ABSTRACT
Short linear motifs, known as LC3-interacting regions (LIRs), interact with mactoautophagy/autophagy modifiers (Atg8/LC3/GABARAP proteins) via a conserved universal mechanism. Typically, this includes the occupancy of 2 hydrophobic pockets on the surface of Atg8-family proteins by 2 specific aromatic and hydrophobic residues within the LIR motifs. Here, we describe an alternative mechanism of Atg8-family protein interaction with the non-canonical UBA5 LIR, an E1-like enzyme of the ufmylation pathway that preferentially interacts with GABARAP but not LC3 proteins. By solving the structures of both GABARAP and GABARAPL2 in complex with the UBA5 LIR, we show that in addition to the binding to the 2 canonical hydrophobic pockets (HP1 and HP2), a conserved tryptophan residue N-terminal of the LIR core sequence binds into a novel hydrophobic pocket on the surface of GABARAP proteins, which we term HP0. This mode of action is unique for UBA5 and accompanied by large rearrangements of key residues including the side chains of the gate-keeping K46 and the adjacent K/R47 in GABARAP proteins. Swapping mutations in LC3B and GABARAPL2 revealed that K/R47 is the key residue in the specific binding of GABARAP proteins to UBA5, with synergetic contributions of the composition and dynamics of the loop L3. Finally, we elucidate the physiological relevance of the interaction and show that GABARAP proteins regulate the localization and function of UBA5 on the endoplasmic reticulum membrane in a lipidation-independent manner.
Abbreviations: ATG: AuTophaGy-related; EGFP: enhanced green fluorescent protein; GABARAP: GABA-type A receptor-associated protein; ITC: isothermal titration calorimetry; KO: knockout; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; NMR: nuclear magnetic resonance; RMSD: root-mean-square deviation of atomic positions; TKO: triple knockout; UBA5: ubiquitin like modifier activating enzyme 5
Introduction
Autophagy is a broad set of lysosome-based catabolic processes that degrade cellular components (reviewed in [Citation1–Citation4]). While it was initially described as a non-selective bulk degradation pathway for recycling cellular metabolites to maintain cellular homeostasis, it is now well recognized that autophagy can specifically target and clear misfolded and/or aggregated proteins, dysfunctional or damaged organelles and intracellular pathogens [Citation5–Citation7]. At the core of this pathway are ubiquitin-like proteins (UBLs) of the Atg8/LC3/GABARAP family (Atg8-family proteins) that, being covalently linked to phosphatidylethanolamine (PE) by a specific ubiquitin-like conjugation cascade, are incorporated into the inner and outer membrane of the double-membrane autophagosome. The Atg8–PE conjugates perform various cellular functions of which the regulation of the autophagosome formation and closure, as well as recruitment of cargo via selective autophagy receptors (SARs), such as SQSTM1/p62, are the most prominent ones [Citation8–Citation10]. There is only 1 member of the Atg8-family in yeast; however, the mammalian genome encodes at least 6 homologs: MAP1LC3A (microtubule-associated protein light chain 3 alpha; LC3A), LC3B, LC3C, GABARAP (GABA type A receptor-associated protein), GABARAPL1 (GABA type A receptor associated protein like 1) and GABARAPL2/GATE-16. Although all 6 mammalian Atg8-family proteins are essential for autophagy, it is not fully known whether they are functionally redundant or have distinct roles in and outside the autophagy pathway [Citation11]. LC3 proteins were initially thought to be important in early phases of autophagy initiation and recruitment of SARs, whereas GABARAPs were shown to be crucial for later stages, such as membrane fusion and transport [Citation12]. However, this functional subdivision may be incomplete, since GABARAPs were also shown to be crucial in the early stages of autophagy via the activation of the ULK1/2 complex and have very distinct roles outside the autophagy pathway [Citation13]. They were, for example, shown to be involved in intra Golgi transport and Golgi reassembly or serve as signaling scaffolds and in membrane recruitment [Citation14–Citation17].
Independent of their functions, Atg8-family proteins interact with SARs and other proteins via short linear motifs designated as LC3-interacting regions (LIRs), LC3 recognition sequences (LRSs), or AIMs (Atg8-interaction motifs) (AIMs) [Citation18–Citation20]. The core of the canonical LIR motif represents a consensus of 4 residues Θ-X-X-Γ, where Θ is an aromatic residue (W/F/Y), Γ is a large hydrophobic residue (L/V/I) and X could be any residue. Although most LIRs known so far fit in this classical definition, there are some examples of atypical LIRs, such as those in SARs NDP52 and TAX1BP1, which lack the class-defining aromatic residue Θ [Citation21,Citation22]. It is commonly accepted that all LIRs form an intermolecular β-sheet with β-strand β2 in Atg8-family proteins. Additionally, the side chain of the aromatic residue Θ binds deeply inside a hydrophobic pocket (HP1) on the Atg8-family protein surface, formed between the Atg8-family proteins α-helix α2 and the β-strand β2 [Citation23]. The side chain of the hydrophobic residue Γ occupies the second hydrophobic pocket (HP2) between the β-strand β2 and the α-helix α3 (reviewed in [Citation3,Citation19]).
Recently, we reported that UBA5 (ubiquitin like modifier activating enzyme 5), the E1-like enzyme for the UBL UFM1 (ubiquitin fold modifier 1) possesses a double-specific LIR/UFIM motif at the C terminus [Citation24]. This short motif has an amino acid sequence, which is distinct from both canonical and non-canonical LIR motifs. The specific feature of the LIR/UFIM is that it not only recognizes the cognate substrate of UBA5, UFM1, but also binds mammalian Atg8-family proteins with a clear preference for GABARAPs. Following the activation by the E1-like enzyme UBA5 [Citation25], UFM1 can be covalently attached to target proteins [Citation26] in an ubiquitin-like process termed ufmylation, which also requires the E2-like UFC1 and E3-like UFL1 enzyme. Ufmylation occurs in almost all eukaryotic organisms (except fungi) where it plays various biological roles, e.g. in the endoplasmic reticulum (ER) stress response, hematopoiesis and erythroid lineage differentiation [Citation27–Citation30]. Additionally, this pathway has been implicated in the development of cancer [Citation31].
We and others identified GABARAPs as major interaction partners of UBA5 [Citation24,Citation26,Citation32]. However, it remained unclear how this highly unusual UBA5 LIR/UFIM interacts with GABARAPs and what the biological role of this interaction is. In the present study, we solved the structures of the complexes of GABARAPL2 and GABARAP with the UBA5 LIR/UFIM (hereafter UBA5 LIR) by solution NMR and by X-ray crystallography. We identified a novel mode of interaction mediated by a conserved tryptophan residue (W341) N-terminal of the UBA5 LIR core sequence that docks into a newly-described hydrophobic pocket on the surface of GABARAP/GABARAPL2. Using peptide arrays, we showed that W341 is crucial for the binding of the core UBA5 LIR/UFIM motif to GABARAP proteins. We observed that the binding of UBA5 LIR to GABARAPs is accompanied by structural rearrangements in side chains of key GABARAPs residues close to the gate-keeping K46 residue. Such conformational changes in side chain geometry are also unique and were not observed in any other Atg8:LIR complexes described to date. Finally, we investigate the biological relevance of this interaction and identify GABARAP proteins as potent regulators of ufmylation by recruiting UBA5 to the ER membrane.
Results
Structural analysis of GABARAP proteins in complex with the UBA5 LIR peptide reveals a new molecular mechanism of Atg8:LIR interactions
We previously showed that UBA5 has an unusual LIR motif consisting of the core sequence EWGIELVSE, which predominantly interacts with GABARAP proteins [Citation24] and does not align with other known LIR motifs [Citation3]. To understand the molecular mechanism of this interaction, we solved the NMR solution structure of GABARAPL2 in complex with a peptide spanning residues 333–348 of UBA5 (.). Additionally, we determined the crystal structure of GABARAP in complex with the length-optimized UBA5 LIR peptide ()) using a chimeric construct consisting of the GABARAP sequence (residues 3–116) C-terminally fused to the UBA5 LIR peptide (residues 337–350) via a short A-M-G linker. Crystals of this UBA5337-350-GABARAP3-117 construct diffracted to 1.30 Å resolution. The complex structures (overviewed in Table S1 and Table S2) were similar to each other and can be superimposed over the backbone atoms of structured regions with an RMSD of 1.2 Å.
