ABSTRACT
Macroautophagy/autophagy is an essential cellular response in the fight against intracellular pathogens. Although some viruses can escape from or utilize autophagy to ensure their own replication, the responses of autophagy pathways to viral invasion remain poorly documented. Here, we show that peste des petits ruminants virus (PPRV) infection induces successive autophagic signalling in host cells via distinct and uncoupled molecular pathways. Immediately upon invasion, PPRV induced a first transient wave of autophagy via a mechanism involving the cellular pathogen receptor NECTIN4 and an AKT-MTOR-dependent pathway. Autophagic detection showed that early PPRV infection not only increased the amounts of autophagosomes and LC3-II but also downregulated the phosphorylation of AKT-MTOR. Subsequently, we found that the binding of viral protein H to NECTIN4 ultimately induced a wave of autophagy and inactivated the AKT-MTOR pathway, which is a critical step for the control of infection. Soon after infection, new autophagic signalling was initiated that required viral replication and protein expression. Interestingly, expression of IRGM and HSPA1A was significantly upregulated following PPRV replication. Strikingly, knockdown of IRGM and HSPA1A expression using small interfering RNAs impaired the PPRV-induced second autophagic wave and viral particle production. Moreover, IRGM-interacting PPRV-C and HSPA1A-interacting PPRV-N expression was sufficient to induce autophagy through an IRGM-HSPA1A-dependent pathway. Importantly, syncytia formation could facilitate sustained autophagy and the replication of PPRV. Overall, our work reveals distinct molecular pathways underlying the induction of self-beneficial sustained autophagy by attenuated PPRV, which will contribute to improving the use of vaccines for therapy.
Abbreviations: ACTB: actin beta; ANOVA: analysis of variance; ATG: autophagy-related; BECN1: beclin 1; CDV: canine distemper virus; Co-IP: coimmunoprecipitation; FIP: fusion inhibitory peptide; GFP: green fluorescent protein; GST: glutathione S-transferase; HMOX1: heme oxygenase 1; hpi: hours post infection; HSPA1A: heat shock protein family A (Hsp70) member 1A; HSP90AA1: heat shock protein 90 kDa alpha (cytosolic), class A member 1; IFN: interferon; IgG: immunoglobulin G; INS: insulin; IRGM: immunity related GTPase M; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MeV: measles virus; MOI: multiplicity of infection; MTOR: mechanistic target of rapamycin kinase; PI3K: phosphoinositide-3 kinase; PIK3C3: phosphatidylinositol 3-kinase catalytic subunit type 3; SDS: sodium dodecyl sulfate; siRNA: small interfering RNA; SQSTM1/p62: sequestosome 1; UV: ultraviolet.
Introduction
Peste des petits ruminants (PPR) is a highly contagious disease that mainly affects goats and sheep, although it also occasionally affects small or even large wild ruminants [Citation1–Citation3]. PPR virus (PPRV) has a linear negative-stranded RNA genome and belongs to the genus Morbillivirus in the family Paramyxoviridae [Citation4]. The PPRV genome encodes six structural proteins (the nucleocapsid protein [N], phosphoprotein [P], fusion protein [F], matrix protein [M], haemagglutinin-neuraminidase protein [HN], and large protein [L]) and two non-structural proteins (C and V) in the order 3ʹ-N-P/C/V-M-F-HN-L-5ʹ [Citation5]. Its clinical signs usually include pyrexia, mucopurulent oculonasal discharges, diarrhoea, stomatitis and pneumonia [Citation6,Citation7]. Notably, PPRV infection often causes foetal mummification, abortions late in pregnancy, or birth of dead or weak lambs that die within a few days [Citation8,Citation9]. PPRV has spread to more than 70 countries in Africa, the Near and Middle East, and Asia and is currently threatening more than 1.7 billion sheep and goats [Citation10], causing significant economic losses [Citation5,Citation11]. Disease control is mostly achieved through clinical or laboratory-based diagnosis coupled with vaccination and/or slaughter. All vaccines currently in use are live attenuated strains of PPRV [Citation4,Citation12]. The Nigeria/75 (N75) vaccine has been shown in different studies to protect against virus isolates of all 4 lineages in most countries [Citation12–Citation14]. However, we lack information on the intracellular responses and signalling mechanisms that occur in sheep/goats treated with the N75 vaccine. Viruses have been found to interact very strongly with specific pathways that are usually involved in the control of infections [Citation15]. The autophagy-related proteome is one of the major targets of RNA viruses [Citation16], which may indicate the importance of autophagy during the course of viral infection.
Cell invasion by pathogens is dependent on their ability to bind to cellular receptors that promote their uptake [Citation17]. Upon intracellular pathogen invasion, autophagy can be activated as an innate immune mechanism to control infection [Citation17–Citation19]. Like all morbilliviruses, PPRV has been established to exhibit lymphatic and epithelial tropism [Citation20]. NECTIN4 is mainly expressed in epithelial tissues and is encoded by multiple haplotypes in different sheep breeds worldwide [Citation21]. Recently, NECTIN4 has been identified as an epithelial receptor for measles virus (MeV), canine distemper virus (CDV), phocine distemper virus and PPRV, which has shed light on the mode of entry of these viruses [Citation21–Citation25]. Increasing evidence suggests that pathogen invasion can initiate autophagy through pathogen receptors [Citation17,Citation18,Citation26]. However, whether NECTIN4 is involved in the regulation of autophagy induced by pathogen invasion is unknown. The MTOR kinase-dependent signalling pathway controls autophagy [Citation27]. Activation of the PI3K-AKT-MTOR and AKT-tuberous sclerosis complex (TSC)-MTOR pathways inhibits autophagy, whereas loss of signalling through this cascade removes the negative regulation of MTOR [Citation27]. A previous study revealed that upon virus recognition, a direct connection between HSP90AA1 (a receptor) and the AKT-MTOR pathway triggers autophagy [Citation18]. Inhibition of the PI3K-AKT-MTOR signalling pathway can affect the autophagic reaction induced by infection with viruses, such as coxsackievirus B3 [Citation28], foot-and-mouth disease virus [Citation29,Citation30], avian influenza A virus [Citation31], and avian reovirus [Citation32]. Therefore, a close relationship exists between autophagy and the MTOR signalling pathway during viral invasion.
Interestingly, research on the kinetics of autophagy induction upon MeV infection has revealed that two successive waves of autophagy occur [Citation33,Citation34]. The induction of the first (early) wave of autophagy has been demonstrated to occur during MeV entry into cells [Citation17,Citation33]. Subsequently, a second wave of autophagy is induced that requires viral replication and expression of the non-structural MeV protein C [Citation33]. Both autophagic wave signals can be observed upon infection with MeV vaccine strains. Vaccine delivery/attenuated MeV infection induces a specific pathway of early autophagy induction involving CD46 (CYT-1)-GOPC, which initiates autophagosome formation by interacting with BECN1 [Citation17]. Increasing evidence suggests that several members of the Morbillivirus genus, including MeV [Citation33], CDV [Citation35] and PPRV [Citation36,Citation37], can utilize an autophagic mechanism to facilitate replication. Importantly, several MeV proteins have been reported to be able to interact with autophagy-associated proteins, which could contribute to modulation of the late wave of autophagy [Citation16,Citation38,Citation39]. The human IRGM (immunity related GTPase M) protein has been shown to be widely targeted by RNA viruses, several of which can exploit autophagy in human cells to improve their replication [Citation16,Citation39]. Several RNA viruses that modulate autophagy to efficiently replicate have been found to target IRGM [Citation16]. Furthermore, infection of murine neuroblastoma cells with the Edmonston strain of MeV (Ed-MeV) results in HSPA1A induction [Citation40]. CDV also induces HSPA1A expression in the brain following naturally occurring infection [Citation41]. Previous studies have suggested that heat shock proteins might play a regulatory role in autophagy [Citation42,Citation43]. Acetylated HSPA1A and TRIM28 (tripartite motif containing 28) mediated PIK3C3 sumoylation are required for autophagosome creation during autophagy [Citation44]. Knockdown of HSPA1A blocks BECN1-PIK3C3 complex formation and inhibits TRIM28 binding to PIK3C3 and autophagic vesicle formation [Citation44]. However, the roles of HSPA1A and IRGM in PPRV-induced autophagy remain unknown.
