Summary
The mechanisms that coordinate transport and clearance of synaptic autophagosomes are poorly understood. We performed forward genetic screens in C. elegans for mutants that abnormally accumulate autophagosomes in neurons, and identified ATG-4.2, a cysteine protease previously thought to be largely redundant with ATG-4.1. We find that atg-4.2 mutant animals, but not atg-4.1 mutant animals, have specific defects in autophagic maturation, resulting in accumulated autophagic vacuoles containing synaptic materials in the neuronal cell bodies. Defects in autophagic clearance are enhanced in animals also mutant for unc-16/Mapk8ip3/Jip3, which we find regulates retrograde transport of autophagosomes. This study demonstrates that ATG-4 isoforms can be specialized to perform distinct functions, from autophagosome biogenesis (through ATG-4.1, and also ATG-4.2) to autophagic vacuole clearance (primarily through ATG-4.2). Our study also highlights how dysfunction in distinct steps of the macroautophagy/autophagy process (transport and clearance) can result in enhanced autophagosome accumulation as seen in neurodegenerative diseases.
Article
Autophagy is a cellular degradation process, which is conserved from yeast to mammals, and which also occurs in specialized cell types, such as neurons. In the nervous system, autophagy at the synapse has been correlated with synaptic activity. In our study we demonstrate that in vivo in C. elegans the number of synaptic autophagosomes predictably varies depending on the firing state of the neuron, which we can manipulate by altering physiological stimuli that promote neuronal responses, by genetically inhibiting synaptic transmission or by chemo-genetically altering the response state of the neuron. We also observe that autophagic structures in neuronal cell bodies contain synaptic cargos. These observations raise questions regarding how biogenesis, transport and clearance are regulated across the polarized cell biology of the neuron.
To address these questions, we monitor autophagosomes by visualizing LGG-1 or LGG-2 localization in single neurons in vivo. LGG-1/2 (C. elegans orthologs of Atg8 in yeast, and LC3 and GABARAP in mammals) associates with both immature and mature autophagic structures. In previous work, we observed dynamic formation of autophagosomes at presynaptic sites and net retrograde transport of autophagosomes towards the cell body. We also found that the transmembrane protein ATG-9 is transported by the kinesin UNC-104/KIF1A to presynaptic sites, and that ATG-9 is required for local autophagosome formation.
To then investigate how synaptic autophagosomes transport, mature, and degrade in a polarized neuron, we used a combination of forward and reverse genetic screening, live imaging, biochemistry and electron microscopy [Citation1]. We first screened animals with mutations in candidate molecules associated with neuronal transport. We found that animals with mutations in the motor adaptor protein UNC-16/MAPK8IP3 display abnormal accumulation of autophagic vacuoles in the neurite due to a defect in retrograde transport. We then used unc-16 mutants as a sensitized background to uncover genetic interactors that would lead to an enhanced accumulation of autophagic structures in the neurite. We hypothesized that such mutants might disrupt processes involved in autophagosome maturation or degradation. Indeed we found a mutant that displays enhanced accumulation of autophagic structures, and uncovered the causative lesion in a poorly understood autophagy cysteine protease gene, atg-4.2.
The Atg4 family of cysteine proteases is a core component of the autophagic machinery, performing 2 cleavage functions. First they cleave a pro-form of Atg8/LGG-1/2/GABARAP/LC3 to allow their conjugation onto the phagophore membrane during autophagosome biogenesis (priming). Later Atg4s cleave Atg8s to remove them from the autophagosome to allow for autophagic maturation and degradation (delipidation). In yeast there is one Atg4 protein that performs both of these functions, whereas in mammals there are 4 ATG4s with different affinities for the 7 LC3/GABARAP family members. Previous biochemical studies of the mammalian isoforms have shown that different isoforms have different affinities for priming or delipidation activity. These findings raise the possibility that different isoforms may act in different steps (priming or delipidation) of the autophagy process ().