Figure 1. The structure of UBA5 LIR in complex with GABARAP proteins reveals a new mechanism of interaction. (a) NMR solution structure of the complex between GABARAPL2 (representative conformer is shown as a gray cartoon) and UBA5 LIR peptide (all 20 conformers are shown; residues 340–348 that interact with GABARAPL2 in red; N-terminal unstructured residues 333–339 in gray). UBA5 side chains I343, L345 and V346 (red sticks) are allocated to the 2 hydrophobic pockets HP1 and HP2 of GABARAPL2 (beige and turquoise, respectively). The side chain of the conserved UBA5 W341 is placed in the cleft between α-helix α1 and loop L3 of GABARAPL2 (HP0, blue). (b) Crystal structure of GABARAP (gray cartoon) in complex with UBA5 LIR peptide (red). Similar, to the complex with GABARAPL2, side chains of UBA5 I343, L345 and V346 are placed in the canonical hydrophobic pockets (HP1, beige, and HP2, turquoise). W341 of UBA5 binds in proximity to α-helix α1 of GABARAP (HP0, blue). (c) Comparison of the Atg8-family protein binding mechanisms for a canonical LIR and UBA5 LIR. In cases of canonical LIRs (left plot, LC3B:SQSTM1/p62 LIR complex as an example; gray and red, respectively), side chains of residues Θ (W340, red sticks) and Γ (L343, red sticks) of the core LIR sequence (Θ-X-X-Γ) are binding to the 2 hydrophobic pockets (HP1, beige; and HP2, turquoise) on the surface of Atg8 proteins. In the GABARAP:UBA5 LIR complex (right plot), HP1 is shallow and covered by I343 (red sticks) and L345 (red sticks) hydrophobic side chains. The UBA5 W341 side chain fits into the new hydrophobic pocket HP0 (blue). LC3B and GABARAP residues contributing to each hydrophobic pocket are indicated. (d) Mutational analysis of hydrophobic residues in UBA5 LIR (W341, I343, L345 and V346). Relative affinities of interaction between GABARAPL2 and mutated UBA5 LIR were determined by normalization to the binding of WT UBA5 LIR. All values are mean ± SD of 3 independent experiments. For each residue, the binding pocket on Atg8-family proteins is indicated. (e) WebLogo plot generated on the sequences of all UBA5 LIR mutations that resulted in an enhancement in their binding to GABARAPL2 and LC3B. Charged amino acids are colored in red, polar residues in green. The amino acid composition of UBA5 and p62 LIRs at each position is shown. The binding pockets on Atg8-family proteins for each residue are highlighted.
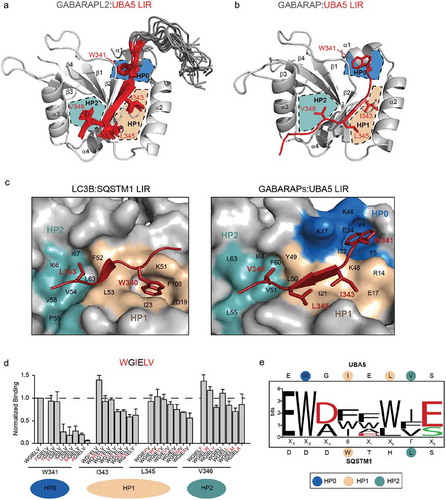
Both structures show the formation of the usual intermolecular β-sheet involving the UBA5 LIR peptide and the β-strand β2 of GABARAP and GABARAPL2. However, we observed a significant rearrangement of GABARAP and GABARAPL2 surfaces involved in contacts with the UBA5 LIR. This includes the 2 hydrophobic pockets, HP1 and HP2, that normally accommodate the Θ and Γ residues of the canonical LIR sequence (illustrated for the canonical p62 LIR interaction to LC3B, ), left plot). While the HP2 of GABARAP is occupied by the V346 side chain of UBA5 and forms a well-pronounced ‘pocket’, HP1 does not adopt a typical ‘pocket’ form but rather resembles a flat hydrophobic surface. The hydrophobic side chains of UBA5 residues I343 and L345 cover this surface (), right plot). The most important and dramatic changes are however near the end of the α-helix α1, where a new hydrophobic pocket is formed (), right panel). This pocket, which we name HP0, is formed by the hydrophobic moieties of the K46, K47 and K48 side chains of GABARAP, and the hydrophobic residues V4, Y5, I32 and V33. The HP0 accommodates the side chain of the conserved UBA5 residue W341 and its conformation is additionally stabilized by an intermolecular hydrogen bond between UBA5 W341 Nε1 and GABARAP E8 Oε1, (Figure S1(a)). Similar surface alterations occur on the surface of GABARAPL2 upon UBA5 LIR binding (Figure S1(b)), indicating a common mechanism of UBA5 LIR recognition and surface adjustment to adopt the specific distribution of hydrophobic residues for all GABARAP proteins.
To analyze the importance of each hydrophobic residue within the core UBA5 LIR motif for the binding to GABARAP proteins, we performed peptide arrays, in which we mutated residues W341, I343, L345 and V346 of UBA5 to other hydrophobic amino acids (W, F, Y, I, L, V, M and A). In agreement with our structural analysis, mutations of I343 and L345, which occupy HP1, as well as V346, which occupies HP2, results in a moderate decrease in binding affinity of up to 30% ()). Thus, these UBA5 LIR residues do not form the critical specific contacts to GABARAPs and could be substituted by any hydrophobic residues without drastic effects. In contrast, W341 appears to be crucial for binding: only aromatic amino acids are tolerated at this position and mutations to any other aliphatic residue abrogated the interaction almost completely ()). This is in line with our previous results [Citation24], where mutating W341 to alanine (W341A) resulted in a loss of binding of the full-length UBA5 to GABARAPs in pull-down experiments, while the mutations I343A, L345A and V346A only partially reduced the interaction.
We also probed the role of each non-hydrophobic residue within the UBA5 LIR core sequence (EWGIELV) for the binding affinity of UBA5 LIR towards GABARAPL2 and LC3B (Figures S2–S3). Data obtained with the peptide array indicate that the UBA5 LIR amino acid composition is well adapted for the binding to the GABARAPL2 protein. There were no individual amino acid substitutions that could significantly enhance the affinity of GABARAPL2 to mutated UBA5 LIR (maximally ~30% improvement). The affinity of LC3B to the mutated UBA5 LIRs was enhanced in some cases, showing ~50% of improvement. According to these results, all non-hydrophobic residues within UBA5 LIR peptides can be substituted to a number of analogous or alternative amino acids (amino acid preferences for each UBA5 LIR positions are given in Figure S2(c)). For example, a G at position 342 in UBA5 could be substituted not only with the similar small residue A, but also with Q, C, N, D and E residues to maintain the same affinity to both LC3B and GABARAPL2. Positively charged residues (K, R, and H) in this position decrease the affinities of the interactions, similar to the substitution with P and S. Aromatic residues (W, F, and Y) at this position also increase the affinity of both LC3B and GABARAPL2 for the UBA5 LIR. However, this is probably due to generation of a canonical LIR sequence within the UBA5 LIR motif: EWWIELV. Interestingly, the E344 of UBA5 cannot be efficiently substituted for E analogs (Q, D or N), indicating the importance of the unique geometry of the side chain of E344 for intermolecular hydrogen bonding with K/R47 in GABARAP proteins. Alanine (A) and aromatic residues (W, F and Y) at this position are the only substitutes that not only maintain but also enhance the affinities of the mutated UBA LIR to both LC3B and GABARAPL2. Introducing aromatic residues at this position does not generate a canonical LIR and therefore, the binding enhancement has to be mediated by another non-canonical mechanism.
Since the mutational analysis of individual residues in the UBA5 LIR in some cases showed an increase in the binding affinity to GABARAPL2 and LC3B, we aimed at identifying novel peptides that would have an increased affinity towards these proteins. Therefore, we combined several of the affinity-increasing mutations and created a library of 56 different peptides, which we analyzed for their affinity for GABARAPL2 and LC3B (Figure S3(a)). We categorized the individual peptides into three different categories: 1. peptides, which fit to the canonical LIR core and have a negatively charged residue (D or E) prior to Θ; 2. peptides with the canonical LIR core but without the negatively charged residue prior to Θ; 3. peptides, which have no resemblance to any LIR reported so far (Figure S3(a)). At first, all peptides were screened for their binding to LC3B and GABARAPL2 compared to the binding capacity of the wild-type UBA5 LIR (Figure S3(b)). Out of all 56 peptides tested, only 18% (10 of 56) showed an increased affinity towards GABARAPL2 but 53% (30 of 56 peptides) showed an enhanced affinity to LC3B (Figure S3(b)). Interestingly, the composition of the peptides, which showed an enhanced affinity to GABARAPL2 and LC3B, is similar, although the absolute number of the peptides varied. Of these affinity-enhancing peptides, ~25% aligned to the canonical LIR sequence and ~18% showed no similarity to canonical or non-canonical LIR motifs. More than half of the peptides (55% for GABARAPL2, 60% for LC3B) that showed an enhanced affinity to GABARAPL2 and LC3B had similarities to the canonical LIR sequence but lacked the negatively charged residues N-terminal to the LIR core (Figure S3(a–b)). To verify this enhanced binding, we analyzed the peptides, which showed the strongest enhancement of the binding to GABARAPL2 and LC3B, in more detail via isothermal titration calorimetry (ITC) and found that all peptides displayed LIR-like affinities with KD values of 1–5 µM (Figure S3(c–d), Table S5). The preferred optimized sequence of the peptides can be described as E-W-[D/A]-I-F-W-[I/V]-E ()), which differs from the canonical LIR motif [W/F/Y]-X1-X2-[L/I/V] and the previously identified GABARAP Interaction Motif (GIM; [W/F]-[V/I]-X2-V) [Citation33].
