ABSTRACT
Damaged or redundant peroxisomes and their luminal cargoes are removed by pexophagy, a selective autophagy pathway. In yeasts, pexophagy depends mostly on the pexophagy receptors, such as Atg30 for Pichia pastoris and Atg36 for Saccharomyces cerevisiae, the autophagy scaffold proteins, Atg11 and Atg17, and the core autophagy machinery. In P. pastoris, the receptors for peroxisomal matrix proteins containing peroxisomal targeting signals (PTSs) include the PTS1 receptor, Pex5, and the PTS2 receptor and co-receptor, Pex7 and Pex20, respectively. These shuttling receptors are predominantly cytosolic and only partially peroxisomal. It remains unresolved as to whether, when and how the cytosolic pools of peroxisomal receptors, as well as the peroxisomal matrix proteins, are degraded under pexophagy conditions. These cytosolic pools exist both in normal and mutant cells impaired in peroxisome biogenesis. We report here that Pex5 and Pex7, but not Pex20, are degraded by an Atg30-independent, selective autophagy pathway. To enter this selective autophagy pathway, Pex7 required its major PTS2 cargo, Pot1. Similarly, the degradation of Pex5 was inhibited in cells missing abundant PTS1 cargoes, such as alcohol oxidases and Fox2 (hydratase-dehydrogenase-epimerase). Furthermore, in cells deficient in PTS receptors, the cytosolic pools of peroxisomal matrix proteins, such as Pot1 and Fox2, were also removed by Atg30-independent, selective autophagy, under pexophagy conditions. In summary, the cytosolic pools of PTS receptors and their cargoes are degraded via a pexophagy-independent, selective autophagy pathway under pexophagy conditions. These autophagy pathways likely protect cells from futile enzymatic reactions that could potentially cause the accumulation of toxic cytosolic products.
Abbreviations: ATG: autophagy related; Cvt: cytoplasm to vacuole targeting; Fox2: hydratase-dehydrogenase-epimerase; PAGE: polyacrylamide gel electrophoresis; Pot1: thiolase; PMP: peroxisomal membrane protein; Pgk1: 3-phosphoglycerate kinase; PTS: peroxisomal targeting signal; RADAR: receptor accumulation and degradation in the absence of recycling; RING: really interesting new gene; SDS: sodium dodecyl sulphate; TCA, trichloroacetic acid; Ub: ubiquitin; UPS: ubiquitin-proteasome system Vid: vacuole import and degradation
Introduction
The 2 major functions of single-membrane bounded peroxisomes are to scavenge reactive oxygen species and to catalyze fatty acid beta-oxidation [Citation1]. In addition to their roles in a variety of secondary metabolic processes, peroxisomes play critical roles in pathogen defense and the immune response [Citation2].
From fungi to plants and mammals, the peroxisomal lumen contains many matrix proteins, which are encoded by nuclear genes, synthesized by ribosomes in the cytosol, and imported into the peroxisome lumen. The majority of peroxisomal matrix proteins are targeted to the peroxisomal matrix by virtue of peroxisomal targeting signals (PTSs). Most such proteins contain, at their C-terminal end, a PTS1 signal, which is recognized by the receptor, Pex5 [Citation3]. A minority of peroxisomal matrix proteins harbor an N-terminal PTS2 signal, which is captured by the receptor, Pex7, and its co-receptor, Pex18 and Pex21 in Saccharomyces cerevisiae or Pex20 in Pichia pastoris and other yeast species [Citation4–Citation8]. The import of PTS1 cargo-receptor complexes is through a minimal peroxisomal translocon comprised of Pex5 and Pex14 [Citation9,Citation10].
In sharp contrast with mitochondria and chloroplasts, the peroxisomal receptors shuttle between the cytosol and peroxisomal lumen [Citation7,Citation11,Citation12]. After cargo import and release into the peroxisome lumen, Pex5 and Pex20 are subject to mono-ubiquitination, which enables receptor recycling back to the cytosol – a process that is dependent on the ubiquitin-conjugating enzyme, Pex4, the peroxisomal Really Interesting New Gene (RING) subcomplex (comprised of Pex2, Pex10 and Pex12), and the AAA ATPases, Pex1 and Pex6 [Citation13–Citation15]. The shuttling of Pex7 depends on Pex5 and Pex20 and is probably dependent on ubiquitination [Citation16]. The entry of peroxisomal cargo, along with their receptors, is fully blocked in the absence of Pex14, one of the 3 components of the docking complex at the peroxisome membrane. Because P. pastoris Pex5 and Pex20 accumulate in the peroxisome matrix in the absence of the RING subcomplex, it has been suggested that their export from peroxisomes is a necessary step in the peroxisomal matrix protein import cycle [Citation17].
In P. pastoris, peroxisome proliferation is induced by switching the carbon source in the cell growth medium from glucose to either methanol or oleate [Citation18,Citation19]. However, peroxisomes become redundant or superfluous in yeast cells when they are shifted from peroxisome proliferation conditions to peroxisome repression conditions, in the presence of glucose accompanied by nitrogen depletion. As a consequence, they are degraded by pexophagy, a selective autophagy pathway. During peroxisome proliferation, peroxisomes are already tagged by Atg30, via its interactions with the peroxisomal membrane proteins (PMPs), Pex3, Pex14, and Atg37 [Citation20,Citation21]. S. cerevisiae Atg36, a functional homolog of Atg30, tags peroxisomes for degradation through interaction with Pex3 [Citation22].
When peroxisomes become redundant or damaged, the pexophagy receptors are activated by multiple phosphorylation events, inducing the recruitment of the scaffold protein, Atg11, and the ubiquitin-like protein, Atg8, leading to the formation of a double-membrane structure (pexophagosome, by analogy to the autophagosome formed during non-selective autophagy) around the peroxisome targeted for degradation [Citation18]. Like autophagosome formation, this pexophagosome generation requires the core autophagy machinery. The outer membrane of the pexophagosome fuses with the vacuole, delivering the pexophagic body into the vacuole lumen for degradation and recycling of the constituent macromolecules. Atg30 and Atg36 homologs have not been yet identified in higher eukaryotic cells. However, in mammals, NBR1 acts as a pexophagy receptor [Citation23,Citation24]. Unlike in yeasts, ubiquitination is necessary for pexophagy in mammals. During starvation, the E3 ligase, PEX2, ubiquitinates PEX5 on the peroxisome surface, which serves as a signal to recruit NBR1 for peroxisome degradation [Citation25].