Although autophagosome formation is observed upon infection of hosts with a wide range of viruses, few molecular details of the interactions of viruses with the autophagic machinery are known. Nevertheless, Morbillivirus species have evolved molecular strategies to counteract autophagy, escaping this process or even exploiting it to improve their own infectivity [Citation33]. We previously reported that autophagy not only promotes PPRV replication in caprine endometrial epithelial cells (EECs) but also acts as a potential mechanism to inhibit PPRV-induced apoptosis [Citation37]. The PPRV-H, PPRV-N and PPRV-C proteins significantly promote the autophagy pathway [Citation37]. Furthermore, we previously showed that PPRV enters EECs via the caveolae-mediated endocytosis pathway [Citation45]. The NECTIN4 protein is located on the surfaces of EECs and is significantly upregulated following PPRV infection [Citation45,Citation46]. In this study, we sought to determine how PPRV and viral proteins regulate autophagy during the course of infection and how the virus exploits the cellular autophagic machinery for replication. We report here that the PPRV receptor NECTIN4 can be physically connected to the autophagic machinery via a unique molecular intermediate. Furthermore, we describe a role for IRGM and HSPA1A in both PPRV-dependent autophagy induction and virus production. Enhanced understanding of the two successful and distinct waves of autophagy induced by attenuated PPRV may be important for controlling viral infection and for enabling better use of PPRV-derived vaccines for therapy.
Results
An attenuated PPRV strain induces two successive but distinct waves of autophagy in EECs
We previously reported that autophagy enhances PPRV replication and inhibits caspase-dependent apoptosis in vitro [Citation37]. We also showed that the binding and entry of PPRV into EECs profoundly affects early cellular gene expression [Citation46]. LC3-II is widely used as a marker of autophagy [Citation47]. To gain insight into how different PPRV infection stages modulate the autophagy process, we tested autophagy kinetics upon infection of cells by PPRV at a multiplicity of infection (MOI) of 3 from 0 to 24 h post infection (hpi). Strikingly, we found that PPRV infection induced two successive waves of autophagic flux (). Compared to that in mock-infected cells, the amount of intracellular LC3-II in PPRV-infected EECs was strongly increased at 1.5 hpi (the first wave) ( and S1); however, this wave was transient, and the LC3-II expression immediately returned to the basal level. Furthermore, we found that intracellular LC3-II expression was induced at 9 hpi (the second wave) and was sustained at an extremely significantly elevated level for up to 24 hpi (). In addition to LC3, we examined SQSTM1 (sequestosome 1), a target of autophagic degradation [Citation47], and found that its expression was reduced during the first wave of autophagy beginning at 1.5 hpi and during the second wave of autophagy from 9 hpi to 24 hpi (), suggesting that increases in autophagic flux occurred at these time points after infection. Moreover, the viral N protein was detectable at 9 hpi, and its levels sharply increased at 12 hpi (). The viral titers also increased rapidly at 12 hpi (). Therefore, for subsequent experiments, 1.5 h and 12 h post PPRV infection were considered the optimal time points for evaluation of the early and late waves of autophagosome accumulation, respectively.
Figure 1. Characterization of PPRV-triggered autophagosome accumulation. (A and B) EECs were mock-infected or infected with PPRV (MOI = 3) for 1.5, 3, 6, 9, 12 or 24 h. At the end of the infection period, the LC3, SQSTM1, PPRV-N, and ACTB (loading control) expression levels were analyzed by immunoblotting with specific antibodies. (C) The LC3-II levels relative to the ACTB levels were determined by densitometry. (D) The SQSTM1 levels relative to the ACTB levels were determined by densitometry. (E) The PPRV-N levels relative to the ACTB levels were determined by densitometry. (F) EECs were infected with PPRV (MOI = 3) for 1.5, 3, 6, 9, 12 or 24 h. The viral titers were measured using the TCID50 method. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; *P < 0.05; ***P < 0.001; #P > 0.05.
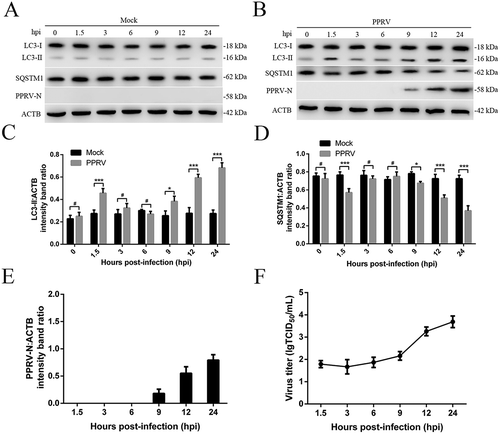
To determine whether PPRV infection regulated the two distinct waves of autophagy, transmission electron microscopy (TEM) was performed for ultrastructural analysis of EECs infected with UV-irradiated PPRV or PPRV. We found that the numbers of single- and double-membrane autophagosome-like vesicles were increased in the cytoplasm of both the UV-irradiated PPRV and PPRV-infected EECs at 1.5 hpi (), whereas similar vesicles were rarely observed in the mock-infected EECs (). We also found that the number of cytoplasmic membrane vesicles was significantly increased in the PPRV-infected EECs at 12 hpi and that many recognizable cytoplasmic contents or partially degraded organelles were sequestered in these vesicles (). Strikingly, compared with the replication-competent PPRV-infected EECs, neither the mock-infected cells nor the cells inoculated with UV-irradiated PPRV exhibited an increase in the number of autophagosome-like vesicles at 12 hpi (). Moreover, in agreement with the TEM results, our immunoblotting results showed that LC3-II expression was upregulated in both the UV-irradiated PPRV and PPRV-infected EECs compared to the mock-infected EECs at 1.5 hpi following SQSTM1 degradation (). LC3-II upregulation and SQSTM1 degradation were detected only in the PPRV-infected EECs at 12 hpi; the UV-irradiated PPRV-treated EECs did not exhibit LC3-II upregulation or SQSTM1 degradation at 12 hpi (). These data indicated that PPRV replication was required for the induction of autophagy during the late wave but not during the early wave.
Figure 2. PPRV induces two waves of autophagy via distinct and uncoupled molecular pathways. (A and B) EECs were mock-infected or infected with PPRV or UV-irradiated PPRV (MOI = 3) for 1.5 or 12 h. The cell samples were fixed and processed for electron microscopy analysis. Scale bars, 1 μm. Magnified views of the autophagosome-like vesicles are enclosed by black square frames. Scale bars, 200 nm. (C) EECs were mock-infected or infected with PPRV or UV-irradiated PPRV (MOI = 3) for 1.5 or 12 h. The cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-PPRV-N and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in the mock-, UV-irradiated PPRV- and PPRV-infected EECs were determined by densitometry. (D) EECs were treated with si-control, si-ATG5 or si-GOPC and infected with PPRV (MOI = 3) for 1.5 or 12 h. The cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-PPRV-N and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in the siRNA-transfected cells were determined by densitometry. (E) EECs were treated with chloroquine and infected with PPRV (MOI = 3) for 1.5, 3 or 6 h. The cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1 and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in the treated cells were determined by densitometry. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; #P > 0.05.