In summary, UBA5 LIR binds GABARAP proteins via a novel molecular mechanism, which includes the formation of 3 hydrophobic pockets on the surface of GABARAP proteins. W341 of UBA5 plays a key role in this interaction as it occupies a specific hydrophobic pocket HP0 on GABARAP surfaces and stabilizes the LIR:GABARAP complexes via a specific intermolecular hydrogen bond (to GABARAP E8 side chain).
Rearrangements of GABARAP residues around the gating K46 mediate HP0 formation and are crucial for the binding of UBA5 to GABARAP proteins
To understand the molecular basis of the newly described UBA5 LIR:GABARAP interaction, we compared the X-ray structure of the UBA5 LIR:GABARAP complex (also with reference to the NMR structure of the UBA5 LIR:GABARAPL2 complex) with the structures of the free GABARAP (PDB 1GNU [Citation34]) and GABARAP in complex with the canonical LIR peptide of PLEKHM1 (PDB 5DPS [Citation33]). We found that the global structure of GABARAP remains the same in all 3 states – RMSD values for backbone atoms do not exceed 0.6 Å, excluding the loops L3 and L4 (Figure S4(a)). The only significant difference in the backbone arrangements of GABARAP proteins was observed for the C-terminal part of the loop L3, starting with D45 and ending with K49 (start of the β-strand β2), thus making it the only relevant GABARAP region with significant changes in side chains conformations upon UBA5 binding (Figure S4(b)). This region harbors the invariant K46 of GABARAP proteins (corresponding to positions 49 in LC3A/LC3B and position 55 in LC3C) that was reported to function as a universal gate-keeper, regulating the entrance of the aromatic residues of canonical LIRs into the HP1 on the surfaces of all LC3/GABARAP proteins upon interaction with LIRs. The side chain of this residue was shown to undergo a ~ 90° rotation upon canonical LIR binding and changes its orientation from a ‘LIR-free’ to the ‘LIR-bound’ state [Citation35].
Upon binding of the UBA5 LIR, the side chain of the GABARAP’s K46 (in coordination with the changes in the backbone geometry) undergoes a large 180° rotation and ~5 Å displacement towards the position of K47 of GABARAP ()). In this new position, the side chain of K46 contributes to the building of HP0 with its aliphatic moiety. This rearrangement is accompanied by a conformational flip-back of the neighboring K47 side chain towards the K46 side chain in the ‘LIR-bound’ state. In other words, these 2 residues swap their relative positions, to form the HP0 and to facilitate the entry of the side chain of UBA5’s W341. The backbone and side chain positions of the next residue, the invariant K48 (K51 in LC3B), remain unchanged.
Figure 2. Rearrangements of the K46 and K/R47 side chains in GABARAP proteins upon interaction to UBA5 LIR mediate the new binding mechanism. (a) Section of the GABARAP:UBA5 LIR complex structure representing rearrangements of the GABARAP K46 and K47 side chains upon binding to the UBA5 LIR. The polypeptides are shown as cartoons (GABARAP gray, UBA5 LIR – red). Upon UBA5 LIR binding, K46 of GABARAP (gray sticks) undergoes a 180° rotation compared to the unbound conformation (orange sticks) to allow the entrance of the UBA5 W341 side chain (red sticks) into HP0. Simultaneously, K47 also undergoes a −180° rotation in comparison to the unbound state to avoid steric clashes. Thus, GABARAP K46 and K47 exchange their positions upon UBA5 LIR binding. Similar rearrangements are observed for the GABARAPL2 residues K46 and R47 (right blot). (b) The rearrangement of GABARAP K47 allows the formation of an additional intermolecular interaction to the UBA5 E344 side chain. In case of the GABARAPL2:UBA5 complex, the side chain of R47 of GABARAPL2 (gray sticks) can also interact with E344 (red sticks), but additionally is able to coordinate with the side chain of E340 of UBA5. These interactions stabilize the complex structure in addition to the formation of intermolecular β-sheet and fixation of the UBA5 aromatic/hydrophobic residues into the HP0, HP1 and HP2 on surface of the GABARAP proteins. (c) ITC results for the binding of GABARAPL2 R47 mutants to UBA5 LIR. The GABARAPL2 R47K mutation reduces slightly both the affinity and enthalpy of its binding to UBA5 LIR, indicating a preference for an arginine in this position (GABARAPL2) over a lysine (GABARAP and GABARAPL1). The ITC data correlate with the geometry of the intermolecular hydrogen bond described above. Accordingly, the GABARAPL2 mutation R47A reduces the affinity and enthalpy of UBA5 LIR binding, showing thermodynamic parameters of interaction close to that for LC3 proteins [Citation24]. The top diagram in each ITC profile displays the raw measurements and the bottom diagram shows the integrated heat per titration step. Best fit is presented as a solid line. Corresponding KD values are shown.
![Figure 2. Rearrangements of the K46 and K/R47 side chains in GABARAP proteins upon interaction to UBA5 LIR mediate the new binding mechanism. (a) Section of the GABARAP:UBA5 LIR complex structure representing rearrangements of the GABARAP K46 and K47 side chains upon binding to the UBA5 LIR. The polypeptides are shown as cartoons (GABARAP gray, UBA5 LIR – red). Upon UBA5 LIR binding, K46 of GABARAP (gray sticks) undergoes a 180° rotation compared to the unbound conformation (orange sticks) to allow the entrance of the UBA5 W341 side chain (red sticks) into HP0. Simultaneously, K47 also undergoes a −180° rotation in comparison to the unbound state to avoid steric clashes. Thus, GABARAP K46 and K47 exchange their positions upon UBA5 LIR binding. Similar rearrangements are observed for the GABARAPL2 residues K46 and R47 (right blot). (b) The rearrangement of GABARAP K47 allows the formation of an additional intermolecular interaction to the UBA5 E344 side chain. In case of the GABARAPL2:UBA5 complex, the side chain of R47 of GABARAPL2 (gray sticks) can also interact with E344 (red sticks), but additionally is able to coordinate with the side chain of E340 of UBA5. These interactions stabilize the complex structure in addition to the formation of intermolecular β-sheet and fixation of the UBA5 aromatic/hydrophobic residues into the HP0, HP1 and HP2 on surface of the GABARAP proteins. (c) ITC results for the binding of GABARAPL2 R47 mutants to UBA5 LIR. The GABARAPL2 R47K mutation reduces slightly both the affinity and enthalpy of its binding to UBA5 LIR, indicating a preference for an arginine in this position (GABARAPL2) over a lysine (GABARAP and GABARAPL1). The ITC data correlate with the geometry of the intermolecular hydrogen bond described above. Accordingly, the GABARAPL2 mutation R47A reduces the affinity and enthalpy of UBA5 LIR binding, showing thermodynamic parameters of interaction close to that for LC3 proteins [Citation24]. The top diagram in each ITC profile displays the raw measurements and the bottom diagram shows the integrated heat per titration step. Best fit is presented as a solid line. Corresponding KD values are shown.](/cms/asset/353d53aa-d7ec-46ac-ab7d-492b687be599/kaup_a_1606637_f0002_oc.jpg)
The new position of K47 of GABARAP allows the formation of an intermolecular hydrogen bond between the amino group of K47 and the carboxyl group of the UBA5 E344 side chain ()). According to the NMR structure of GABARAPL2 in complex with the UBA5 LIR, the same rearrangements occur with the GABARAPL2 K46 and R47 residues ()). The intermolecular hydrogen bond between the GABARAPL2 R47 Nη2 and UBA5 E344 Oε2 shows a more favorable geometry and possibly allows multiple intermolecular hydrogen bonds between these residues. Therefore, the difference in quality of the intermolecular hydrogen bond of K47 (GABARAP) and R47 (GABARAPL2) to UBA5’s E344 explains the higher affinity of UBA5 for GABARAPL2. Indeed, mutation of R47 of GABARAPL2 to a lysine (K) residue weakens the affinity of GABARAPL2 for the UBA5 LIR by more than 2 fold, as measured by ITC, mostly by reducing the favorable enthalpy contribution ()). Accordingly, mutation of R47 to an alanine (R47A) in GABARAPL2 leads to a significant loss in the binding affinity, highlighting the importance of the K/R47-mediated intermolecular hydrogen bond for the binding affinity of UBA5 LIR to GABARAP proteins ()).
Taken together, we show that the binding of the non-canonical LIR motif in UBA5 induces extensive rearrangements of the K46 and K/R47 backbone and side chain positions in the 2 GABARAP proteins. These rearrangements result in the formation of i) the new hydrophobic pocket HP0 and ii) the intermolecular hydrogen bond between the residues K/R47 in GABARAPs and E344 in the UBA5 LIR.
Mutational analysis of LC3B and GABARAPL2 reveals that the GABARAPs K/R47 residue is crucial for the specific binding of UBA5 LIR to GABARAP proteins
Despite the high sequence similarity among human Atg8-family proteins, there is a growing number of LIR motifs being identified that preferentially bind to either the LC3 or the GABARAP protein subfamily or even to individual members within these subfamilies [Citation17,Citation21,Citation33,Citation36,Citation37]. Understanding the molecular basis of this specificity is crucial for elucidating unique functions of each of the 6 human Atg8-family proteins.