Figure 1. The schematic illustrates a model for synaptic autophagy across the cell biology of the neuron. Formation: Autophagosomes form at presynaptic sites dependent on neuronal activity, likely engulfing synaptic substrates including synaptic vesicle proteins and active zone proteins. Priming: Autophagy cysteine protease isoforms, primarily ATG-4.1, and also ATG-4.2, cleave the C-terminal end of LGG-1 to expose a terminal glycine residue, which is then conjugated onto the phagophore membrane at a phosphatidylethanolamine (PE) phospholipid to promote autophagosome formation. Transport: Autophagic vacuoles at the synapse are transported in a net retrograde fashion along the axon towards the cell body. Delipidation: The ATG-4.2 isoform, but not the ATG-4.1 isoform, cleaves LGG-1/2 off of the autophagosomal membrane to promote autophagosome maturation. Degradation: Autophagic vacuoles then fuse with acidic lysosomes and are degraded.
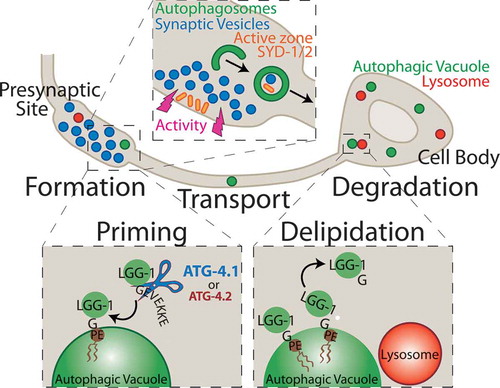
In C. elegans, there are 2 Atg4 enzymes, ATG-4.1 and ATG-4.2. We therefore examined whether atg-4.1 and atg-4.2 acted similarly. We found that atg-4.1 mutant animals do not display autophagic accumulation, which was surprising because at the time ATG-4.2 was thought to be redundant with ATG-4.1. To complement our in vivo studies, we performed electron microscopy and found that atg-4.2, but not atg-4.1, mutant animals accumulate multilamellar structures in C. elegans neurons.
Are these 2 Atg4 proteases performing different functions in vivo? We examined atg-4.1; atg-4.2 double mutant animals and observed a severe loss of LGG-1 puncta, suggesting that atg-4.1 and atg-4.2 are redundant for LGG-1 priming. However, our in vivo genetic data indicate that ATG-4.2 might be specialized for delipidation. We purified ATG-4.2 and tested its cleavage efficiency toward soluble LGG-1 (priming) versus lipidated LGG-1 (delipidation), and found that ATG-4.2 was more efficient at cleaving the lipidated substrate. Together these data are consistent with a model where ATG-4.1 and ATG-4.2 are partially redundant in priming, but ATG-4.2 is specialized for delipidation of LGG-1/2.
Why do autophagosomes accumulate in atg-4.2 mutant animals? We examined if the atg-4.2 mutant defects in delipidation of LGG-1/2 also affect the autophagic maturation process. Using a tandem label strategy to monitor LGG-1 acidification and a lysosome marker, LAAT-1, to monitor autolysosome formation, we determined that autophagosomes in atg-4.2 mutant animals fail to undergo maturation and fusion with lysosomes. We then examined if loss of ATG-4.2 function also affects accumulation of autophagosome synaptic cargos. We monitored cargo acidification by fusing the tandem label to the synaptic protein SYD-1 and found that atg-4.2 mutant animals accumulate non-acidified synaptic cargo in the neuronal soma. These results suggest that autophagosomes with synaptic cargos are transported to the cell body where their maturation and clearance is then dependent on delipidation of LGG-1/2 by ATG-4.2. Furthermore, our findings reveal that LGG-1/2 needs to be removed from the autophagic membrane for autophagosome maturation and cargo degradation.
Is there a benefit to specialized functions for the ATG-4 isoforms in vivo? We speculate that this specialization might provide targets for regulation. For example, regulated inhibition of ATG-4.2 would, in theory, pause autophagic progression without affecting new biogenesis (as priming continues due to partial redundancy with ATG-4.1). Understanding the specialized functions of these proteases could provide important pharmacological targets for regulating specific steps of autophagy, and allow for precise control of autophagy flux in distinct cell types and diseases.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Hill SE, Kauffman KJ, Krout M, et al. Maturation and clearance of autophagosomes in neurons depends on a specific cysteine protease isoform, ATG-4.2. Dev Cell. 2019;49:251–66 e8.