Interaction of CD46 (Cyt-1) with GOPC can stabilize PIK3C3-BECN1 complex activation to facilitate the formation of autophagosomes [Citation17]. A previous study reported that attenuated MeV induces autophagy at 1.5 hpi through the CD46 (CYT-1)-GOPC pathway [Citation33]. To determine whether the molecular mechanism of autophagy induction by PPRV at 1.5 hpi was consistent with that mediated by MeV, we successfully used small interfering RNAs (siRNAs) to reduce the expression of putative key proteins in EECs, specifically ATG5 and GOPC (Fig. S2A). Importantly, immunoblotting revealed that knockdown of GOPC expression did not influence the expression of LC3-II and SQSTM1 in EECs at 1.5 and 12 hpi, showing that the early and late induction of autophagy following PPRV infection did not require the expression of this scaffold protein (). As expected, treatment of cells with si-ATG5 significantly suppressed LC3-II upregulation and SQSTM1 degradation in EECs at 1.5 and 12 hpi (). Moreover, suppression of ATG5 expression strongly reduced viral N protein expression and viral titers in EECs at 12 hpi ( and S2B). These results indicated that infection with an attenuated strain of PPRV induced two successive waves of autophagy that were dependent on ATG5 but not on the scaffold protein GOPC.
Chloroquine inhibits the fusion of autophagosomes and lysosomes and can thus increase the accumulation of LC3-II and SQSTM1 [Citation47,Citation48]. To further validate whether the low level of autophagy observed between the two waves was caused by PPRV, EECs were treated with chloroquine and infected with PPRV for 1.5, 3 or 6 h. Our data showed that chloroquine treatment significantly increased the expression of LC3-II and SQSTM1 in the mock and PPRV-infected EECs at 1.5 hpi (). However, LC3-II and SQSTM1 expression was not significantly increased in the PPRV-treated EECs in the presence of chloroquine at 3 and 6 hpi (). These results indicated that PPRV actively and rapidly inhibited autophagosome formation following the first transient wave of autophagy.
Altogether, the results revealed that PPRV induces two waves of autophagy in EECs via distinct and uncoupled molecular pathways.
The role of the AKT-MTOR pathway in PPRV-induced autophagy in EECs
The AKT and MTOR kinase-dependent signalling pathway controls autophagy [Citation27]. Cell metabolism, proliferation and survival are highly dependent on the phosphorylation statuses of AKT and MTOR [Citation49,Citation50]. To determine whether AKT-MTOR modulated autophagy upon PPRV infection, we tested AKT and MTOR activity in host cells. Compared with that in the mock-infected EECs, AKT and MTOR phosphorylation in the PPRV-infected EECs was downregulated at 1.5 hpi (). However, AKT and MTOR phosphorylation did not remain downregulated at 12 hpi (). These data indicated that PPRV induced autophagy upon inactivation of the AKT-MTOR pathway during the early stage of infection. However, the late wave of autophagy induced by PPRV did not involve inhibition of AKT and MTOR activity.
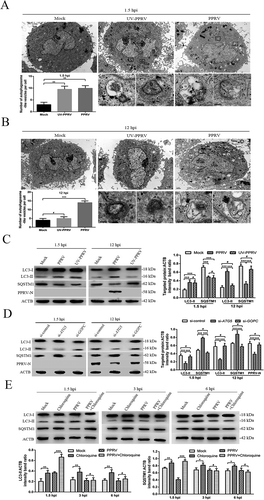
Figure 3. The role of the AKT-MTOR pathway in PPRV-induced autophagy in host cells. (A and B) EECs were mock-infected or infected with PPRV (MOI = 3). The cells were harvested at 1.5, 3, 6, 9, 12 and 24 h and then immunoblotted with anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, and anti-ACTB antibodies. (C) The p-MTOR levels relative to the MTOR levels were determined by densitometry. (D) The p-AKT levels relative to the AKT levels were determined by densitometry. (E and F) EECs were pre-treated with INS (1 μM) for 6 h prior to viral infection. Then, the cells were infected with PPRV or UV-irradiated PPRV (MOI = 3) for 1.5 or 12 h. The cell samples were analyzed by immunoblotting with anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, anti-LC3, anti-PPRV-N and anti-ACTB (loading control) antibodies. (G) The relative target protein levels in the INS-pre-treated cells at 1.5 hpi were determined by densitometry. (H) The relative target protein levels in the INS-pre-treated cells at 12 hpi were determined by densitometry. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; **P < 0.01; ***P < 0.001; #P > 0.05.
To further validate the roles of AKT and MTOR in the PPRV-dependent induction of autophagy, we activated the AKT-MTOR pathway with INS (insulin) before infecting cells with PPRV or treating them with UV-irradiated PPRV. Our data showed that the levels of phosphorylated AKT and MTOR were persistently downregulated in the UV-irradiated PPRV-treated EECs compared to the mock-infected EECs in the absence of INS at 1.5 hpi (). Following INS treatment, no obvious decreases occurred in AKT or MTOR phosphorylation during the early stage in the PPRV-infected or UV-irradiated PPRV-treated cells compared to the mock-infected cells (). Correspondingly, the LC3-II levels in the PPRV-infected and UV-irradiated PPRV-treated EECs did not significantly increase compared to the level in the mock-infected cells (), indicating that AKT-MTOR signalling pathway activation inhibited the first PPRV-induced wave of autophagy. Therefore, our observations confirm that inactivation of the AKT-MTOR pathway during the early stage of PPRV infection induces autophagy. However, we found that p-AKT and p-MTOR were not significantly downregulated in the UV-irradiated PPRV-treated EECs compared to the mock-infected EECs in the absence of INS at 12 hpi (). Importantly, our data showed that the LC3-II levels in the PPRV-infected EECs were significantly higher than those in the mock-infected cells in the absence and presence of INS (), indicating that AKT-MTOR signalling pathway activation by INS did not influence the late wave of autophagy induced by PPRV. Moreover, the viral N protein expression and viral titers did not significantly differ between the groups of cells treated with or without INS ( and S3). Taken as a whole, our findings reveal that the AKT-MTOR pathway plays different roles in the two waves of autophagy induced by PPRV in host cells.
PPRV-H and NECTIN4 are critical for the induction of the first wave of autophagy via the AKT-MTOR pathway
The H protein is responsible for viral attachment to host cells [Citation5]. First, we expressed and purified the fusion proteins His-GST, His-V, His-H, and His-N (). Then, we determined whether the viral proteins could modulate the first wave of autophagosome formation. To this end, GFP-LC3 EECs were treated with 100 ng/mL His-GST, His-V, His-H or His-N, and autophagy was analyzed by tracking GFP+ autophagosomes. We found that only the H protein induced a significant enhancement of LC3 punctate staining signals distributed throughout the entire cytoplasm (), which indicated that the viral H protein facilitated a significant increase in the number of autophagosomes compared to the GST protein. However, both the His-V- and His-N-treated EECs exhibited faint diffuse staining patterns and showed little LC3 punctate accumulation (). Consequently, we examined whether the H protein activated autophagy effectively via the AKT-MTOR pathway. We incubated cells with purified His-GST, His-V, His-H, or His-N fusion proteins for 1.5 h and subsequently examined the endogenous LC3-II and SQSTM1 levels. Our data showed that LC3-II was upregulated and that SQSTM1 was downregulated in a dose-independent manner in the His-H-treated cells (). In contrast, LC3-II and SQSTM1 expression was not changed in the His-V, His-N or His-GST-treated cells (). Moreover, we determined the AKT and MTOR phosphorylation levels 1.5 h after treatment of the cells with the H protein to clarify the signalling pathway involved in the H-induced early wave of autophagy. We found that p-AKT and p-MTOR were decreased significantly in a dose-independent manner in His-H-treated cells and that no detectable changes occurred in His-V, His-N or His-GST-treated cells (), indicating that the H protein inhibited AKT-MTOR phosphorylation. Together, these results confirm that the viral H protein, which is responsible for viral attachment to the host cell, is sufficient to effectively activate the first wave of autophagy by decreasing AKT and MTOR activity.