The UBA5 LIR displays a strong preference towards GABARAP over LC3 proteins [Citation24]. Therefore, we aimed to identify residues in GABARAP proteins that are crucial for this selectivity. Based on our structural findings, we identified 3 regions in human Atg8-family proteins that display subfamily-associated differences in their amino acid sequence ()) and significant structural deviations upon binding of canonical/non-canonical LIR motifs ()). Region I is located at the beginning of the α-helix α1 and is involved in the formation of the HP0 that accommodates the W341 residue of UBA5 (). In this region, GABARAPs mostly contain negatively charged residues (E7D8H9 in GABARAPL2) compared to mostly positively charged residues in LC3s (Q9R10R11 in LC3B). A second region (region II) lies in the loop L3 between the β-strands β1 and β2, where the highest structural diversity between GABARAP and LC3 proteins is observed in their free and LIR-bound states, and where most structural changes occur upon UBA5 LIR binding. NMR-spectroscopy studies indicated different dynamics for this region in GABARAP and LC3 proteins [Citation33]. Therefore, this loop L3 might contribute to the specificity of UBA5 LIR recognition towards both subfamilies. Region III is located at the beginning of the β-strand β2, where 2 highly conserved lysine (K) residues in all Atg8-family proteins (K46 and K48 in human GABARAPs; K49 and K51 in LC3A/B) flank a subfamily-specific residue (K/R47 in GABARAPs; T50 in LC3s). This region is of crucial importance for binding of Atg8-family proteins to LIR motifs and undergoes large conformational rearrangements upon UBA5 binding.
Figure 3. Loop L3 and K/R47 in GABARAP proteins predetermine their specificity to UBA5 LIR over LC3 proteins (a and b). 3 regions in LC3 and GABARAP proteins show large differences in the sequences and the structures of their canonical LIR and UBA5 LIR bound forms. (a) Region I resides around α-helix α1; region II defines the loop L3, which has a different length and composition in between the LC3 and GABARAP subfamily. Region III is defined as the R/K47 residue in GABARAPs (T50 in LC3A/LC3B and T56 in LC3C) that forms an intermolecular hydrogen bond with UBA5 E344 and undergoes large rearrangements upon UBA5 LIR binding. (b) Cartoon representation of the backbone superposition of the structures of GABARAP (blue) in complex with UBA5 LIR (red) and free LC3B (green). The indicated regions I, II and III locate around the binding interface of the UBA5 W341 side chain (red stick) and might be responsible for the selectivity and the novel binding mechanism. (c) ITC profiles representing interactions between LC3B swapping mutants in regions II, III and a combination of both with the WT UBA5 LIR. The top diagram in each ITC profile displays the raw measurements and the bottom diagram shows the integrated heat per titration step. The best fit is presented as a solid line. Corresponding KD values for each mutant are shown. (d) ITC profiles representing interactions between GABARAPL2 swapping mutants in regions II, III and a combination of both with the WT UBA5 LIR. The top diagram in each ITC profile displays the raw measurements and the bottom diagram shows the integrated heat per titration step. The best fit is presented as a solid line. Corresponding KD values for each mutant are shown.
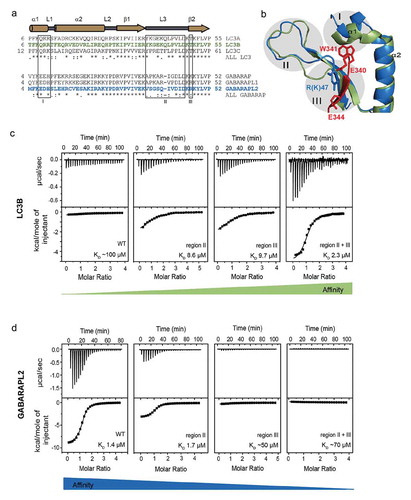
To investigate the contributions of each of these regions (I-III) for the observed specificity of the UBA5 LIR to GABARAP proteins, we created a set of swapping mutants between LC3B (as the weakest interactor) and GABARAPL2 (as the strongest one). In these mutants, we exchanged the individual regions described above between both proteins, applied combined swapping mutations of 2 or 3 regions, and analyzed their binding to the wild-type UBA5 LIR (WT UBA5 LIR) by ITC. Mutations in region I did not significantly alter the affinity and are thus not responsible for the selectivity of the UBA5 LIR (Figure S5(a–b)). An exchange of the LC3B sequence in region II (loop L3) to corresponding residues in GABARAPL2, resulted in an increased affinity of mutated LC3B variants to UBA5 LIR with the KD dropping from 100 µM to 8.6 µM ()). In agreement with this, swapping the region II of GABARAPL2 to that of LC3B, resulted in a decreased affinity of GABARAPL2 towards UBA5, with KD changes from 1.4 µM to 7.4 µM ()), indicating that this region indeed contributes to the specificity of GABARAPs to UBA5 LIR ()). Swapping mutants in region III displayed a significantly reversed specificity to the UBA5 LIR (KD values decreasing from 100 µM to 9.7 µM for LC3B T50R and increasing from 1.4 µM to ~50 µM for GABARAPL2 R47T; ). The combined mutations within regions II and III in both LC3B and GABARAPL2 resulted in almost quantitative exchange of their affinities to the UBA5 LIR with a KD of 2.3 µM for mutated LC3B vs. ~70 µM for mutated GABARAPL2 ()). Swapping mutants including all 3 regions represented similar affinities to the UBA5 LIR as for the aforementioned mutants within regions II and III, indicating again a non-significant role of region I in GABARAP specificity to this non-canonical LIR (Figure S5).
In summary, we identified 2 regions in GABARAP subfamily proteins that are crucial for the specific binding of the non-canonical LIR in UBA5. The first region, loop L3, needs to be of specific length and amino acid composition to allow UBA5 LIR binding. The second region harbors residue R47 in GABARAPL2 (T50 in LC3B) that is pivotal in the specific binding of UBA5 to GABARAP proteins. However, a synergistic contribution of the loop L3 and a conserved ionic interaction between R/K47 and UBA5 is needed for the full selectivity of the binding.
The interaction between UBA5 and GABARAP proteins is crucial for UBA5 localization to ER membranes
Despite the fact that UBA5 interacts with GABARAPL2 [Citation26] and generally with GABARAP-subfamily proteins [Citation24,Citation32], a functional role for this interaction remains undiscovered. As we showed previously [Citation24], the UBA5 LIR binds both GABARAPs and UFM1, with its affinity to GABARAPL2 being significantly higher than to UFM1 (KD values ~1 and ~10 µM, respectively). Although the LIR sequence of UBA5 is crucial for binding to UFM1, it has been shown, that ufmylation in vitro still takes place upon deletion or modification of this sequence [Citation38], although to a lesser extent and with slower kinetics.
Since GABARAPL2 has no effect on the kinetics and abundance of UBA5-UFM1 conjugate formation in vitro and in vivo [Citation24], we investigated a possible role of GABARAPL2 in the regulation of UBA5 activity and efficiency of ufmylation in cells with respect to their compartmentalization. It was previously shown, that overexpressed UFM1 and UFBP1 (known as ufmylation target with an unidentified function) localize to ER membranes [Citation28] and that perturbations in the ufmylation pathway are associated with the ER membrane localization of the targets [Citation31].
This prompted us to investigate whether GABARAP proteins may function as signaling scaffolds by regulating the localization of UBA5, a function that is well described for GABARAP proteins [Citation17]. In contrast to previous studies, where the localization of UBA5 and components of the ufmylation pathway were analyzed upon overexpression, we focused on the localization of the endogenous UBA5. By immunofluorescence microscopy, we identified that UBA5 is distributed in the cytosolic part of a cell, similar to the localization of overexpressed UFM1 [Citation28]. However, a strong proportion of UBA5 co-localized to PDI (protein disulfide-isomerase)-positive structures, indicating, that UBA5 is localized both to the cytoplasm and to the ER (Figure 4(a,b)). In contrast to that, cells with a knockout (KO) of all 3 GABARAP proteins (HeLa GABARAP-TKO) showed a reduced localization of UBA5 to PDI-positive structures, whereas the cytosolic pool did not show any alteration in the absence of GABARAP proteins ()). This was not observed in cells lacking all 3 LC3 proteins, supporting the notion that this is specific for GABARAPs () and S6A). Surprisingly, the role of GABARAPs as recruitment factors of UBA5 to the ER is not dependent on their lipidation status and membrane binding capacities: HeLa ATG7-KO cells show only a very insignificant reduction and UBA5 co-localization with the ER marker PDI () and S6A). Additionally, we observed that overexpressed HA-tagged GABARAPL2, but not HA-LC3B, co-localized with UBA5 in both HeLa wild-type and HeLa ATG7-KO cells (Figure S6(b)).