In yeasts, the PTS receptors are predominantly cytosolic and only partially peroxisomal. According to cell fractionation experiments, about 95% Pex5, 70–90% Pex7, and 70% Pex20 is localized in the cytosol in P. pastoris [Citation4,Citation26,Citation27]. Similar results were observed in other yeast species and human cells [Citation28,Citation29]. It is expected that the peroxisomal pools of Pex5, Pex7 and Pex20 are degraded with the organelles via the Atg30-dependent pexophagy pathway. However, it remains elusive as to whether, when and how the cytosolic pools of peroxisomal receptors and co-receptors are degraded under pexophagy conditions. Moreover, there are likely newly-synthesized peroxisomal matrix proteins that have not been imported into the peroxisome lumen when cells encounter pexophagy conditions, or in mutants (including human patients) impaired in peroxisome biogenesis. The fate of these non-essential peroxisomal proteins is also unknown under pexophagy conditions. However, these proteins, if left in the cytosol, might participate in futile enzymatic reactions and/or generate toxic products that may be harmful to the cells.
In this manuscript, we show that Pex5 and Pex7 are degraded in a manner that is independent of Atg30 under starvation conditions. However, the degradation of Pex5 and Pex7 is blocked in cells deficient in either autophagy or vacuolar proteases, suggesting the involvement of the core autophagy machinery and vacuolar proteolysis. Evidence for the selective degradation of these PTS receptors was gleaned from the finding that the degradation of Pex5 and Pex7 was disabled in the absence of their respective major cargoes under pexophagy conditions, whereas a cytosolic protein, Pgk1 (3-phosphoglycerate kinase), was still degraded under these conditions. However, the degradation of these PTS cargoes was not dependent on their cognate receptors. Other PTS proteins mislocalized to the cytosol were also cleared by an Atg30-independent, selective autophagy. Our results suggest that non-essential, obsolete or mistargeted cytosolic pools of peroxisomal proteins are degraded by a novel, Atg30-independent, selective autophagy pathway under starvation conditions.
Results
PTS receptors and co-receptors are degraded independent of Atg30 under nitrogen starvation conditions
The PTS receptors shuttle between the cytosol and the peroxisome lumen. As previously shown by cell fractionation experiments, only around 5% Pex5, 10–30% Pex7, and 30% Pex20 is localized to peroxisomes in P. pastoris, indicating that PTS receptors are predominantly cytosolic under peroxisome proliferation conditions [Citation4,Citation7,Citation26]. Redundant or superfluous peroxisomes are recognized by Atg30, the pexophagy receptor, and subjected to degradation via selectively autophagy when yeast cells are shifted from peroxisome proliferation to pexophagy conditions [Citation20]. It is expected that PTS receptors associated with peroxisomes are degraded together with the whole organelles.
In order to investigate the fate of the cytosolic pools of PTS receptors under pexophagy conditions, the protein levels of PTS receptors were analyzed. Wild-type cells (WT, PPY12) were grown in oleate medium to mid-log phase, then transferred to glucose medium without nitrogen (SD-N) to activate pexophagy. We used Pot1/thiolase, a PTS2-containing peroxisomal matrix protein to follow pexophagy [Citation20]. In agreement with our previous results, Pot1 was degraded in WT cells under starvation conditions (Figure 1A), but not in cells lacking Atg30 (). In sharp contrast, in replicate samples Pex5 was degraded in both WT and atg30∆ cells, although the degradation rate of Pex5 was slightly slower in atg30∆ cells in comparison with the WT cells (–C). Pex5 would likely not be degraded in atg30∆ cells if the cytosolic Pex5 was associated with peroxisomes under pexophagy conditions. The relatively fast clearance of Pex5 in atg30∆ cells suggests the existence of a new degradation pathway for Pex5 that is independent of the well-known pexophagy pathway.
Figure 1. Atg30-independent degradation of PTS receptors. (A, B) Pex5, Pex7, and Pex20 were degraded in both (A) WT and (B) atg30∆ cells. Peroxisome proliferation were induced by growing cells in oleate medium, followed by adaptation in glucose medium without nitrogen (SD-N) for the indicated times. Crude cell lysates were extracted by TCA precipitation, resolved by SDS-PAGE, and detected with anti-Pot1, anti-Pex5, and anti-HA (for Pex7 and Pex20). (C) Quantification of the degradation of Pex5, Pex7, and Pex20 (from A and B) using ImageJ and expressed as the percentage of the total signal at 0 h. The results represent the mean and standard deviation (SD) of triplicate biological replicates.
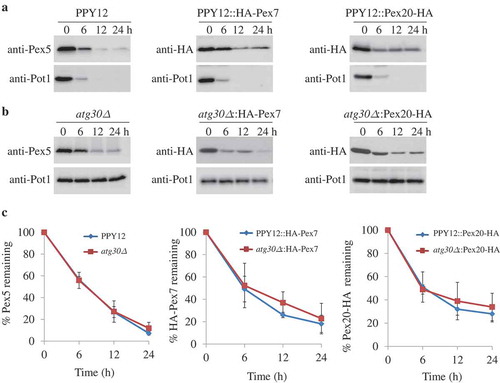
The expression levels of Pex7 and Pex20 were also monitored by expressing HA-Pex7 or Pex20-HA from their endogenous promoters, respectively, in the WT and atg30∆ strains. Our earlier studies showed that the addition of HA tags to Pex7 and Pex20 did not affect their functions [Citation16,Citation27]. Similar to Pex5, in replicate experiments the degradation of Pex7 and Pex20 was also independent of Atg30 under pexophagy conditions (). Therefore, all 3 PTS receptors/co-receptors are degraded under starvation conditions, but they do not follow the known pexophagy pathway.
The PTS receptors, but not the co-receptor, are degraded by selective autophagy under starvation conditions
In P. pastoris, when the recycling of Pex5 and Pex20 is blocked, they can be poly-ubiquitinated and degraded by the alternative receptor accumulation and degradation in the absence of recycling (RADAR) pathway under peroxisome proliferation conditions [Citation27], so we wondered if the RADAR pathway accounted for the degradation of these receptors. The poly-ubiquitination sites of P. pastoris Pex5 and Pex20 are located at positions K22 and K19, respectively [Citation30,Citation31]. The protein levels of Pex5K22R or Pex20K19R, with mutations of the ubiquitin acceptor sites on Pex5 and Pex20, respectively, were analyzed in the atg30∆ strain. As shown by the western blots, a simultaneous block of the RADAR and pexophagy pathways did not significantly affect the degradation of Pex5K22R or Pex20K19R under starvation conditions (Figure 2A–D). To further explore whether the degradation of peroxisomal receptors and the co-receptor follows the proteasome-dependent pathway, ubiquitin (Ub) mutated at position K48 to arginine, UbK48R, was overexpressed to inhibit proteasomal degradation in atg30∆ cells. Cells overexpressing UbK48R did not stabilize the level of Pex5 compared to that in WT cells at 3 separate time points (6, 12, and 24 h) following shift of the cells to pexophagy conditions (,C). Similarly, the protein level of Pex20-HA was not stabilized by overexpressing UbK48R in atg30∆ cells (,D). Therefore, the degradation of Pex5 and Pex20 is also independent of the RADAR and ubiquitin-proteasome system (UPS) pathways.