Figure 4. PPRV-H and NECTIN4 are sufficient to induce the first wave of autophagy via AKT and MTOR dephosphorylation. (A) His-tagged V, H and N were expressed in E. coli BL-21 and purified using Ni-NTA columns. The purified products were separated using SDS-PAGE and analyzed by immunoblotting with an anti-His antibody. (B) EECs were transfected with GFP-LC3 for 24 h and incubated with 100 ng/mL His-GST, His-V, His-H, or His-N for 1.5 h. Autophagy was monitored by evaluating the number of GFP+-LC3 vesicles per cell profile by confocal immunofluorescence microscopy. Scale bars, 20 μm. (C) Corresponding graph representing the numbers of GFP+-LC3 vesicles per cell profile in EECs pre-treated with His-GST, His-V, His-H, or His-N. (D) EECs were cultured in DMEM/F12 supplemented with 2% fetal bovine serum containing His-GST, His-V, His-H or His-N for 1.5 h and analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, and anti-ACTB (loading control) antibodies. The relative target protein levels were determined by densitometry in His-GST, His-V, His-H and His-N pre-treated cells. (E) EECs were treated with si-control or si-NECTIN4 and infected with PPRV (MOI = 3) for 1.5 h. The cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, anti-NECTIN4 and anti-ACTB (loading control) antibodies. The relative target protein levels in the siRNA-transfected cells were determined by densitometry. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; **P < 0.01; ***P < 0.001; #P > 0.05.
To confirm that NECTIN4 mediates PPRV-induced autophagy, we used siRNAs to reduce NECTIN4 expression in EECs (). Importantly, we found that knockdown of NECTIN4 expression disrupted the early induction of autophagy following PPRV infection, showing that the first wave of autophagy required NECTIN4 expression (). As expected, treatment of cells with si-NECTIN4 increased the p-AKT and p-MTOR levels following PPRV infection at 1.5 hpi (). However, reducing NECTIN4 expression did not influence the levels of p-AKT or p-MTOR in the mock-infected EECs (). Altogether, these results indicated that NECTIN4 was also a critical protein for autophagy induction at the early stage of PPRV infection via the AKT-MTOR pathway.
PPRV-H binding to NECTIN4 triggers autophagy via AKT-MTOR dephosphorylation
The first interaction of the host cell with PPRV is mediated by binding of the virus to cell receptors through the viral H protein [Citation5]. To investigate the interactions between PPRV-H and NECTIN4 (the cell receptor), we examined the ability of these proteins to form a complex in co-immunoprecipitation (co-IP) experiments. Our data showed that NECTIN4 was present in the immunoprecipitates obtained with an anti-HA antibody (). Next, we co-expressed exogenous H-HA and NECTIN4-Flag in EECs and expressed NECTIN4-Flag alone as a control to detect the interactions between the H protein and NECTIN4. The results showed that exogenous NECTIN4 and PPRV-H were both present in the immunoprecipitate (). Reciprocal co-IP experiments further confirmed the interaction of the viral H protein and the cell receptor NECTIN4 (). Importantly, we detected endogenous AKT in the immunoprecipitates obtained with the anti-Flag antibody in NECTIN4-Flag-EECs at 1.5 hpi (). We did not detect AKT in the immunoprecipitates obtained with the anti-Flag antibody at 3 hpi (). Moreover, we detected the interaction of AKT and NECTIN4 only in the His-H-treated EECs (). Endogenous AKT was not detected in the immunoprecipitates obtained with the anti-Flag antibody in NECTIN4-Flag-EECs treated with His-GST, His-V, or His-N (). These results further indicated that infection with PPRV for 1.5 h and the presence of the H protein clearly led to the interaction of AKT with the cell receptor NECTIN4 in EECs, confirming that the interaction between AKT and NECTIN4 is critical for the induction of the first wave of autophagy.
Figure 5. PPRV-H binding to NECTIN4 triggers autophagy via AKT-MTOR dephosphorylation. (A) Co-IP assay results demonstrating that endogenous NECTIN4 binds to H-HA in transfected cells. EECs were transfected with pCDNA3.1-H-HA for 48 h and harvested. Cell lysates from transfected cells and untransfected control cells were immunoprecipitated with an antibody against HA and then subjected to immunoblotting. (B) Exogenous NECTIN4-Flag and H-HA coexpression in EECs. Cell lysates from cells co-transfected with NECTIN4-Flag and H-HA or transfected with NECTIN4-Flag alone were immunoprecipitated with an antibody against HA and then subjected to immunoblotting. (C) Results of reciprocal co-IP experiments showing that the anti-Flag antibody precipitated H-HA. (D) EECs were transfected with pCDNA3.1-NECTIN4-Flag for 48 h and then infected with PPRV (MOI = 3) for 1.5 or 3 h. Cell lysates from the transfected cells were immunoprecipitated with an antibody against Flag and then subjected to immunoblotting. (E) EECs were transfected with pCDNA3.1-NECTIN4-Flag for 48 h and then incubated with 100 ng/mL His-GST, His-V, His-H, or His-N for 1.5 h. Cell lysates from the transfected cells were immunoprecipitated with an antibody against Flag and then subjected to immunoblotting. (F) Immunoblotting was performed using anti-LC3 and anti-SQSTM1 antibodies on lysates from EECs cultured in uncoated plates or in plates coated with anti-NECTIN4 or an irrelevant isotype control IgG for 4 h at 37°C. The target protein levels relative to the ACTB levels in cells pre-treated with anti-NECTIN4 or an irrelevant isotype control IgG were determined by densitometry. (G) EECs were transfected with GFP-LC3 for 24 h and cultured in plates coated with the irrelevant isotype control IgG or anti-NECTIN4 for 4 h. Autophagy was monitored by evaluating the number of GFP+-LC3 vesicles per cell profile by confocal immunofluorescence microscopy. Scale bars, 20 μm. The corresponding graph shows the number of GFP+-LC3 vesicles per cell profile among the EECs pre-treated with anti-NECTIN4 or the irrelevant isotype control IgG. (H) EECs were incubated with His-GST, a mixture of His-GST and NECTIN4 (His-GST:NECTIN4 = 1:3), a mixture of His-H and NECTIN4 (His-H:NECTIN4 = 1:3), or His-H for 1.5 h and analyzed by immunoblotting with anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, anti-LC3, and anti-ACTB (loading control) antibodies. The relative target protein levels in the pre-treated cells were determined by densitometry. (I) EECs were incubated with His-GST (100 ng/mL) or His-H (100 ng/mL) and PPRV for 1.5 h at 37°C or 4°C. The cells were analyzed by immunoblotting with anti-p-MTOR, anti-MTOR, anti-p-AKT, anti-AKT, anti-LC3, and anti-ACTB (loading control) antibodies. The relative target protein levels in the pre-treated cells were determined by densitometry. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; **P < 0.01; ***P < 0.001; #P > 0.05.
Interestingly, LC3-II was upregulated and SQSTM1 was downregulated in anti-NECTIN4 antibody-treated EECs, but no obvious changes were observed in the irrelevant immunoglobulin G (IgG)-treated or control cells (). Likewise, the immunofluorescence assay revealed significantly increased autophagosome accumulation in the anti-NECTIN4-treated cells transfected with GFP-LC3 () but not in the irrelevant IgG-treated or control cells. Furthermore, we examined whether cellular NECTIN4 mediated PPRV-H-induced early autophagy via the AKT-MTOR pathway. PPRV-H protein treatment significantly decreased p-MTOR and p-AKT expression and significantly increased LC3-II expression in EECs. However, a similar change was not found in H-NECTIN4 protein complex-treated cells (), indicating that the interaction between the H protein and NECTIN4 played an important role in the induction of early autophagy via the AKT-MTOR pathway. Moreover, EECs treated with the H protein or infected with PPRV and then incubated at 4°C or 37°C exhibited significantly decreased p-MTOR and p-AKT levels compared with the mock-treated EECs (). These data indicated that disruption of the binding of the membrane receptor NECTIN4 to the viral H protein impaired AKT-MTOR signalling activity in a temperature-independent manner.