Figure 4. GABARAPs recruits UBA5 to ER membranes. (a) Localization of endogenous UBA5 in HeLa wild-type and HeLa GABARAP-TKO cells visualized by immunofluorescence. Cells were stained for endogenous UBA5 (red) and endogenous PDI (green) was used as ER marker. Co-localization was determined via the Co-localization Finder software plugin of the ImageJ software and is shown in white. Scale bar: 10 µm. (b) Quantification of co-localization of the endogenous UBA5 and the ER marker PDI in HeLa wild-type and HeLa ATG7-knockout, LC3s-knockout and GABARAPs-TKO. Pearson’s correlation coefficient was determined using the coloc2 software implemented in ImageJ. Results show the evaluation of 50 randomly chosen cells of 3 independent biological experiments. ***, P < 0.001. (c) The microsomal (M) and cytoplasmic (C) fractions of HEK293T cell lines were prepared from indicated genotype cells and subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. Data are representative of 3 separate experiments. (d) Bar graphs indicate the quantitative densitometric analysis of microsomal UBA5 relative to microsomal and cytoplasmic UBA5 in HEK293T cell lines. Statistical analysis was performed using an unpaired t test (Welch test). Bars represent the mean ± SE of 3 separate experiments. *, P < 0.05. (e) Empty vector or MYC-UFL1 together with UFBP1-MYC were transfected into HEK293T cells as indicated. 24 h after transfection, the microsome (M) and cytoplasmic (C) fractions were prepared and subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. (f) Bar graphs indicate the quantitative densitometric analysis of microsomal ufmylated proteins (shown by the line at the left side in figure 5E) relative to the loaded microsomal protein amount (estimated by Ponceau-S staining) in normal conditions (left bars) and upon induction of UFL1/UFBP1 activity (right bars). Statistical analysis was performed using an unpaired t test (Welch test). Bars represent the mean ± SE of 3 separate experiments. **, P < 0.01.
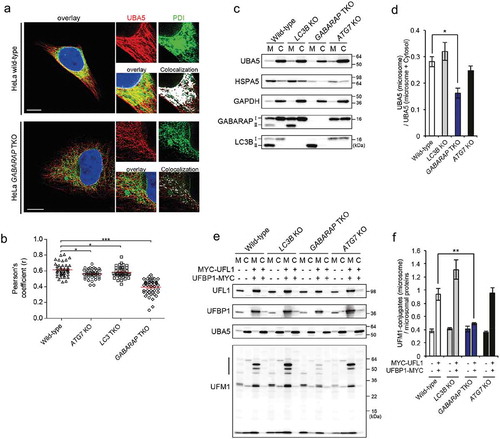
To confirm that the localization of UBA5 to ER membranes is regulated by GABARAP proteins and is independent from GABARAPs lipidation status, we generated MAP1LC3B-, GABARAP-/GABARAPL1-/GABARAPL2-triple and ATG7-knockout HEK293T cells (Figure S6(c)) and performed a cellular fractionation assay to analyze distribution of endogenous UBA5 in the cytosolic and microsomal fractions ()). Notably, since digital PCR analysis indicated, that HEK293T cells hardly express LC3A and LC3C proteins (Figure S6(d)), MAP1LC3B KO (LC3B-KO thereafter) cells represent an almost complete absence of the LC3-family proteins. While non-lipidated forms (GABARAP-I and LC3B-I) were fractionated into both microsomal and cytoplasmic fractions, their lipidated forms (GABARAP-II and LC3-II) were detectable in the microsomal fraction only ()). Consistent with the immunofluorescence analysis, the amount of UBA5 in the microsomal fraction of GABARAP TKO but not LC3B KO cells was significantly smaller than that of the parental HEK293T cells (). Such a reduction was again not observed in the case of ATG7-deficient HEK293T cells (), supporting the idea that free GABARAPs rather than their lipidated forms are critical for the translocation of UBA5 to the ER. Next, we investigated the ufmylation in the microsomal fraction prepared from each HEK293T cell lines. We could not observe any significant differences of the level of microsomal ufmylation among each cell lines, probably due to quite low levels of ufmylation under normal conditions (). As previously reported [Citation39,Citation40], overexpressed UFL1 and UFBP1, which constitute an ER-associated E3-ligase complex for UFM1, promoted ufmylation in the microsomal fraction in HEK293T wild-type cells. Similarly, we observe that the overexpression of the E3 components induced the microsomal ufmylation in wild-type, as well as LC3B KO or ATG7 KO HEK293T cells (). However, such induction was almost completely suppressed by loss of GABARAP family proteins (), indicating dependence of ufmylation on ER-associated UBA5 fraction.
Taken together, we could show that GABARAP proteins regulates the cellular localization of UBA5 by recruiting it to ER membranes and thus serves as a recruitment factor for UBA5. Consequently, this UBA5 localization predetermined the functional activity of the UFL1/UFBP1 E3 complexes within the ER, highlighting the role of GABARAPs as a signaling scaffold in the ufmylation pathway.
Discussion
Atg8-family protein interaction networks are involved in regulating an increasing number of cellular processes, of which autophagy is the best studied one [Citation24,Citation32]. Being covalently conjugated to PE, Atg8-family proteins are localized in the autophagic membranes to generate docking sites for a plethora of proteins. For instance, SQSTM1/p62 and NBR1 with their bound cargo, are recruited to Atg8-decorated membranes to mediate selective autophagy, a critical process that maintains cellular homeostasis. In addition, autophagy-related proteins responsible for PE conjugation (e.g., Atg1, Atg4, Atg7, etc.) recognize and interact with Atg8-family proteins in the cytosol. Besides the autophagy pathway, Atg8-family proteins are involved in non-autophagic processes, interacting with different classes of proteins such as kinases, RABGAPs, ubiquitin-ligases, etc. All these interactions require the presence of an accessible LIR motif in the Atg8 interaction partners. All LIRs known so far (both canonical, containing the spaced aromatic and aliphatic residues, and non-canonical, e.g., lacking the aromatic residue) bind Atg8-family proteins via a well described mechanism. This includes the formation of a parallel intermolecular β-sheet with the β-strand β2 within the Atg8-family protein and with aromatic (W,F,Y) and large hydrophobic (V,L,I) residues at corresponding positions 1 and 4 of the LIR fixed deeply inside the 2 hydrophobic pockets, HP1 and HP2, respectively. This interaction mode, first described in 2008 [Citation20,Citation23] has been valid for all the Atg8:LIR pairs described to date (including canonical or non-canonical LIRs; LIRs generated from a specific three-dimensional protein structure and unusual LIRs obtained from the rational design and/or phage display, etc).
Unusual ‘3 hydrophobic pocket’ binding mechanism
We describe an alternative mechanism of Atg8-family protein interactions with the non-canonical UBA5 LIR. Its sequence differs not only from canonical (extensively analyzed in the publication describing iLIR – software to predict LIR motifs in proteins [Citation41]) but also from any non-canonical LIRs [Citation21–Citation23]. Its binding mechanism still includes the formation of a parallel intermolecular β-sheet. However, 3 (HP0, HP1, and HP2), and not the usual 2, hydrophobic pockets are formed on the UBA5 LIR-bound surface of GABARAP proteins. The positions of HP1 and HP2 on the surfaces of GABARAP and GABARAPL2 in complex with the UBA5 LIR are similar to positions for HP1 and HP2 in the published examples of other LIR complexes, however adopting to distinct features of the UBA5 LIR. The HP0, harboring the W341 side chain, appears to be close to the end of the α-helix α1 after rearrangements of GABARAP’s residues K46 and K/R47 at the end of the loop L3 and beginning of the β-strand β2. This indicates, that the conformational flexibility of the Atg8-family protein structures at the LIR-contacting site described previously [Citation33], is one of the major factors in determining particular LIR recognition and binding.
Specificity of interaction between gabarap-proteins and UBA5 LIR
The results of systematic swapping mutagenesis revealed, that GABARAPs residue K/R47, which is pivotal for the affinity of UBA5 LIR binding to GABARAPs, is also the key determinant of the observed specificity. In all human LC3-proteins, this position is occupied by a threonine (T50 in LC3A/LC3B, T56 in LC3C), and exchange of the threonine (T) to an arginine (R) resulted in a ~ 10-fold enhancement in binding affinity. Interestingly, this T was reported to be phosphorylated in LC3-protein family [Citation42], which could be the basis for further modulation of the LC3 binding specificity to certain LIR types. Residues in the C-terminal loop L3 mediate an additional contribution to the selectivity of UBA5 LIR towards GABARAP proteins. In GABARAP proteins, the loop L3 is 1 amino acid shorter in comparison to the L3 in the LC3s and displays significantly higher dynamic (according to accumulated NMR data [Citation43];).
The presence of an electropositive residue (K or R) at the relative position 47, as well as the shorter length of the L3 loop in human GABARAP proteins is conserved for all known GABARAP-family members in different species. These 2 features of the GABARAPs sequence potentially could serve as a specific identification of the GABARAP-subfamily. A shorter L3 and the presence of R/K at position 47 is conserved in yeast Atg8 and in the C. elegans ortholog LGG-1, but not LGG-2 [Citation44], indicating their close relation to GABARAP- rather than to LC3- subfamily proteins.