Figure 2. Pex5 and Pex7 are removed by selective autophagy. (A, B) Peroxisomes were induced by growing atg30∆ cells expressing Pex5K22R, Pex5K22R and UbK48R, Pex20K19R, or Pex20K19R and UbK48R in oleate medium. Subsequently, cells were starved for pexophagy experiments. Samples were taken at the indicated time points under starvation conditions and analyzed by immunoblots. (C, D) The degradation of Pex5 (from A) and Pex20 (from B) from triplicate biological repeats was quantified in ImageJ and expressed as the percentage of total signal at time 0 h. (E, F) Oleate-grown WT, SMD1163, atg1∆, atg8∆, atg11∆, atg17∆, atg11∆ atg17∆ cells or strains expressing HA-Pex7 were adapted to glucose medium without nitrogen (SD-N). Samples were collected at the indicated time points under starvation conditions for immunoblotting analysis. (G, H) The degradation of Pex5 (from E) and Pex7 (from F) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of total signal at 0 h.
In order to investigate whether the degradation of peroxisomal receptors follows any vacuole-dependent pathways, we checked the protein levels of Pex5, Pex7, and Pex20 in a strain lacking 2 vacuolar proteases, Prb1 and Pep4 (SMD1163). Interestingly, we found that the degradation of Pex5 and Pex7 was fully blocked in the SMD1163 strain compared to WT cells (–H). To further explore whether the degradation of Pex5 and Pex7 is autophagy dependent, their protein levels were checked in strains lacking components of the core autophagy machinery, Atg1 or Atg8. In comparison with the WT cells, stabilization of Pex5 and Pex7 was observed in atg1∆ and atg8∆ cells, indicating that Pex5 and Pex7 are indeed degraded by autophagy (–H). Atg11 and Atg17 function as scaffold proteins in autophagy machinery. The degradation of Pot1 was blocked in atg11∆ or atg17∆, as expected, while the degradation of Pex5 was partially blocked in atg11∆ or atg17∆ cells. However, the degradation of Pex5 was fully blocked when both Atg11 and Atg17 were absent (,G). Similarly, Pex7 was partially blocked in cells lacking Atg11 or Atg17, while fully stabilized in cells lacking both Atg11 and Atg17 (,H).
The cytoplasm to vacuole targeting (Cvt), mitophagy, pexophagy and nucleophagy pathways occur via selective autophagy and are dependent on Atg11. To further explore the involvement of Atg11 in the degradation of Pex5 and Pex7, we shifted cells from oleate to glucose medium (SD) to induce pexophagy, without nitrogen starvation, which activates autophagy. Pex5 and Pex7 were degraded in WT cells, but fully stabilized in cells lacking Atg11 or Atg17 (Fig. S1). Together, these data suggested that Atg11 and Atg17 are required for the selective degradation of Pex5 and Pex7.
However, unlike Pex5 and Pex7, the level of Pex20 was not stabilized in atg1∆, atg8∆, and SMD1163 cells, suggesting that the degradation of Pex20 is independent of autophagy-related pathways (Fig. S2A). We analyzed the turnover rate of Pex20 by treatment with cycloheximide, a protein synthesis inhibitor, in starvation medium. The half-life of Pex20 was around 4 h since its protein level was decreased by 50% at this time point after cycloheximide treatment (Fig. S2B,C). Moreover, the transcription level of PEX20 under starvation conditions was analyzed by RT-PCR and was repressed in starvation medium (Fig. S2D).
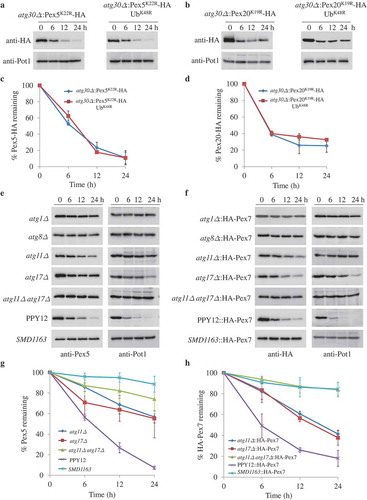
The degradation of Pex5 and Pex7 are independent of each other
Our previous results showed that the recycling of Pex7, and consequently its degradation under peroxisome-proliferation conditions, is dependent on the shuttling pathway of Pex5 and Pex20 [Citation16]. Therefore, we tried to dissect whether the degradation of Pex5 and Pex7 depended on each other and/or Pex20. In the absence of Pex7 and/or Pex20, Pex5 was still degraded upon starvation (Figure 3A,C). Pex2, a PMP, was used as a marker for pexophagy since Pot1 was mislocalized to the cytosol in cells missing either the PTS2 receptor or co-receptor. Likewise, Pex7 was still degraded in pex5∆, pex20∆, and pex5∆ pex20∆ cells (,D). Therefore, although both Pex5 and Pex7 are degraded under pexophagy conditions, their degradation does not depend on each other or on Pex20.
Figure 3. The absence of Pex5 or Pex7 under pexophagy conditions is irrelevant for the turnover of other PTS receptors. (A) Upon starvation, Pex5 was degraded in pex7∆, pex20∆, and pex7∆ pex20∆ cells. (B) Pex7 was degraded in pex5∆, pex20∆, and pex5∆ pex20∆ cells under pexophagy conditions. Pex2, a PMP, was used as a positive control for the degradation of peroxisomes. The protein levels of Pex2, Pex5, and HA-Pex7 were analyzed by immunoblotting with anti-Pex2, anti-Pex5, and anti-HA (for Pex7). (C, D) The degradation of Pex5 (from A) and Pex7 (from B) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of total signal at 0 h.
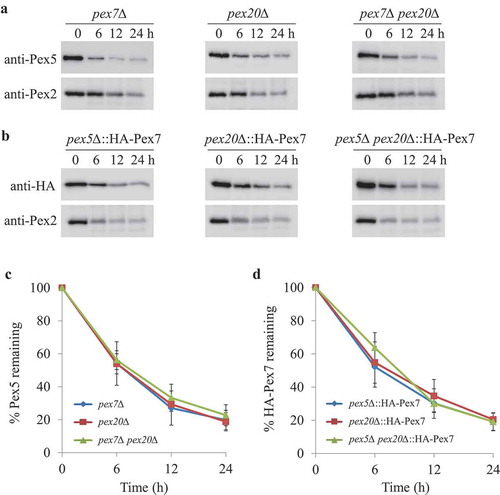
Pex7, but not cytosolic Pgk1, degradation was blocked in cells lacking Pot1
In P. pastoris, Pot1 is a major PTS2 protein. To dissect whether cargo binding to Pex7 modulates Pex7 degradation upon starvation conditions, we analyzed the fate of Pex7 in the absence of Pot1. To our surprise, Pex7 was stabilized in cells deficient in Pot1, indicating that Pex7 needs to form a complex with Pot1 and that they might be degraded together (Figure 4A,B). This dependence of Pex7 degradation on cargo binding was shown independently by using the Pex7A248R mutant, which is unable to bind Pot1 [Citation4]. Upon starvation, Pex2 and Pot1 were degraded in pex7∆ cells expressing Pex7A248R, while the degradation of Pex7A248R was blocked, as expected (,B). In order to investigate whether the Pot1-dependent degradation of Pex7 is specific and selective, we analyzed the fate of Pgk1, a cytosolic protein used as a control of nonselective autophagy [Citation32]. Under starvation conditions, Pgk1-GFP was slowly degraded by non-selective autophagy in WT cells, as demonstrated by its stabilization in SMD1163 cells (Fig. S3). However, in sharp contrast to Pex7, the degradation of Pgk1-GFP was not affected in cells deficient in Pot1. These results suggest that the degradation of Pex7 under starvation conditions requires it to bind its cargo and importantly, its degradation is through a selective mechanism and not via bulk autophagy.