The second wave of autophagy during PPRV infection depends on IRGM and HSPA1A
Recently, the IRGM protein was highlighted for its contribution to autophagy upon viral infection [Citation16,Citation39], which prompted us to test whether this protein is involved in PPRV-induced autophagy. Moreover, the heat shock response, represented by HSPA1A, is an intracellular control mechanism that switches cells from autophagy to the growth and protein synthesis phase [Citation43]. Here, we investigated IRGM and HSPA1A protein expression during PPRV infection. Our results showed that IRGM and HSPA1A expression was upregulated in the mock- and PPRV-infected EECs at 12 hpi (). However, the IRGM and HSPA1A expression levels were not significantly increased in the mock or PPRV-infected EECs at 1.5 hpi (). We also found that PPRV infection did not significantly influence HSP90AA1 or HMOX1 expression (Fig. S4A).
Figure 6. The second wave of autophagy induced by PPRV depends on IRGM and HSPA1A. (A and B) EECs were mock-infected or infected with PPRV (MOI = 3) for 1.5 or 12 h. At the end of the infection period, the IRGM, HSPA1A and ACTB (loading control) expression levels were analyzed by immunoblotting with specific antibodies. The target protein levels relative to the ACTB levels were determined by densitometry. (C-E) EECs were treated with si-control, si-IRGM or si-HSPA1A and infected with PPRV (MOI = 3) for 1.5 or 12 h. The cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-PPRV-N and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in the siRNA-transfected cells were determined by densitometry. The viral titers were measured using the TCID50 method. (F) GFP-LC3 EECs were treated with si-control, si-IRGM or si-HSPA1A and infected with PPRV (MOI = 3) for 12 h. Autophagy was monitored by evaluating the number of GFP+-LC3 vesicles per cell profile by confocal immunofluorescence microscopy. Scale bars, 20 μm. The corresponding graph shows the number of GFP+-LC3 vesicles per cell profile among the siRNA pre-treated EECs. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; #P > 0.05.
To determine whether IRGM was involved in the autophagosome formation observed upon PPRV infection, we reduced IRGM and HSPA1A expression using specific siRNAs. The expression levels of the endogenous proteins were specifically decreased in the cells treated with si-IRGM or si-HSPA1A (Fig. S4B). First, we tested the requirement for IRGM and HSPA1A during PPRV-dependent autophagic modulation by monitoring the upregulation of LC3-II and the degradation of SQSTM1 by immunoblotting. Interestingly, our data showed that LC3-II was upregulated and that SQSTM1 was downregulated (p < 0.05) in the mock-infected EECs upon treatment with si-HSPA1A (). Importantly, we found that knockdown of IRGM or HSPA1A expression did not disrupt the early induction of autophagy following PPRV infection at 1.5 h, showing that the first wave of autophagy did not require the expression of these proteins (). As expected, we found that knockdown of IRGM or HSPA1A expression did significantly disrupt the late induction of autophagy upon PPRV infection at 12 h, showing that the second wave of autophagy required the expression of these proteins (). Moreover, suppression of IRGM or HSPA1A expression strongly reduced viral N protein expression and viral titer (). Moreover, in agreement with the immunoblotting results, the immunofluorescence assay results revealed that autophagosome accumulation was significantly downregulated in both the si-IRGM and si-HSPA1A-treated cells but not in the si-control-treated cells at 12 hpi (). Altogether, these data indicated that IRGM and HSPA1A played important roles in the induction of autophagy in the second wave but not in the first wave upon PPRV infection.
Viral proteins interact with IRGM and HSPA1A and induce autophagosome accumulation via a molecular process involving these proteins
Previous studies have revealed an unexpected role for IRGM in virus-induced autophagy and have suggested that several different families of RNA viruses might use common strategies to manipulate autophagy and improve viral infectivity [Citation16,Citation51]. Moreover, we previously reported that the PPRV-H, PPRV-N and PPRV-C proteins increase LC3-II levels and highly co-localize with LC3-positive fluorescent puncta in host cells [Citation37]. To further validate the molecular mechanisms of IRGM and HSPA1A in both PPRV-induced autophagy and viral particle formation, we first analyzed whether IRGM interacted with viral proteins in host cells. In transfected EECs, we found that endogenous IRGM interacted with PPRV-C but not with PPRV-N (). We also showed that endogenous HSPA1A interacted with PPRV-N but not with PPRV-C using a co-IP assay (). Importantly, we confirmed the physical and direct interaction between HSPA1A and PPRV-N using GST pulldown analysis (). In agreement with the co-IP analysis results, the immunofluorescence assay results revealed that IRGM co-localized with PPRV-C and that HSPA1A co-localized with PPRV-N (). Thus, HSPA1A and IRGM are new intracellular partners of viral proteins that may induce the second wave of autophagy.
Figure 7. Physical interaction and co-localization of IRGM and HSPA1A with viral proteins. (A and B) Co-IP assay results demonstrating that endogenous IRGM binds C-HA but not N-HA in transfected cells. EECs were transfected with pCDNA3.1-C-HA or pCDNA3.1-N-HA for 48 h and harvested. Cell lysates from the transfected cells and from untransfected control cells were immunoprecipitated with an antibody against HA and then subjected to immunoblotting. (C and D) Co-IP assay results demonstrating that endogenous HSPA1A binds N-HA but not C-HA in transfected cells. EECs were transfected with pCDNA3.1-C-HA or pCDNA3.1-N-HA for 48 h and harvested. Cell lysates from the transfected cells and from untransfected control cells were immunoprecipitated with an antibody against HA and then subjected to immunoblotting. (E) GST pulldown assay results demonstrating the physical and direct interaction between HSPA1A and PPRV-N. Glutathione beads conjugated to GST or a GST-HSPA1A fusion protein were incubated with recombinant N-HA protein. After washing, the proteins were eluted from the beads, and SDS-PAGE was performed. Expression of the N protein was detected by immunoblotting with an anti-HA antibody. GST and GST-HSPA1A protein expression was confirmed by immunoblotting with an anti-GST antibody. (F) IRGM co-localizes with PPRV-C, and HSPA1A co-localizes with PPRV-N. EECs were transfected with pCDNA3.1-C-HA or pCDNA3.1-N-HA for 48 h. The transfected cells were fixed and processed for indirect immunofluorescence analysis using antibodies against HA (green), IRGM (red) and HSPA1A (red). The cell nuclei were counterstained with Hoechst 33342 solution. Scale bars, 20 μm.