Interestingly, the swapping mutations at the end of the α-helix α1 do not significantly affect the binding specificity. Only minor modulations of KD values were observed for binding of mutated LC3B and GABARAPL2 proteins to the WT UBA5 LIR (Figure S5, Table S4). This region has prominent sequential features in LC3 and GABARAP proteins (Q9R10R11 instead of E7D8H9 residues for LC3B and GABARAPL2, respectively) and is involved in formation of an intermolecular hydrogen bond between GABARAP E8 Oε1 in this region and UBA5 W341 Nε1 that stabilizes the complex structure. Taking into account that UBA5 W341 can be mutated to F or Y without a significant loss of affinity to GABARAPL2 ()), we can assume that this region is rather flexible and could adopt multiple conformations to facilitate effective docking of the aromatic side chains into HP0. This conformation flexibility for GABARAPs N-terminal regions was reported previously [Citation43] and is important for interactions of Atg8-family proteins with proteins in and outside of autophagy pathways (reviewed in [Citation9]).
Do GABARAP family proteins predetermine membrane localization of UBA5?
UBA5 serves as an E1-like enzyme that activates UFM1 and initiates the ufmylation cascade (reviewed in [Citation25]). Ufmylation has been shown to be involved in many cellular pathways, such as the ER stress response, fatty acid metabolism, hematopoiesis, G-protein coupled receptor biogenesis and erythroid differentiation [Citation27–Citation30,Citation45,Citation46], and also linked to several diseases such as cancer, ischemic heart disease and diabetes [Citation25,Citation31]. Mutations in the UBA5 gene can cause neuronal disorders [Citation47–Citation49] and mutations in other proteins of the ufmylation cascade (UFM1 and UFC1) affect brain development [Citation50]. It has been shown that UBA5 is a novel pancreatic cancer target [Citation51]. However, many components, targets, and the exact physiological functions of ufmylation are still largely unknown.
In this work, we were able to show, that the membrane localization of UBA5 is strongly dependent on GABARAP proteins. It is commonly accepted, that the highly soluble UBA5 is localized in the cytosol (observation mostly made using overexpressed UBA5), however, the best characterized ufmylation target (UFBP1) and the E3 enzyme responsible for its ufmylation (UFL1), are both located at the ER-membrane [Citation28]. The molecular details on how and whether UBA5 is also recruited to membranes, especially to the ER, remained unclear. Here, we addressed this question following the reported UBA5 interaction with GABARAP proteins, which can be localized to membranes. We looked at the subcellular localization of UBA5 in HeLa cells and found that knockout of all 3 GABARAP, but not LC3 proteins, resulted in a reduction of ER-localized UBA5 (). Interestingly, this was not observed for lipidation-deficient ATG7-knockout cells, indicating a lipidation-independent mechanism. This finding was further supported by analysis of the UBA5 content in membrane and cytosolic fractions in LC3B-, GABARAPs- and ATG7-deficient cells. Importantly, non-lipidated GABARAPs (GABARAP-I) are also found in the microsomal fraction, suggesting that GABARAP-I per se can tether UBA5 to the ER. Consequently, we could show that this reduced localization of UBA5 to the ER in the absence of GABARAP proteins directly results in reduced ufmylation of targets at these sites.
Therefore, we propose that GABARAP-family proteins act as ER-recruitment factors for UBA5 and subsequently play a critical role in the regulation of the ufmylation pathway in ER structures. Interestingly, the identified target of the ufmylation, UFBP1 (ubiquitin-fold binding protein 1, also known as DDRGK1), was also shown to play a role in the localization of UFM1 to ER membranes [Citation28]. However, the physiological role of the different pools of UFM1 remain elusive. Additionally, UFBP1 was recently shown to be a critical regulator of ER homeostasis during the ER stress [Citation52]. Understanding the exact role of GABARAPs as well as the function of the different cellular pools of UBA5, UFM1 and ufmylated targets will help to elucidate the physiological function and regulation of this conserved pathway and its involvement in many diseases as well as help to identify its relevant targets. However, to carefully link functional activity of the UBA5 with membrane-localization mediated by GABARAP proteins, a tool is needed that allows the visualization of all individual LC3 and GABARAP subfamily members on endogenous level. This is the challenging task, and the new peptides derived from UBA5 LIR might help to approach it.
Is the LIR docking site the only site that mediates interaction of atg8-family proteins to their partners?
Structural and functional data obtained over the last few years indicate that functionally relevant interactions between Atg8-family proteins and their partners are mediated by a significantly broader set of Atg8 recognition elements and by altered binding mechanisms. α-helical and combined β-stranded/α-helical structures are shown to be equally well (or even more efficiently) recognized by Atg8-family proteins [Citation53–Citation55]. It was also shown that sequences surrounding the canonical LIR motif could dramatically enhance their affinity for Atg8-family proteins [Citation56–Citation59]. Moreover, there are examples of a through-space coordination of specific residues fixed by 3D structure to occupy HP1 and HP2 [Citation60]. Some interactions between Atg8-family proteins and their interaction partners could not be explained in terms of the usual LIR model [Citation61–Citation63], indicating that interfaces other than the LIR docking site (HP1/HP2) on the surface of Atg8-family proteins can be involved. Taken into account that we showed a lipidation-independent ER membrane localization of GABARAP proteins and their ability to recruit UBA5 to the ER via the UBA5 LIR, such alternative binding site(s) on the surface of Atg8-family proteins could play a more general role in the ability of LC3/GABARAP proteins to bind and recruit other proteins to cell membranes.
Materials and methods
Preparation of the E. coli expression constructs for NMR, X-ray and ITC experiments
For the expression of human LC3 and GABARAP proteins for ITC and NMR experiments, plasmids with appropriate modified Ub-leaders in pET vectors were used [Citation64]. The gene, encoding UBA5 LIR/UFIM peptide for NMR spectroscopy, was provided as a synthetic oligonucleotide (Eurofins Genomics GmbH) and cloned into the pET39_Ub19_ vector [Citation64] by NcoI-BamHI restriction sites. After TEV cleavage, the resulting peptide has the amino-acid sequence GAM-333EIIHEDNEWGIELVSE348, where the first three residues (GAM) are due to a cloning artefact. For X-ray crystallography, the peptide sequence was optimized to avoid non-interacting residues in it (based on NMR data analysis). Corresponding gene was cloned in the same pET39_Ub19_ vector, the resulting peptide (GAM-337EDNEWGIELVSEVSE351) was also used in all the ITC experiments. The chimeric constructs of the UBA5 LIR attached to the human GABARAP were prepared by inserting the oligonucleotide sequence corresponding to the UBA5 LIR337-351 peptide and glycine-methionine-serine linker into the NcoI site of the existed GABARAPs expression constructs, placing the UBA5 LIR337-351 at the N-terminus of the mature chimeric protein. For the generation of LC3B, GABARAPL2 and UBA5 LIR mutants, a site-directed mutagenesis was used with standard overlapping primers encoding targeted nucleotide(s) substitution. As initial constructs, the pETm60_Ub3_LC3B5-119, pET39_Ub19_GABARAPL23-116 and pET39_Ub19_UBA5337-351 were used.
Protein expression and purification
All proteins and peptides in this work were expressed in E. coli T7 Express (NEB) strain in LB or M9 (for stable 13C and 15N isotope labelling) media. Isolation and purification procedures were performed as described previously [Citation17,Citation24,Citation37]. Before data collection, all proteins and peptides were equilibrated in an appropriate buffer (50 mM Na2HPO4 [Carl Roth, 2370.1], pH 7.0, 100 mM NaCl [Carl Roth, P029.3]), and supplied with 5 mM protease inhibitors (Roche, 4693132001). For structural NMR experiments, 13C15N-labelled GABARAPL2 was concentrated to ~800–900 µM to which the non-labelled UBA5 LIR peptide was added to 2 fold molar excess. Vice versa, 13C15N-labelled UBA5 LIR was concentrated to ~400–500 µM to which the non-labelled GABARAPL2 was added to 2 fold molar excess. Mutated UBA5 LIR peptides, showing a high degree of hydrophobicity, were isolated with the buffers containing 8 M urea (Carl Roth, 2317.2) until the final size-exclusion chromatography step. GST-tagged LC3B and GABARAPL2 were expressed and purified in a similar manner using GST-affinity beads.