Figure 4. Degradation of Pex7 depends on Pot1, which is selectively degraded when mislocalized. (A) Peroxisomes were induced by growing pot1∆ expressing HA-Pex7 and pex7∆ expressing HA-Pex7A248R in oleate medium. Subsequently, cells were transferred to starvation medium for pexophagy experiments. Samples were taken at the indicated time intervals under starvation condtions and analyzed by immunoblotting with anti-HA, anti-Pot1, and anti-Pex2 antibodies. (B) The degradation of Pex7 (from A) from triplicate biological repeats was quantified in ImageJ and expressed as the percentage of total signal at time 0 h. (C) Fluorescence analysis of WT, pex7∆, pex7∆ atg11∆, and pex7∆ atg11∆ atg17∆ strains expressing Pot1-GFP under starvation conditions in the presence of the vacuolar stain, FM 4–64. The differential interference contrast (DIC) and GFP images are shown. (D) Strains shown in (C) were used for GFP cleavage assays. Samples were analyzed by immunoblotting with anti-GFP. (E) The full-length Pot1-GFP and the cleaved GFP under starvation conditions (from D) were quantified using tripliciate biological repeats and ImageJ and expressed as the ratio between cleaved GFP and total GFP signals at each time point. *: non-specific band.
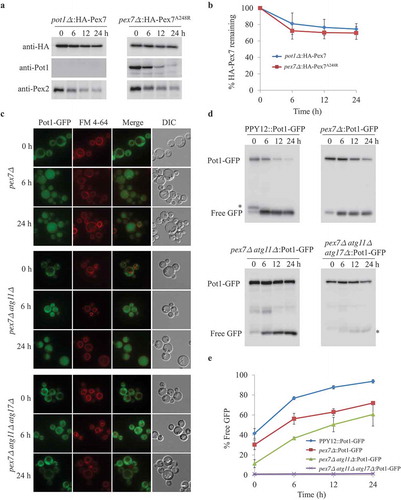
Selective degradation of mislocalized Pot1
To further characterize whether the cytosolic pool of Pot1 is degraded under starvation conditions, we used a strain lacking Pex7, in which Pot1 was mistargeted to the cytosol. To investigate whether mislocalized Pot1 was degraded by autophagy under starvation conditions, the fate of Pot1, which was fused to GFP (Pot1-GFP), was monitored using fluorescence microscopy. In oleate medium, Pot1-GFP was imported into peroxisomes in the WT cells, but was mislocalized to the cytosol in pex7∆ cells (, S4A). Upon transfer to SD-N medium, GFP appeared in the vacuole in pex7∆ cells, a hallmark of autophagy () and a significant fraction of the Pot1-GFP was degraded, as judged by the release of free GFP in the vacuole (,E). In pex7∆ atg1∆ and in pex7∆ atg11∆ atg17∆ cells, but not in pex7∆ atg30∆ or pex7∆ atg11∆ cells, by contrast, the GFP fluorescence remained cytosolic and outside the vacuole after cells were starved for 24 h (, S4A). However, we found that the GFP fluorescence was still largely cytosolic in pex7∆ atg11∆ cells, but not in pex7∆ and pex7∆ atg30∆ cells starved for 6 h, indicating the requirement of Atg11 in the degradation of cytosolic Pot1. Moreover, upon transfer to starvation medium, the degradation of Pot1-GFP was monitored by immunoblotting. The appearance of free GFP and a decrease in the Pot1-GFP levels were detected in pex7∆, pex7∆ atg30∆ cells, but in contrast, no free GFP appeared and the protein levels of Pot1-GFP were stabilized in pex7∆ atg1∆ and in pex7∆ atg11∆ atg17∆ cells (,E, S4B,C). Interestingly, the appearance of free GFP in pex7∆ atg11∆ and pex7∆ atg17∆ cells was slower in comparison with that in pex7∆ and pex7∆ atg30∆ cells, suggesting the involvement of Atg11 in the degradation of the cytosolic pool of Pot1 (,E,S4B,C).
If cytosolic Pot1 is degraded by autophagy, it should be found in autophagosomes. To confirm this point, we explored the fate of Pot1-GFP in pex7∆ prb1∆ pep4∆ mutants, in which autophagosomes should be stabilized in the vacuole due to the lack of vacuolar proteases, Prb1 and Pep4. We found Pot1-GFP-labeled punctate structures in the vacuole of pex7∆ prb1∆ pep4∆ cells after starvation for 3 h (Fig. S5). More and larger Pot1-GFP labeled punctate structures were observed in the vacuole with longer starvation times. These results indicate that in the absence of Pex7, cytosolic Pot1 is degraded in the vacuole by an autophagy-related pathway under starvation conditions, while Pex7 depends on Pot1 to enter this Atg30-independent degradation pathway. This asymmetrical dependence of the degradation of cytosolic Pex7 on Pot1, but not vice versa, is a strong indication of selectivity.
Block in degradation of Pex5 when it cannot bind abundant PTS1 cargoes
Unlike the existence of only one major PTS2 protein (Pot1), there are dozens of PTS1 proteins in P. pastoris. To investigate whether any specific PTS1 proteins bound to cytosolic Pex5 to be degraded together under starvation conditions, we performed Co-immunoprecipitation experiments using tandem HA-tagged Pex5, followed by mass spectrometry. In order to arrest the autophagic degradation, we transformed Pex5-HA into ypt7∆ cells, in which the fusion between the autophagosomes and vacuole is blocked [Citation33]. We found that around 40 proteins, including Pex14, Aox1, Aox2, and Fox2, could be pulled down with Pex5 (Table S1). The AOX1 and AOX2 genes encode alcohol oxidases, which are non-canonical PTS1 proteins bound to the N-terminal part of Pex5 [Citation34]. AOX1 and AOX2 are essential for the metabolism of methanol, and they are expressed at very low levels in oleate medium. Fox2 is a canonical, PTS1-containing, hydratase-dehydrogenase-epimerase protein that is involved in the peroxisomal fatty acid beta-oxidation pathway [Citation35].