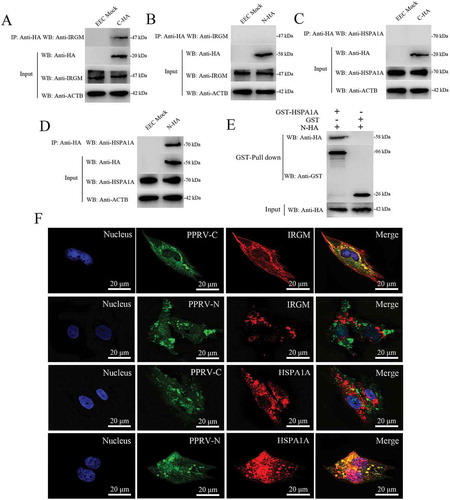
Then, we determined whether the viral proteins that were able to interact with IRGM and HSPA1A could modulate autophagosome formation. To this end, we expressed PPRV-C and PPRV-N in GFP-LC3 EECs and analyzed autophagy by tracking GFP+ autophagosomes. We found that each of these viral proteins induced a significant increase in the number of autophagosomes in the viral protein-transfected cells compared with the pCDNA3.1-HA-GST-transfected cells, which were used as controls (). Interestingly, compared to transfection with only PPRV-C, we found that co-transfection with PPRV-C and PPRV-N significantly increased the number of autophagosomes (). Importantly, impairment of IRGM or HSPA1A expression decreased the number of autophagosomes observed upon overexpression of each of the two viral proteins (). However, we did not detect endogenous IRGM in the immunoprecipitates obtained with the anti-Flag antibody in the mock or PPRV-infected EECs at 12 hpi (Fig. S6). Furthermore, in agreement with the immunofluorescence assay results, our data showed that knockdown of IRGM or HSPA1A expression significantly weakened the observed LC3-II upregulation and SQSTM1 degradation in EECs upon overexpression of both PPRV-C and PPRV-N (). Therefore, our results showed that the PPRV-C and PPRV-N proteins promoted autophagosome accumulation via a molecular process involving IRGM and HSPA1A.
Figure 8. The PPRV-C and PPRV-N proteins modulate autophagy via IRGM and HSPA1A. (A) Overexpression of PPRV-C, PPRV-N or PPRV-C+ PPRV-N modulates autophagosome formation. GFP-LC3 EECs were transfected with a GST-encoding vector (control) or a vector encoding the PPRV-C and/or PPRV-N proteins. Twenty-four h after transfection, the number of autophagic vesicles was determined by confocal immunofluorescence microscopy. Scale bars, 20 μm. (B) The corresponding graph shows the number of GFP+-LC3 vesicles per cell profile among the transfected EECs. (C) PPRV-C and PPRV-N modulate autophagosome formation partly via IRGM-HSPA1A. GFP-LC3 EECs were treated with si-control, si-IRGM or si-HSPA1A 24 h prior to transfection with a vector encoding PPRV-C or PPRV-N. After an additional 24 h, the cells were fixed, and the number of autophagosomes was determined by confocal immunofluorescence microscopy. Scale bars, 20 μm. (D) The corresponding graph shows the number of GFP+-LC3 vesicles per cell profile among the siRNA-pre-treated and transfected EECs. (E and F) EECs were pre-treated and transfected as described in C. Cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in siRNA-pre-treated and transfected EECs were determined by densitometry. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; **P < 0.01; ***P < 0.001.
Syncytia formation facilitates sustained autophagy induced by PPRV in EECs
Fusion inhibitory peptide (FIP) can inhibit syncytia formation without blocking virus particle entry into host cells [Citation33]. To further validate the role of syncytia formation in the PPRV-dependent induction of sustained autophagy, we investigated whether FIP regulated the autophagy induced by PPRV in EECs for 48 h. Following FIP treatment, we found that the number of autophagosomes decreased significantly in PPRV-infected EECs but that no detectable changes occurred in mock-infected or rapamycin-treated EECs (). Furthermore, in agreement with the immunofluorescence assay data, our data showed that inhibition of syncytia formation significantly weakened LC3-II upregulation and SQSTM1 degradation in PPRV-infected EECs (). Importantly, suppression of syncytia formation strongly reduced viral N protein expression and viral titer (). Thus, these data indicated that in the course of PPRV infection, syncytia formation is sufficient to maintain sustained autophagy.
Figure 9. Syncytia formation facilitates sustained autophagy in PPRV-infected EECs. (A) GFP-LC3 EECs treated with DMSO or rapamycin (100 nM) or infected with PPRV (MOI = 3) were further cultured in the absence or presence of 10 μg/mL FIP for 48 h. The number of autophagic vesicles was determined by confocal immunofluorescence microscopy. Scale bars, 20 μm. (B) Corresponding graph showing the number of GFP+-LC3 vesicles per cell profile of FIP-treated EECs. (C) EECs were treated as described in A. Cell samples were analyzed by immunoblotting with anti-LC3, anti-SQSTM1, anti-PPRV-N and anti-ACTB (loading control) antibodies. The target protein levels relative to the ACTB levels in the FIP-treated EECs were determined by densitometry. (D) EECs were treated with FIP and infected with PPRV for 48 h. The viral titers were measured using the TCID50 method. The data represent the mean ± SD of three independent experiments. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; #P > 0.05.
Collectively, our findings revealed the molecular mechanism of sustained autophagy induced by PPRV ().
Figure 10. Proposed model of the two waves of autophagy induced by PPRV via distinct pathways. PPRV induces a first transient wave of autophagy via a mechanism involving its cellular receptor NECTIN4 and an AKT-MTOR-dependent pathway. Soon after infection, new autophagic signalling is initiated that requires viral replication and protein expression. The IRGM-HSPA1A pathway is required for the second wave of autophagy induced by PPRV infection.
Discussion
Upon intracellular pathogen invasion, autophagy can be induced as an innate immune mechanism to control infection. Previous studies have suggested that autophagy not only serves a protective function for cell survival under stress but also plays a role in cellular defence against pathogen infection [Citation48,Citation52,Citation53]. Thus, the survival of viruses is tightly linked to their ability to counteract autophagy-associated antiviral defences [Citation30,Citation54–Citation56]. Accordingly, pathogens have developed strategies to avoid or hijack autophagy for their own benefit. An increasing number of viruses belonging to the family Paramyxoviridae have been reported to utilize an autophagic mechanism to facilitate replication, including MeV [Citation33], PPRV [Citation36,Citation37], human respiratory syncytial virus [Citation57], CDV [Citation35] and Newcastle disease virus [Citation58]. Among these pathogens, attenuated MeV induces two successive waves of autophagy via distinct molecular pathways [Citation33]. Here, we showed that the kinetics of autophagy induction upon PPRV infection similarly indicates two waves of autophagy. The first wave of autophagic flux induction was observed very early after PPRV binding and entry. Interestingly, we found that the return of autophagy to basal levels after the first wave was caused by PPRV, which indicated that PPRV has developed a way to escape autophagy-dependent degradation. Revealing the mechanism of this degradation escape process will require further detailed study. Subsequently, a second wave of autophagic flux was very significantly induced by PPRV at 12 hpi following expression of the viral N protein, which indicated that the second wave of autophagy was required for viral replication. The occurrence of two different waves suggests that two distinct molecular mechanisms are involved in the induction of autophagy during the course of PPRV infection. We mainly focused on elucidating the molecular pathways underlying the induction of these two waves of autophagy in response to PPRV infection.
Immediately upon infection, attenuated MeV induces a first transient wave of autophagy via a pathway involving its cellular receptor CD46 and the scaffold protein GOPC [Citation17]. GOPC was first identified as a Golgi-associated protein and has been reported to be involved in intracellular trafficking and to promote receptor recycling and degradation [Citation59,Citation60]. Notably, CFTR (cystic fibrosis transmembrane conductance regulator) binds to GOPC and causes lysosomal degradation, indicating that cellular receptors might be connected to autophagy via GOPC [Citation60]. Therefore, in this study, we initially investigated GOPC and confirmed that GOPC did not regulate the first wave of autophagy, indicating that PPRV might not induce autophagy through the CD46 (CYT-1)-GOPC pathway.