Nuclear magnetic resonance spectroscopy
All NMR experiments were performed at 298 K on Bruker Avance spectrometers operating at proton Larmor frequencies of 600, 700, 800, 900 or 950 MHz. Spectrometers were equipped with cryogenically cooled {13C/15N}1H triple-resonance probes. Spectra were processed with TopSpin 3.2 (Bruker) and analyzed using the Sparky 3.114 software (UCSF). Backbone and aliphatic side chain resonances of 13C15N labelled GABARAPL2 in complex with non-labelled UBA5 LIR333-348 peptide, as well as 13C15N-labelled UBA5 LIR333-348 peptide in complex with non-labelled GABARAPL2 were assigned using [15N-1H]-TROSY versions of 3D HNCACB, HNCO, HN(CA)CO, (H)CC(CO)NH-TOCSY, and H(CCCO)NH-TOCSY experiments [Citation65–Citation67]. Acceleration of longitudinal 1H relaxation between scans in the HNCACB, HNCO and HN(CA)CO experiments was achieved in the Band-Selective Excitation Short-Transient (BEST) manner [Citation68–Citation70]. Assignments of the aromatic side chains were performed with a (H)CB(CGCC-TOCSY) Haro experiment [Citation71] and verified with a 3D constant-time NOESY-[13C,1H]-TROSY experiment optimized for aromatic CH groups [Citation72,Citation73]. Distance restraints for structure calculations were obtained from 3D 15N-separated NOESY and 13C-separated NOESY spectra (additionally, as optimized versions for arginine and aromatic side chains, respectively) with mixing times of 70 ms. Intermolecular distance restraints between non-labeled and 13C,15N-labeled polypeptides were obtained from a 3D F1-13C/15N-filtered NOESY-[13C]-HSQC experiment [Citation74] recorded with a mixing time of 100 ms. Structure calculations were performed with the program CYANA [Citation75] using combined automated NOE assignment and structure calculation by torsion angle dynamics [Citation76]. The 20 conformers with the lowest final CYANA target function values were subjected to restrained energy refinement using the program OPALp [Citation77]. The 20 energy-refined conformers have been deposited in the Protein Data Bank with accession code 6H8C. The chemical shift assignments have been deposited in the BioMagResBank (BMRB) database with accession code 34307.
Crystallization and data processing
The purified chimera construct of GABARAP3-117 C-terminally fused to UBA5337-350 via a short A-M-G linker was dialyzed into a crystallization buffer (50 mM TRIS [Carl Roth, 4855.3], pH 8.0, 100 mM NaCl) and concentrated to ~0.75 mM. The crystallization was performed using the sitting-drop vapor diffusion method at 293 K and crystals were obtained in 1.6 M Na2HPO4, 0.1 M HEPES [Carl Roth, 6763.3], pH 7.5, as a reservoir solution. Diffraction data were collected at Swiss Light Source, beam line PXIII and processed with XDS [Citation78]. The crystal structure of the complex was determined by molecular replacement using the GABARAP crystal structure (PDB: 1GNU [Citation34]) as a search model. Manual model building and refinement were done with Coot, CCP4 software suite and Phenix [Citation79–Citation81]. The structure has been deposited in the Protein Data Bank with the accession code 6HB9.
Isothermal titration calorimetry (ITC)
All titration experiments were performed at 25°C using a MicroCal VP-ITC microcalorimeter (Malvern Instruments Ltd., UK). LC3 and GABARAP proteins (wild-type and mutants) equilibrated into a buffer containing 50 mM TRIS, pH 7.5, 100 mM NaCl and used at concentration of ~20–25 µM. The UBA5 LIR peptides (wild-type and mutants) were equilibrated into the same buffer and titrated to LC3/GABARAPs at concentrations of ~ 400 µM. All ITC data were analyzed with the MicroCal software implemented in Origin 7.0 and fitted using a ‘one-site’ binding model. The proteins and peptides concentrations were calculated from the UV-absorption at 280 nm by Nanodrop spectrophotometer (Thermo Fisher Scientific, DE, USA).
Peptide array
The peptide array was performed similar as described previously [Citation33,Citation82]. Briefly, biotinylated peptides (~50 nmole) were ordered (JPT, Germany) and solubilized in PBS (gibco, 14190094) containing 15% DMSO (Carl Roth, 994.2). 2 µl of the peptide solution was mixed with 100 µl PBST (PBS + 0.1% Tween 20 [Carl Roth, 9127.3]) with 5% BSA (Carl Roth, T844.2) and were then immobilized in streptavidin coated 96-well plates (Thermo Fisher Scientific, 436014) overnight at 4°C. After 3 washing steps with PBST, 1 µM GST-LC3B or GST-GABARAPL2 in 100 µl PBST were added and incubated for at least 1 h at 4°C. After another 3 washing steps, bound GST-LC3B/GABARAPL2 was detected by anti-GST-HRP antibody (GE healthcare, RNV1236-V, 1:10,000 in PBST, 1 h at 4°C). Afterwards, wells were again washed 3 times with PBST buffer and HRP signal was detected by the addition of homemade ECL solution and direct detection in ELISA plate reader.
Cell culture
All cell lines used in this study were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS, penicillin-streptomycin and sodium-pyruvate; and cultured at 37°C in a humidified incubator containing 5% CO2. HeLa LC3- and GABARAP-triple knockout cells [Citation83] were a gift from Dr. M. Lazarou (Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Melbourne, Australia). Wild-type HEK293T were acquired from ATCC (CRL-3216). To generate LC3B-, GABARAPs- and ATG7-KO HEK293T cells, each LC3B, GABARAPs and ATG7 guide RNA designed by CRISPR Design tool (http://crispr.mit.edu/) was subcloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene. 42230, deposited by Feng Zhang Lab), a human codon-optimized SpCas9 and chimeric guide RNA expression plasmid. HEK293T cells were co-transfected with the pX330 and pEGFP-C1 (Clontech Laboratories, 6084–1) vectors, and cultured for 2 days. Thereafter, the GFP-positive cells were sorted and expanded. Loss of LC3B, of GABARAPs and of ATG7 was confirmed by heteroduplex mobility assay followed by immunoblot analysis with anti-LC3B, anti-GABARAPs and anti-ATG7 antibodies, respectively. HEK293T cells at subconfluence were transfected with MYC-UFL1 together with UFBP1-MYC using Polyethylenimine, Linear (MW 25,000) (Polysciences, Inc., 23966–2). A test for mycoplasma contamination was performed for all cell lines monthly.
Immunofluorescence staining
For the staining of endogenous UBA5, HeLa cells lines were plated on 18 mm coverslips in 12-wells. 24 h after seeding, cells were fixed with 4% PFA in PBS for 10 min at room temperature. Cells were then permeabilized with PBSt (PBS + 0.1% Triton X-100 [Carl Roth, 3051.4]) and blocked with 5% BSA in PBSt for 1 h at room temperature. Cells were then incubated with primary antibodies for the staining of endogenous UBA5 (Novus biologicals, NBP1-82087, 1:100) and PDI (abcam, ab2792, 1:100) overnight at 4°C. After intensive washing, cells were incubated with secondary antibody (anti-rabbit-Alexa Fluor 555, Life Technologies, A31572, and anti-mouse Alexa Fluor 647, Life Technologies, A31571, 1:100 each) for 2 h at room temperature. After other washing steps, cells were mounted on glass slide holders with homemade mounting medium containing DAPI. For the staining of overexpressed HA-tagged GABARAPL2/LC3B, cells were transfected with the corresponding plasmids 24 h prior to the staining and proceeded with as described above and stained for endogenous UBA5 and HA-tagged proteins (HA-antibody [Biolegends, 901502, 1:50]) All images were acquired on a Leica TCS SP5 confocal microscope using the LAS X software, which was also used for image generation. Images were then processed with the DeconvolutionLab2 software implemented in ImageJ. Quantification of co-localization was performed using the coloc2 software implemented in ImageJ.
Cellular fractionation assay
To prepare cytosolic and microsome fractions, cell pellet was suspended in fractionation buffer (50 mM TRIS pH 7.5 [FUJIFILM Wako Pure Chemical Corporation, 011–16381], 0.3 M sucrose [FUJIFILM Wako Pure Chemical Corporation, 196–00015] and protease inhibitors [Merck, 05056489001]), and the cell suspension was passed through a 26 gauge needle 10 times using 1 mL syringe. After centrifugation at 8,000 xg for 10 min, the supernatant was further centrifuged at 100,000 xg for 1 h. The resultant pellet was dissolved in fractionation buffer with 0.2% NP-40 [Nacalai tesque, 18558–54] and it was used as microsomal fraction. The supernatant was used as cytoplasmic fraction. The samples were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride (PVDF) membranes. Antibodies against LC3B (Cell Signaling Technology, 2775), GABARAP (Medical & Biological Laboratories Co., PM037), GABARAPL1 (Cell Signaling Technology, 26632S), GABARAPL2 (Medical & Biological Laboratories Co., PM038), BiP (Cell Signaling Technology, 3177) and GAPDH (Merck Millpore, MAB374) were purchased from the indicated suppliers. Anti–UBA5, anti-UFL1, and anti-UFBP1 polyclonal antibodies were described previously [Citation39,Citation40]. The immunoreactive bands were detected by LAS-4000 (GE Healthcare UK Ltd.). The quantitative densitometric analysis of UBA5 and ufmylation was carried out using Multi Gauge Version 3.2 Image software (Fuji Film, Tokyo, Japan). Statistical analysis was performed using an unpaired t test (Welch test). The data represents the means ± SE of 3 separate experiments.
Digital PCR
cDNA was synthesized as described above in ‘Reverse-transcriptase PCR and quantitative real-time PCR. Absolute quantification was performed using the QuantStudio 3D Digital PCR System (Thermo Fisher Scientific) and analyzed with the QuantStudio 3D AnalysisSuite Cloud Software (Thermo Fisher Scientific). The sequences of primers and probes were purchased from Thermo Fisher Scientific (Hs01076567_g1 for LC3A, Hs00797944_s1 for LC3B, Hs01374916_m1 for LC3C, Hs00925899_g1 for GABARAP, Hs00740588_mH for GABARAPL1 and Hs00371854_m1 for GABARAPL2).