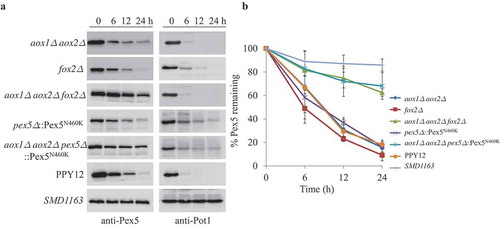
Because we found the PTS2 cargo protein, Pot1, was essential for the degradation of Pex7 under starvation conditions, we asked whether Aox1, Aox2 and Fox2 are also important components of the Pex5 degradation pathway. Although the degradation of Pex5 was not blocked in aox1∆ aox2∆ or fox2∆ cells, its degradation was severely blocked in the aox1∆ aox2∆ fox2∆ triple mutant (). To further confirm these results, we replaced endogenous Pex5 with Pex5N460K, a mutant that cannot bind to canonical PTS1 proteins [Citation31], in WT and aox1∆ aox2∆ cells. While non-canonical PTS1 proteins could still bind to Pex5N460K in the pex5∆::Pex5N460K strain, almost no PTS1 proteins bound to Pex5N460K in the aox1∆ aox2∆ pex5∆::Pex5N460K strain. Interestingly, the degradation of Pex5N460K was only severely slowed down in aox1∆ aox2∆ pex5∆::Pex5N460K, but not in the pex5∆::Pex5N460K strain (), suggesting Pex5 also depends on its cargoes to enter the degradation pathway.
Figure 5. Pex5 is degraded together with its cargoes. (A) Peroxisomes were induced by growing WT, SMD1163, aox1∆ aox2∆, fox2∆, aox1∆ aox2∆ fox2∆ or pex5∆ and aox1∆ aox2∆ pex5∆, each expressing the Pex5N460K mutant in oleate medium. Subsequently, cells were transferred to starvation medium for pexophagy experiments. Samples were taken at the indicated time points under starvation conditions and analyzed by immunoblotting with anti-Pex5 and anti-Pot1. (B) The degradation of Pex5 (from A) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of the total signal at 0 h.
In order to investigate whether the cargo-dependent degradation of Pex5 is specific and selective, we analyzed the fate of Pgk1 in the aox1∆ aox2∆ fox2∆ triple mutant. In sharp contrast to Pex5, the degradation of Pgk1-GFP was not affected in cells deficient in Aox1, Aox2, and Fox2 (Fig. S3). Once again, the requirement of the cargoes only for Pex5, but not Pgk1, degradation during pexophagy conditions is evidence of a selective pathway to degrade the cytosolic pool of PTS receptor-cargo complexes.
Selective degradation of cytosolic PTS1 proteins
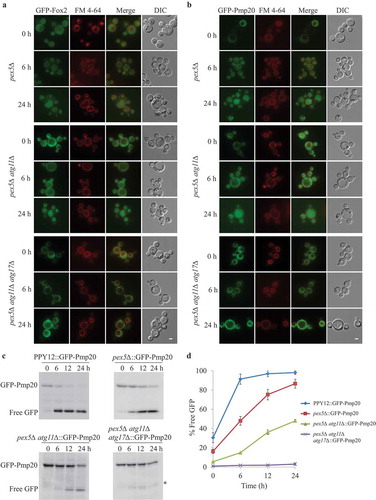
To elucidate whether the formation of PTS1-Pex5 complex is a prerequisite for the degradation of cytosolic localized Pex5 and their cargoes, we investigated the fate of PTS1 cargoes in the absence of Pex5. In pex5∆ cells, both canonical and non-canonical PTS1 proteins remained cytosolic, as expected. As shown by fluorescence microscopy, GFP-labeled Fox2 was targeted to peroxisomes in WT cells, but mislocalized to the cytosol in pex5∆ cells (Figure 6A, S6A). Upon starvation, peroxisomal GFP-Fox2 was degraded in the vacuole in WT cells, while the cytosolic, mislocalized GFP-Fox2 was targeted to the vacuole in pex5∆ cells for degradation as well. When Atg1 or both Atg11 and Atg17, but not Atg30, was missing in pex5∆ cells, GFP-Fox2 remained in the cytosol upon starvation (, S6). Moreover, when only Atg11 was missing in pex5∆ cells, GFP-Fox2 remained in the cytosol for up to 6 h of starvation, indicating the degradation of cytosolic GFP-Fox2 occurred via selective autophagy. As shown by fluorescence microscopy and GFP-cleavage assays, we found that another canonical PTS1 cargo, P. pastoris Pmp20/peroxiredoxin, was degraded in the vacuole of pex5∆ and pex5∆ atg30∆ cells, but it remained cytosolic in pex5∆ atg1∆ and in pex5∆ atg11∆ atg17∆ cells upon starvation (–D, S7). Pmp20 remained in the cytosol in pex5∆ atg11∆ cells, for up to 6 h of starvation, but was degraded more slowly than in pex5∆ cells. Pmp20 was degraded in the vacuole when pex5∆ atg11∆ cells were continuously starved for 24 h ().
Figure 6. Cytosolic Fox2 and Pmp20 are degraded by selective autophagy. (A, B) Oleate-grown pex5∆, pex5∆ atg11∆, and pex5∆ atg11∆ atg17∆ expressing GFP-Fox2 (A) or GFP-Pmp20 (B) from their endogenous promoters were transferred to starvation medium for pexophagy experiments in the presence of the vacuolar stain, FM 4–64. Fluorescence pictures were taken at 0, 6, and 24 h after starvation. (C) WT, pex5∆, pex5∆ atg11∆, and pex5∆ atg11∆ atg17∆ expressing GFP-Pmp20 were used for GFP cleavage assays. Samples were analyzed by immunoblotting with anti-GFP. A slightly longer exposure time for the pex5Δ atg11∆::GFP-Pmp20 blot to see the free GFP band. (D) The full-length GFP-Pmp20 and free GFP under starvation conditions (from C) were quantified using tripliciate biological repeats and ImageJ and expressed as the ratio between cleaved GFP and total GFP signals at each time point. *: non-specific band.
The fate of cytosolic GFP-Fox2 in the pex5∆ prb1∆ pep4∆ mutant was explored to investigate whether any autophagosome-like structures formed under starvation conditions. GFP-Fox2 labeled punctate structures were observed in the vacuole of pex5∆ prb1∆ pep4∆ starved for 3 h (Fig. S5). More and larger GFP-Fox2-labeled punctate structures were observed in the vacuole with longer starvation times. These data show that the PTS1 cargoes analyzed, when rendered cytosolic (as is the case in peroxisome biogenesis mutants and human peroxisome biogenesis disorders), are also degraded by Atg30-independent, selective autophagy.