Many adenocarcinomas express NECTIN4 on their cell surfaces, making them obvious targets for oncolytic and immune therapies based upon recombinant morbilliviruses [Citation61,Citation62]. NECTIN4 plays a key role in viral spread from immune to epithelial cells and is a member of the group of poliovirus receptor-like proteins (PVRLs), which are adhesion receptors of the immunoglobulin superfamily [Citation63,Citation64]. A previous study reported high NECTIN4 expression levels in sheep epithelial tissues [Citation21]. We previously found that EECs also express a NECTIN4 receptor and that the levels and distributions of goat NECTIN4 correlate well with PPRV infection [Citation45,Citation46]. The high NECTIN4 expression in EECs supports a role for this protein in PPRV epithelial pathogenesis. Throughout our study, caprine EECs were used for the experiments, and our results revealed that NECTIN4 played an important role in the first wave of autophagy. Our findings confirmed a possible relationship between NECTIN4 and autophagy upon PPRV infection and suggested that NECTIN4 might be available to initiate autophagy and optimize cellular innate defences against invading pathogens. NECTIN4 has been biochemically shown to support binding to MeV H through its V domain, leading to viral entry [Citation22]. A crystallographic structure of the interaction between the ligand-binding domains of NECTIN4 and MeV H has identified three binding interfaces [Citation65]. Our data also suggest that goat NECTIN4 can interact with PPRV-H to recruit endogenous AKT, which triggers the first wave of autophagic flux. We report here that the pathogen receptor NECTIN4 is physically connected to the autophagic machinery via a unique molecular intermediate.
Despite the key role of autophagy in the control of pathogens, the molecular events leading to the selective induction of this machinery upon viral infection are still largely unknown. The MTOR pathway is an important integrator of nutrient-sensing signals that coordinate cell proliferation in all mammalian cells, and this pathway regulates cell metabolism and innate immune responses by regulating the AKT kinase [Citation66,Citation67]. A rotavirus-encoded virus-like small RNA has been found to trigger autophagy by targeting IGF1R (insulin like growth factor 1 receptor) via the PI3K-AKT-MTOR pathway [Citation68]. Previous findings have also suggested that the PI3K-AKT-MTOR signalling pathway participates in the autophagy induced by coxsackievirus group B type 3 infection [Citation28]. Importantly, we found in this study that MTOR and AKT were inactivated in a manner dependent on the NECTIN4 receptor during the first wave of autophagy in the early stage of PPRV infection. Infectious bursal disease virus invasion has also been found to initiate the HSP90AA1-AKT-MTOR signalling pathway to induce early autophagy [Citation18]. Our research revealed that NECTIN4 regulated a unique AKT-MTOR molecular cascade pathway during the early stage of PPRV infection to activate the autophagy pathway. As shown in , our findings demonstrate, for the first time, a connection between the NECTIN4 receptor and an AKT-MTOR-dependent signalling pathway upon viral invasion.
Notably, we found that the second wave of autophagy induced by PPRV was required for viral replication. PPRV can successfully replicate in caprine EECs [Citation37,Citation46], which explains the clinical occurrence of abortion in PPRV-infected goats [Citation8,Citation9]. We previously suggested that autophagy may enhance PPRV replication in host cells [Citation37]. Here, we have highlighted a previously unidentified role of the C and N proteins in autophagy induction upon PPRV infection; these proteins are ultimately used by the virus to improve its infectivity. Many immunity-related GTPases (IRGs) are stably expressed in mammalian cells, and these proteins play important roles in defences against intracellular pathogens [Citation39]. Importantly, previous studies have suggested that MeV infection-induced autophagy is dependent on IRGM expression [Citation16]. IRGM has also been shown to be targeted by at least two MeV proteins (C and N) [Citation16]. Therefore, we focused on whether the second wave of autophagy induced during PPRV infection was dependent on IRGM. We found that IRGM interacted with PPRV-C but not with PPRV-N. However, both the C and N proteins promoted autophagosome accumulation via a molecular process involving IRGM. These data demonstrate that viral proteins might facilitate the interaction between IRGM and its autophagic partners by promoting IRGM re-localization to or stabilization with autophagy-associated proteins involved in the initiation of autophagosome formation. Thus, IRGM plays an indispensable role in the late wave of autophagy induced by PPRV infection. However, although IRGM contributes to autophagy, the mechanisms underlying this contribution during PPRV infection are unknown. Recently, IRGM was found to regulate ULK1 (unc-51 like autophagy activating kinase 1) and promote the formation of autophagosomal membrane initiation complexes [Citation69,Citation70]. IRGM can interact with several autophagy-associated proteins involved in the initiation of autophagy, including ATG5, ATG10, SH3GLB1 (SH3 domain containing GRB2 like 1, endophilin B1), and BECN1, which may contribute to the induction of autophagy upon viral infection [Citation16,Citation70]. However, further investigation is needed to conclusively explain how the interaction between viral proteins and IRGM leads to the induction of autophagy.
The major inducible protein HSPA1A is strongly induced by viral invasion and by the phylogenetically conserved febrile response, which suggests that HSPA1A induction plays a protective role in the host [Citation71]. Previous studies have demonstrated that the heat shock response, from the top of the regulatory cascade down to the execution of the response by HSPA1A, controls autophagy. This finding connects the two ends of homeostatic systems in eukaryotic cells and indicates that the induction of HSPA1A results in inhibition of autophagy [Citation42,Citation43]. However, panobinostat-induced formation of autophagic vesicles is prevented by hspa1a knockout in mouse embryonic fibroblasts [Citation44]. Moreover, exposure to the chemotherapeutic agent carboplatin increases autophagy, as indicates by increases in LC3 puncta and LC3 and ATG7 protein expression in human breast adenocarcinoma cells, and these effects are prevented by knockdown of HSF1 (heat shock transcription factor 1) [Citation72]. Therefore, HSPA1A and HSF1 have been shown to be required for autophagosome formation. Here, we found that PPRV upregulated HSPA1A expression and that knockdown of HSPA1A expression significantly weakened the induction of autophagy upon PPRV infection. Our study thus demonstrates, for the first time, that the late wave of autophagy is correlated with HSPA1A expression at the beginning of efficient viral replication. For both MeV and CDV, the viral N protein and/or nucleocapsid interact with HSPA1A [Citation73,Citation74]. We have shown here that HSPA1A interacts with PPRV-N but not with PPRV-C. Interestingly, reducing HSPA1A expression increased the number of autophagosomes in mock-infected EECs, indicating that HSPA1A plays a role as a negative regulatory factor of autophagy in normal cells. However, knockdown of HSPA1A expression significantly inhibited virus-induced autophagy. These data indicated that the interaction of HSPA1A with the N protein might disrupt the original function of HSPA1A. Importantly, HSPA1A also plays an essential role in PPRV-C-induced autophagy, indicating that IRGM and HSPA1A have an underlying relationship that regulates the late wave of autophagy. Indeed, autophagy induction involving IRGM and HSPA1A is ultimately exploited by PPRV and favours viral infectious particle production. MeV and PPRV can exploit autophagy to protect infected cells from cell death, which otherwise can limit viral replication and the propagation of numerous infectious viral particles [Citation33,Citation37]. Overriding cell death enables infected cells to survive and support further viral spread. The molecular mechanism by which IRGM and HSPA1A are linked with autophagy to regulate viral replication is largely unknown. In a previous study, HSPA1A was found to serve as a sensor of MeV infection that enhanced the antiviral state through type I interferon (IFN), indicating a role for HSPA1A in eliciting innate immunity, a finding with potentially broad relevance [Citation71]. Furthermore, IRGM targeting to mitochondria could allow viruses to limit type I IFN production in a manner similar to that of two other mitochondrial proteins, MFN2 (mitofusin 2) and TUFM (tu translation elongation factor, mitochondrial), which have been shown to block type I IFN production while inducing autophagy [Citation75–Citation77]. The contributions of different autophagic signals during PPRV infection remain to be investigated.
Previous studies have suggested that MeV infection very strongly induces syncytia formation [Citation33]. The formation of syncytia might contribute to the induction of autophagy and to efficient cell-to-cell spread upon morbillivirus infection [Citation33,Citation56]. Moreover, we previously reported that PPRV infection can also facilitate syncytia formation in host cells [Citation46]. Here, we have shown that reducing syncytia formation significantly weakens the sustained autophagy following PPRV infection. Moreover, viral N protein expression and increases in viral titers also required syncytia formation. These data further confirm that syncytia formation plays an important role in PPRV-induced autophagy and viral replication.