Supplemental Material
Download MS Word (3.2 MB)Acknowledgments
We are indebted to Sabrina Habisov (Germany) for the help in sample preparation; and Birgit Schäfer for her work on cell cultures maintenance. We thank R. Ishimura (Niigata University) for his excellent technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741.
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93.
- Rogov V, Dötsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178.
- Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017;86:193–224.
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822.
- Kirkin V, McEwan DG, Novak I, et al. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269.
- Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841.
- Ohsumi Y, Ichimura Y, Kirisako T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492.
- Shpilka T, Weidberg H, Pietrokovski S, et al. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226.
- Abdollahzadeh I, Schwarten M, Gensch T, et al. The Atg8 family of proteins—modulating shape and functionality of autophagic membranes. Front Genet. 2017;8:1–7.
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178.
- Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. Embo J. 2010;29:1792–1802.
- Joachim J, Jefferies HBJ, Razi M, et al. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol Cell. 2015;60:899–913.
- Sagiv Y, Legesse-Miller A, Porat A, et al. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. Embo J. 2000;19:1494–1504.
- Muller JMM, Shorter J, Newman R, et al. Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J Cell Biol. 2002;157:1161–1173.
- Leil TA. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci. 2004;24:11429–11438.
- Genau HM, Huber J, Baschieri F, et al. CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol Cell. 2015;57:995–1010.
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145.
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385.
- Ichimura Y, Kumanomidou T, Sou Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857.
- von Muhlinen N, Akutsu M, Ravenhill BJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–342.
- Tumbarello DA, Manna PT, Allen M, et al. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of salmonella typhimurium by autophagy. PLoS Pathog. 2015;11:e1005174.
- Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218.
- Habisov S, Huber J, Ichimura Y, et al. Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. J Biol Chem. 2016;17:9025–9041.
- Daniel J, Liebau E. The ufm1 cascade. Cells. 2014;3:627–638.
- Komatsu M, Chiba T, Tatsumi K, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. Embo J. 2004;23:1977–1986.
- Tatsumi K, Yamamoto-Mukai H, Shimizu R, et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun. 2011;2:181.
- Lemaire K, Moura RF, Granvik M, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One. 2011;6:e18517.
- Zhang Y, Zhang M, Wu J, et al. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS One. 2012;7:e48587.
- Cai Y, Pi W, Sivaprakasam S, et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015;11:e1005643.
- Yoo HM, Kang SH, Kim JY, et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol Cell. 2014;56:261–274.
- Behrends C, Sowa ME, Gygi SP, et al. Network organization of the human autophagy system. Nature. 2010;466:68–76.
- Rogov VV, Stolz A, Ravichandran AC, et al. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep. 2017;18:1382–1396.
- Knight D, Harris R, Msb M, et al. The X-ray crystal structure and putative ligand-derived peptide binding properties of γ-aminobutyric acid receptor type a receptor-associated protein. J Biol Chem. 2002;277:5556–5561.
- Suzuki H, Tabata K, Morita E, et al. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure. 2014;22:47–58.
- Ichimura Y, Takagi K, Yang Y, et al. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 2014;15:557–565.
- Stadel D, Millarte V, Tillmann KD, et al. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol Cell. 2015;60:89–104.
- Bacik J-P, Walker JR, Ali M, et al. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme. J Biol Chem. 2010;285:20273–20280.
- Tatsumi K, Sou Y, Tada N, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2010;285:5417–5427.
- Ishimura R, Obata M, Kageyama S, et al. A novel approach to assess the ubiquitin-fold modifier 1-system in cells. FEBS Lett. 2017;591:196–204.
- Jacomin A-C, Samavedam S, Promponas V, et al. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–1953.
- Wilkinson DS, Jariwala JS, Anderson E, et al. Phosphorylation of LC3 by the Hippo Kinases STK3/STK4 is essential for autophagy. Mol Cell. 2015;57:55–68.
- Schwarten M, Stoldt M, Mohrlüder J, et al. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem Biophys Res Commun. 2010;395:426–431.
- Wu F, Watanabe Y, Guo X-Y, et al. Structural basis of the differential function of the two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in autophagy. Mol Cell. 2015;60:914–929.
- Gannavaram S, Connelly PS, Daniels MP, et al. Deletion of mitochondrial associated ubiquitin fold modifier protein Ufm1 in leishmania donovani results in loss of β-oxidation of fatty acids and blocks cell division in the amastigote stage. Mol Microbiol. 2012;86:187–198.
- Chen C, Itakura E, Weber KP, et al. An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLoS Genet. 2014;10:e1004082.
- Duan R, Shi Y, Yu L, et al. UBA5 mutations cause a new form of autosomal recessive cerebellar ataxia. PLoS One. 2016;11:1–12.
- Colin E, Daniel J, Ziegler A, et al. Biallelic Variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am J Hum Genet. 2016;99:695–703.
- Muona M, Ishimura R, Laari A, et al. Biallelic variants in UBA5 link dysfunctional UFM1 ubiquitin-like modifier pathway to severe infantile-onset encephalopathy. Am J Hum Genet. 2016;99:683–694.
- Nahorski MS, Maddirevula S, Ishimura R, et al. Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain. 2018;141:1943–1945.
- Roberts AM, Miyamoto DK, Huffman TR, et al. Chemoproteomic screening of covalent ligands reveals UBA5 as a novel pancreatic cancer target. ACS Chem Biol. 2017;12:899–904.
- Liu J, Wang Y, Song L, et al. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat Commun. 2017;8:14186.
- Weiergräber OH, Stangler T, Thielmann Y, et al. Ligand binding mode of GABAA receptor-associated protein. J Mol Biol. 2008;381:1320–1331.
- Keown JR, Black MM, Ferron A, et al. A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5α and mammalian autophagy-related ATG8 proteins. J Biol Chem. 2018;293:18378–18386.
- Real E, Rodrigues L, Cabal GG, et al. Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat Microbiol. 2018;3:17–25.
- Cheng X, Wang Y, Gong Y, et al. Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins. Autophagy. 2016;12:1330–1339.
- Olsvik HL, Lamark T, Takagi K, et al. FYCO1 contains a C-terminally extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy. J Biol Chem. 2015;290:29361–29374.
- Li J, Zhu R, Chen K, et al. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat Chem Biol. 2018;14:778–787.
- Sakurai S, Tomita T, Shimizu T, et al. The crystal structure of mouse LC3B in complex with the FYCO1 LIR reveals the importance of the flanking region of the LIR motif. Acta Crystallogr Sect F Struct Biol Commun. 2017;73:130–137.
- Noda NN, Satoo K, Fujioka Y, et al. Structural basis of atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44:462–475.
- Lin L, Yang P, Huang X, et al. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J Cell Biol. 2013;201:113–129.
- Marshall RS, Li F, Gemperline DC, et al. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in arabidopsis. Mol Cell. 2015;58:1053–1066.
- Kumar S, Jain A, Farzam F, et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol. 2018;5:997–1013.
- Rogov VV, Rozenknop A, Rogova NY, et al. A universal expression tag for structural and functional studies of proteins. ChemBioChem. 2012;13:959–963.
- Salzmann M, Wider G, Pervushin K, et al. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc. 1999;4:884.
- Logan T, Olejniczak E, Xu R, et al. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3:225–231.
- Montelione GT, Lyons BA, Emerson SD, et al. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114:10974–10975.
- Schanda P, Van Melckebeke H, Brutscher B. Speeding up three-dimensional protein NMR experiments to a few minutes. J Am Chem Soc. 2006;28:9042–9043.
- Farjon J, Boisbouvier J, Schanda P, et al. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J Am Chem Soc. 2009;131:8571–8577.
- Solyom Z, Schwarten M, Geist L, et al. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR. 2013;55:311–321.
- Löhr F, Hänsel R, Rogov VV, et al. Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J Biomol NMR. 2007;37:205–224.
- Pervushin K, Riek R, Wider G, et al. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J Am Chem Soc. 1998;25:6394–6400.
- Brutscher B, Boisbouvier J, Pardi A, et al. Improved sensitivity and resolution in 1H−13C NMR experiments of RNA. J Am Chem Soc. 1998;46:11845–11851.
- Zwahlen K, Legault P, Vincent SFJ, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA Complex. J Am Chem Soc. 1997;29:6711–6721.
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378.
- Güntert P, Buchner L. Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR. 2015;62:453–471.
- Koradi R, Billeter M, Güntert P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput Phys Commun. 2000;124:139–147.
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132.
- Emsley P, Lohkamp B, Scott WG, et al. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501.
- Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242.
- Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221.
- Stolz A, Putyrski M, Kutle I, et al. Fluorescence-based ATG8 sensors monitor localization and function of LC3/GABARAP proteins. Embo J. 2017;36:549–564.
- Nguyen TN, Padman BS, Usher J, et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016;215:857–874.