Cue5 and Vid24 are not involved in the turnover of cytosolic peroxisomal proteins
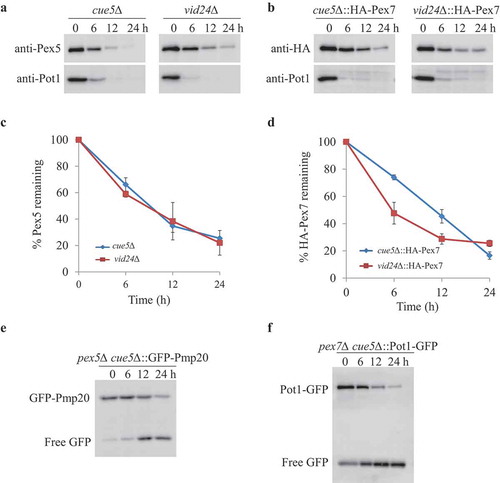
Aggrephagy and the vacuole import and degradation (Vid) pathways are 2 known autophagy-related pathways involved in the degradation of cytosolic proteins [Citation36,Citation37]. In S. cerevisiae, ubiquitinated protein aggregates are degraded by Cue5-mediated aggrephagy pathway [Citation36]. In order to investigate whether cytosolic peroxisomal receptors and proteins are degraded by the aggrephagy pathway, we deleted the CUE5 gene in P. pastoris. However, neither the degradation of Pex5 and Pex7 in cue5∆ cells, nor the degradation of cytosolic Pot1 in pex7∆ cue5∆ cells, respectively, was affected, suggesting that the aggrephagy pathway is irrelevant to the degradation of PTS receptors and cytosolic PTS2 proteins (Figure 7). The cytosolic Pmp20 was degraded slightly slower only at the 6 h time point of starvation in pex5∆ cue5∆ cells compared to pex5∆ cells by the GFP cleavage assay, but this delay was not obvious under microscopy analysis (Fig. S7A), suggesting that the aggrephagy pathway plays a minor role, if at all, in the degradation of cytosolic Pmp20.
S. cerevisiae Vid24 is an essential protein of the Vid pathway that is involved in the degradation of the key gluconeogenic enzyme, fructose-1,6-bisphosphatase, when cells are shifted from medium containing poor carbon sources to fresh glucose [Citation37]. However, the degradation of Pex5 and Pex7 was not affected in the absence of Vid24 in P. pastoris (,B). Therefore, our data indicate that cytosolic peroxisomal proteins are degraded by means of a novel selective autophagy pathway instead of by the aggrephagy or Vid pathways.
Figure 7. Aggrephagy and Vid pathways were not involved in removing Pex5, Pex7 and cytosolic peroxisomal cargoes. (A, B) oleate-grown cue5Δ and vid24∆ (A) or cue5∆ and vid24∆ cells expressing HA-Pex7 from its native promoter (B) were shifted to starvation medium for pexophagy experiments. (C, D) The degradation of Pex5 and Pex7 (from A and B) was quantified in ImageJ and expressed as the percentage of the total signal at time 0 h. The results represent the mean and standard deviation (SD) of triplicate biological repeats. (E, F) Oleate-grown pex5∆ cue5∆ cells expressing GFP-Pex20 (E) and pex7∆ cue5∆ cells expressing Pot1-GFP (F) were transferred to stavation medium for GFP cleavage assays. Samples were taken at the indicated time points under starvation condtions and analyzed by immunoblotting with anti-GFP.
Discussion
Previous studies have shown that like many other organelles, peroxisomes, along with their matrix and membrane constituents, are turned over by pexophagy when they are either redundant or dysfunctional, and that this process is important for peroxisome homeostasis [Citation18,Citation20].
Independent of pexophagy, at the peroxisomal membrane there exists a quality control pathway that prevents a logjam when PTS receptor or co-receptor recycling from the peroxisome membrane back to the cytosol is impaired, via the activation of the RADAR pathway, which involves poly-ubiquitination of these receptors and co-receptors and their subsequent degradation by the UPS [Citation14,Citation15,Citation27,Citation38].
However, since the PTS receptors and co-receptors that shuttle cytosolic, PTS-containing cargoes to and from the peroxisomes are predominantly cytosolic [Citation4,Citation26,Citation27], we addressed the fate of this cytosolic pool of PTS receptors and co-receptors during pexophagy. Additionally, in many human peroxisome biogenesis disorders, in which peroxisomal matrix protein import is compromised, most of the proteins destined for the peroxisome matrix would remain in the cytosol, where they might produce toxic byproducts and/or engage in futile metabolic reactions that would be detrimental to the cells [Citation39]. Aging cells, or those subjected to oxidative stress, have also been shown to exhibit reduced peroxisomal matrix protein import, which would enhance the cytosolic pool of these cargoes [Citation40,Citation41]. We also addressed the fate of these cytosolic peroxisomal matrix proteins.
Data presented in this study show that the cytosolic pools of Pex5, Pex7, and Pex20 are also degraded under pexophagy conditions, but not by pexophagy because they were not dependent on Atg30 (). Although these proteins are polyubiquitinated [Citation14,Citation15,Citation27,Citation38], the degradation of their cytosolic pools is not affected by perturbations in the UPS pathway, for example by overexpression of UbK48R or by expression of mutants that prevent polyubiquitination of these proteins (). Instead, Pex5 and Pex7 are turned over, independent of each other, by a new Atg30-independent, autophagy pathway ( and ). Several independent lines of evidence support this view. First, the degradation of Pex5, Pex7, and Pex20 is independent of Atg30 (). Second, the degradation of Pex5 and Pex7 requires Atg11 (, S1), whose requirement in other selective autophagy pathways suggests that selective autophagy may be involved in the degradation of cytosolic Pex5 and Pex7. This point is reinforced by the requirement of Atg17 for Pex5 and Pex7 degradation (). Finally, the dependence of Pex5 and Pex7 degradation on their respective PTS1 and PTS2 cargoes is strong evidence that these receptors are degraded selectively ( and ). In contrast to Pex5 and Pex7, the level of Pex20 was not stabilized in atg1∆ cells, suggesting that the degradation of Pex20 is independent of autophagy (Fig. S2).
Our studies also show that peroxisomal matrix proteins that mislocalize to the cytosol are degraded by Atg30-independent and Atg11-dependent selective autophagy ( and , S4, S6, S7), but their turnover does not require their cognate PTS receptors, suggesting that the PTS receptors hitch a ride via their cargoes, as they are destroyed.
