Y, et al. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 2016;7:20855–20868.
- Yamaguchi H, Woods NT, Dorsey JF, et al. SRC directly phosphorylates Bif-1 and prevents its interaction with Bax and the initiation of anoikis. J Biol Chem. 2008;283:19112–19118.
- Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750.
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010;5:758–772.
- Lucocq JM, Berger EG, Warren G. Mitotic golgi fragments in HeLa cells and their role in the reassembly pathway. J Cell Biol. 1989;109:463–474.
- Xiang Y, Seemann J, Bisel B, et al. Active ADP-ribosylation factor-1 (ARF1) is required for mitotic golgi fragmentation. J Biol Chem. 2007;282:21829–21837.
- Nguyen TN, Padman BS, Usher J, et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016;215:857–874.