Selective degradation of cytosolic pools of peroxisomal receptors
In addressing what proteins are necessary for the Atg30-independent degradation of cytosolic Pex5 and Pex7 under pexophagy conditions, we found that they require both the core and selective autophagy machineries (). Under peroxisome proliferation conditions, the import of peroxisomal matrix proteins by PTS receptors is reasonably efficient. Although there are only low levels of peroxisomal matrix proteins in the cytosol, which are probably newly-synthesized or damaged cargoes, the respective PTS cargoes are essential for the degradation of cytosolic Pex5 and Pex7 ( and ). The degradation of Pex7 is blocked if it cannot bind to Pot1, a PTS2 protein, or if no Pot1 exists in the cytosol (), suggesting that Pex7 is degraded together with its cargo. This cargo-dependent degradation of cytosolic PTS receptors is also true for Pex5 since it was stabilized in the absence of its major PTS1 cargoes Aox1, Aox2, and Fox2, or when it could not bind to the majority of PTS1 cargoes due to mutations in its cargo-binding domain ().
Clearance of mislocalized cytosolic PTS cargoes
Somewhat surprising was the finding that while PTS receptor degradation was cargo-dependent, the converse was not true, in that neither the cytosolic pools of Pot1 nor Fox2 and Pmp20 required their cognate PTS receptors for their turnover ( and ). We therefore asked whether these PTS cargoes might be degraded by an alternative pathway. There are at least 2 known autophagy-related pathways implicated in the vacuolar degradation of cytosolic proteins, the aggrephagy and Vid pathways [Citation36,Citation37]. However, neither the degradation of Pmp20 nor Pot1 was blocked in pex5∆ cue5∆ or pex7∆ cue5∆ cells ().
In the absence of functional human PEX7, as seen in the case of human patients with rhizomelic chondrodysplasia punctata, or as we show in the absence of yeast Pex7, its cargo, Pot1, is mislocalized to the cytosol [Citation42]. The cytosolic Pot1 was degraded in cells deficient in either Pex7 () or Atg30 (Fig. S4), but was partially stable in pex7∆ atg11∆ and fully stabilized in pex7∆ atg1∆ cells (, S4). The Atg1 requirement suggests the involvement of autophagy, while the Atg11 dependence is reminiscent of other selective autophagy pathways. Therefore, an Atg11-dependent, but Atg30-independent, selective autophagy pathway is involved in the degradation of cytosolic Pot1.
Similarly, PTS1 proteins are mislocalized to the cytosol in the absence of Pex5, or in Zellweger syndrome patients expressing a non-functional PEX5 [Citation43]. The degradation of mislocalized cytosolic PTS1 proteins, Fox2 and Pmp20, also partially depended on an Atg11-dependent, selective autophagy pathway (, S6, S7A). It is likely that a selective autophagy receptor, similar to the soluble Cvt receptor Atg19, targets the cytosolic PTS1 proteins for degradation.
In summary, our data indicate the existence of 2 selective autophagy pathways related to peroxisomes. The first one is the Atg30-dependent, canonical pexophagy pathway, and the second one is an Atg30-independent and Atg11-dependent, non-canonical pathway that remove the cytosolic pool of peroxisome-related proteins that is described here. Under pexophagy conditions, these pathways act together to maintain the homeostasis of proteins that either reside in peroxisomes or are destined for peroxisomes.
Materials and methods
Yeast strains and culture conditions
P. pastoris strains used in this study represent genomic integrations and are listed in Table S2. Growth media components were as follows: YPD medium (2% glucose, 2% Bactopeptone, and 1% yeast extract), nitrogen starvation medium (0.67% yeast nitrogen base without ammonium sulfate and amino acids, 2% glucose), YNM medium (0.67% yeast nitrogen base without amino acids, 0.05% yeast extract, 0.5% [v/v] methanol), YNO medium (0.67% yeast nitrogen base without amino acids, 0.05% yeast extract, 0.2% oleic acid, 0.02% Tween 80), supplemented with the appropriate Complete Supplement Mixture of amino acids (Sunrise science, 84-1001-100). Yeast cells were grown in YPD overnight at 30°C, then diluted in YPD and grown to the early-mid exponential phase. Cells were washed in sterile distilled water, inoculated in oleate medium at an OD600 of 0.3–0.6 and grown for approximately 16 h at 30°C. Then, cells were washed twice with sterile distilled water and inoculated into glucose medium with nitrogen (SD) or glucose medium without nitrogen (SD-N) medium to induce peoxphagy. For the cycloheximide treatment, a final concentration of 5 mg/ml cycloheximide (MedChemExpress, HY-12320) was added to SD-N medium.
Plasmid construction
A DNA fragment encoding GFP fused to the C-terminal end of PpPot1 was amplified using pJCF701 (lab stock) as template and cloned in pMY69 or pMY72 at ClaI/AflII sites, creating pCLM362 and pCLM340, respectively. A DNA fragment containing the promoter of PGK1 and its CDS was amplified from PPY12 genomic DNA and cloned in pWD3 at XmaI/SphI sites, creating pCM301. A linear DNA fragment containing the Nourseothricin expression cassette was amplified from pAG25 and cloned in pCM301 at the SmaI site, creating pCLM571. The promoter of PpPMP20 and GFP-PMP20 were amplified from PPY12 genomic DNA and pCLM361 plasmid (lab stock), respectively. The two DNA fragments were assembled together in pMY72 at the BspHI/AflII sites using In-Fusion HD Cloning (Takara, 638910), creating pCLM363.”
To generate FOX2 and ATG11 knock-out constructs, the 3′ and 5′ flanking regions of the genes were amplified from PPY12 genomic DNA, respectively, and fused together by overlap PCR with a BamHI site in between. The 3′-UTR-BamHI-5′-UTR fragments were transferred to pMYZeo at the EcoRI/SalI by In-Fusion HD Cloning (Takara, 638910). Similarly, to generate PEX20 and ATG1 knockout constructs, 3′-UTR-BamHI-5′-UTR fragment of PEX20 and 3′-UTR-SalI-5′-UTR fragment of ATG1 were amplified by overlap PCR using PPY12 genomic DNA as template and cloned in pMYZeo at the KpnI/HindIII and EcoRI/SphI by In-Fusion HD Cloning, respectively. To generate the PEX5 knock-out construct, the 5′-UTR and 3′-UTR regions of PEX5 were amplified from PPY12 genomic DNA and transferred to the pAG25 vector at the HindIII/BamHI and SacI/SpeI sites, respectively. Similarly, to generate the PEX7 knock-out construct, the 5′-UTR and 3′-UTR regions of the PEX7 gene were transferred to the pAG25 vector at the HindIII/BamHI and SacI/HpaI sites, respectively. The FOX2, PEX5, PEX7, PEX20, and ATG1 knock-out constructs were linearized and integrated into the corresponding gene loci in P. pastoris genome. To generate the pex5∆ atg30∆ and pex7∆ atg30∆ double mutants, a linear DNA fragment containing the Zeocin expression cassette flanked by the 5′ and 3′ regions of the ATG30 gene was amplified from the atg30∆ strain and introduced into the pex5∆ or pex7∆ mutant to replace the endogenous ATG30 gene.
All plasmids used in this study are listed in Table S3.
Pexophagy assay
In P. pastoris, peroxisome proliferation was induced by incubation of cells in oleate medium for overnight and transferred to SD or SD-N medium at an OD600 of 0.5 or 2, respectively, to induce pexophagy. Equal volumes of samples were taken at the indicated time points. Samples were precipitated with trichloroacetic acid (TCA, 12.5%) and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by immunoblot analysis with the indicated antibodies.
Co-immunoprecipitation assay
To induce peroxisome proliferation, ypt7Δ cells expressing Pex5-HA from its endogenous promoter were pre-cultured in YPD medium overnight and then shifted to oleate medium for around 16 h. Subsequently, cells were transferred to SD-N medium to induce pexophagy. 250 OD600 of cells was harvested and resuspended in 4 mL of Zymolyase solution (Nacalai, 07663–91) after cells were starved for 1 h. The following co-immunoprecipitation was performed as described previously [Citation44]. Sigma-Aldrich EZview Red Anti-HA (Sigma, E6779) was used to pull down Pex5-HA. The eluate was separated by SDS-PAGE and used for mass spectroscopy analysis at the UC San Diego core facility.
Immunoblotting
For analysis of protein levels, equal amounts of samples were removed and TCA precipitated. Samples were subject to SDS-PAGE and followed by western blotting with anti-GFP (mouse, 1:2,000; Roche, 11,814,460,001), anti-GFP (mouse, 1:2,000; Utibody, UM3002, for detection of GFP-Fox2 only), anti-HA (rat, 1:2,000; Roche, 11,867,423,001), anti-Pex2 (rat; 1:5,000; Subramani Laboratory), anti-Pex5 (rabbit, 1:10,000; Subramani Laboratory), anti-Pex14 (rabbit, 1:10,000; Subramani Laboratory), anti-Pot1 (rabbit, 1:5,000; Subramani Laboratory). Image Studio Ver5.2 software was used to examine if the blots were oversaturated.
Fluorescence microscopy
Pexophagy induced yeast cells were analyzed by fluorescence microscopy. Images were taken using a Zeiss Axio Imager Z2 fluorescence microscope coupled to a cooled CCD monochrome camera (AxioCam HRm, Zeiss) and analyzed using ZEN pro software.
Supplemental Material
Download Zip (2.1 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Materials
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193(1):7–16.
- Dixit E, Boulant S, Zhang Y, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–681.
- Terlecky SR, Nuttley WM, McCollum D, et al. The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. Embo J. 1995;14(15):3627–3634.
- Elgersma Y, Elgersma-Hooisma M, Wenzel T, et al. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J Cell Biol. 1998;140(4):807–820.
- Marzioch M, Erdmann R, Veenhuis M, et al. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. Embo J. 1994;13(20):4908–4918.
- Zhang JW, Cai X, Lazarow PB. Peb1p (Pas7p) is an intra-peroxisomal receptor for the N-terminal, type 2, peroxisomal targeting signal of thiolase. Ann N Y Acad Sci. 1996;804:654–655.
- Leon S, Goodman JM, Subramani S. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim Biophys Acta. 2006;1763(12):1552–1564.
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143(7):1859–1869.
- Ma C, Schumann U, Rayapuram N, et al. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell. 2009;20(16):3680–3689.
- Meinecke M, Cizmowski C, Schliebs W, et al. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12(3):273–277.
- Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105(2):187–196.
- Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167(4):599–604.
- Grimm I, Saffian D, Platta HW, et al. The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta. 2012;1823(1):150–158.
- Platta HW, El Magraoui F, Baumer BE, et al. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29(20):5505–5516.
- Liu X, Subramani S. Unique requirements for mono- and polyubiquitination of the peroxisomal targeting signal co-receptor, Pex20. J Biol Chem. 2013;288(10):7230–7240.
- Hagstrom D, Ma C, Guha-Polley S, et al. The unique degradation pathway of the PTS2 receptor, Pex7, is dependent on the PTS receptor/coreceptor, Pex5 and Pex20. Mol Biol Cell. 2014;25(17):2634–2643.
- Zhang L, Leon S, Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol Biol Cell. 2006;17(2):690–699.
- Farré JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17(9):537–552.
- Oku M, Sakai Y. Pexophagy in yeasts. Biochim Biophys Acta. 2016;1863(5):992–998.
- Farré JC, Manjithaya R, Mathewson RD, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14(3):365–376.
- Nazarko TY, Ozeki K, Till A, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014;204(4):541–557.
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. Embo J. 2012;31(13):2852–2868.
- Deosaran E, Larsen KB, Hua R, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126(4):939.
- Kim PK, Hailey DW, Mullen RT, et al. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105(52):20567–20574.
- Sargent G, van Zutphen T, Shatseva T, et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol. 2016;214(6):677–690.
- Gould SJ, Kalish JE, Morrell JC, et al. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135(1):85–95.
- Leon S, Zhang L, McDonald WH, et al. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172(1):67–78.
- Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135(6):1763–1774.
- Titorenko VI, Smith JJ, Szilard RK, et al. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142(2):403–420.
- Léon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J Biol Chem. 2007;282(10):7424–7430.
- Ma C, Hagstrom D, Polley SG, et al. Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J Biol Chem. 2013;288(38):27220–27231.
- Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6(6):794–797.
- van Zutphen T, Veenhuis M, van der Klei IJ. Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha. Autophagy. 2011;7(8):863–872.
- Gunkel K, van Dijk R, Veenhuis M, et al. Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery. Mol Biol Cell. 2004;15(3):1347–1355.
- Hiltunen JK, Wenzel B, Beyer A, et al. Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the FOX2 gene and gene product. J Biol Chem. 1992;267(10):6646–6653.
- Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–563.
- Chiang MC, Chiang HL. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998;140(6):1347–1356.
- El Magraoui F, Brinkmeier R, Schrotter A, et al. Distinct ubiquitination cascades act on the peroxisomal targeting signal type 2 co-receptor Pex18p. Traffic. 2013;14(12):1290–1301.
- Waterham HR, Ferdinandusse S, Wanders RJ. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta. 2016;1863(5):922–933.
- Terlecky SR, Koepke JI, Walton PA. Peroxisomes and aging. Biochim Biophys Acta. 2006;1763(12):1749–1754.
- Walton PA, Brees C, Lismont C, et al. The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim Biophys Acta. 2017;1864(10):1833–1843.
- Braverman N, Steel G, Obie C, et al. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet. 1997;15(4):369–376.
- Schliebs W, Saidowsky J, Agianian B, et al. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J Biol Chem. 1999;274(9):5666–5673.
- Farré JC, Carolino K, Stasyk OV, et al. A new yeast peroxin, Pex36, a functional homolog of mammalian PEX16, functions in the ER-to-peroxisome traffic of peroxisomal membrane proteins. J Mol Biol. 2017;429(23):3743–3762.