Autophagy is among the first lines of cellular defence against invading viruses [Citation17]. However, viruses may utilize the membranes of autophagosome-like vesicles to promote their replication [Citation33,Citation37,Citation78]. Therefore, molecular analysis of the interplay between autophagy and viruses as well as the consequences of this interplay for viral and cellular biology may be important for the development of methods to control viral infections. Our work describes the induction of successive molecular pathways that contribute to autophagy in response to infection with attenuated PPRV. Although the early wave of autophagy induction is triggered by the NECTIN4-H complex through AKT-MTOR pathway inactivation, the later wave seems to be the result of a direct interaction of viral proteins with IRGM and HSPA1A and is sustained during viral replication. The second wave is exploited by attenuated PPRV to promote the production of infectious viral particles. Our findings will provide many insights to enhance understanding of the interplay between autophagy and PPRV and support the identification of many potential molecular targets of morbilliviruses among autophagy-associated proteins.
Materials and methods
Cell lines and virus
Caprine EECs were immortalized by transfection with human telomerase reverse transcriptase (hTERT), and we have previously confirmed that the secretory function of these cells is consistent with that of primary EECs [Citation79,Citation80]. The cells were cultured in Dulbecco’s modified Eagle medium/F-12 Ham’s medium (DMEM/F12; Hyclone, SH30023) supplemented with 10% fetal bovine serum (Gibco, 10270–106), 100 IU/mL penicillin, and 10 μg/mL streptomycin (Hyclone, SV30010) at 37°C in 5% CO2. The PPRV attenuated strain Nigeria 75/1 was obtained from our laboratory culture collection. The viral stock was prepared by collecting infected cell supernatant when a cytopathic effect (CPE) was apparent in approximately 80% of the cells. To determine the viral titers, cells cultivated in 96-well plates were inoculated with 10-fold serial dilutions of the virus and incubated at 37°C for 5–7 d. The viral titers were estimated with the Reed and Muench method [Citation81] and are expressed as the 50% tissue culture infective dose (TCID50)/mL. The MOI was confirmed according to the viral titers of the respective cell lines. UV inactivation of PPRV was performed by irradiation with UV light for 1 h at room temperature. The infectivity of UV-irradiated PPRV was confirmed by detecting the viral titers as described above, and the MOIs were the same as those for the untreated viruses.
Antibodies and reagents
An anti-PPRV-N monoclonal antibody was provided by the China Animal Health and Epidemiology Centre (Qingdao, China). The following primary antibodies were used: anti-NECTIN4 (Abcam, ab155692), anti-SQSTM1/p62 (Abcam, ab101266), anti-HSPA1A (Abcam, ab79852), anti-ATG5 (Abcam, ab228668), anti-IRGM (Abcam, ab93901), anti-GOPC (Cell Signaling Technology, 8576), anti-p-AKT (Ser473; Cell Signaling Technology, 9271), anti-AKT (Cell Signaling Technology, 4691), anti-p-MTOR (Ser2448; Cell Signaling Technology, 2971), anti-MTOR (Cell Signaling Technology, 2983), anti-HA (Cell Signaling Technology, 3724), anti-HMOX1 (Protein tech, 10701–1-AP), anti-HSP90AA1 (Protein tech, 13171–1-AP), anti-ACTB (TransGen Biotech, HC201), anti-Flag (TransGen Biotech, HT201), anti-HA (TransGen Biotech, HT301), anti-His (TransGen Biotech, HT501), anti-GST (TransGen Biotech, HT601), anti-LC3B (Sigma-Aldrich, L7543). Secondary antibodies: HRP-conjugated mouse anti-rabbit IgG (conformation-specific) (Cell Signaling Technology, 5127), PE-conjugated goat anti-rabbit IgG (TransGen Biotech, HS121), HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich, A9917), HRP-conjugated goat anti-rabbit IgG (Sigma-Aldrich, A0545), fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Sigma-Aldrich, F9887), tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse IgG (Sigma-Aldrich, T7782). Chemicals and reagents: chloroquine (Sigma-Aldrich, C6628), rapamycin (Sigma-Aldrich, R0395), INS (insulin; Sigma-Aldrich, I0305000), FIP (Bachem, H-9430), GST protein (Protein tech, Ag25094). GST-HSPA1A and His-NECTIN4 expression and purification were confirmed by the supplier Zoonbio Biotechnology (Fig. S5A and B).
Plasmid construction and viral protein expression and purification
The Capra hircus LC3B gene (GenBank accession number: XM_018061829.1) was PCR-amplified from total cellular RNA of EECs with gene-specific primers and cloned into the pEGFP-N1 vector (Clontech, 6085–1). The Capra hircus NECTIN4 (GenBank accession number: XM_005677180.3) and HSPA1A (GenBank accession number: JN604433.1) genes were cloned into the pCDNA3.1 (+) vector (Invitrogen, V790-20). The PPRV V, H and N genes were amplified from PPRV genomic cDNA and cloned into the pCDNA3.1 (+) (Invitrogen, V790-20) and pET30a (Novagen, 69909–3) vectors. His-V, His-H, and His-N were expressed in E. coli BL-21 and purified using Ni-NTA resin (TransGen Biotech, DP101). The purified proteins were eluted with elution buffer containing 8 M urea, which was removed by dialysis against a buffer (50 mM NaH2PO4 and 300 mM NaCl) with gradually decreasing urea concentrations (6 M, 3 M, and buffer alone). The concentrations of the purified proteins were measured with a BCA assay. The three purified proteins were detected using immunoblotting with the appropriate antibodies.
Viral infection and cell treatment
According to the requirements of the different experiments, EECs were infected with PPRV at an MOI of 3 or mock-infected with phosphate-buffered saline (PBS; 8.4 mM Na2HPO4, 1.5 mM KH2PO4, 136.9 mM NaCl, 2.7 mM KCl) at 37°C for the indicated durations. UV-irradiated PPRV was added to the EECs as required. For the MTOR activation experiments, the cells were pre-treated with INS (1 μM) for 6 h prior to viral infection. For experiments involving inhibition of the fusion of autophagosomes and lysosomes, the EECs were treated with chloroquine (100 μM) and infected with PPRV for 1.5, 3 or 6 h. For the NECTIN4 receptor stimulation experiment, the EECs were precoated with 20 μg/mL anti-NECTIN4 polyclonal antibody or irrelevant isotype control IgG for 4 h at 37°C. For the viral protein stimulation experiment, the EECs were precoated with His-GST, His-V, His-H, or His-N for 1.5 h at 37°C. For the syncytia formation experiment, EECs were treated with FIP (10 μg/mL) and infected with PPRV for 48 h. The cells were harvested and lysed with lysis buffer, and immunoblotting was performed using the appropriate antibodies to monitor autophagy.
Transfection and gene silencing with siRNAs
SMARTpool siRNAs targeting NECTIN4, ATG5, GOPC, IRGM, and HSPA1A were designed using online design tools (Block-iT RNAi Designer; Invitrogen). We selected the three short target sequences with the best scores for each gene. The siRNA for each gene was synthesized by Ribo Biotechnology. We selected the most efficient siRNA for each gene for the experiment (Table S1). EECs grown to 80% confluence in 6-well cell culture plates were transfected with 6 μg/well plasmids or 300 nM siRNA using TurboFect Transfection Reagent (Thermo Fisher Scientific, R0531) as previously described [Citation37,Citation45]. Then, the cells were incubated in Opti-MEM (Gibco, 31985–088) at 37°C for 48 h. The reaction mixture was discarded, and the cells were infected with PPRV at an MOI of 3. Following 1 h of PPRV absorption, the cells were incubated in fresh medium until harvested or until the culture medium was collected at the indicated time point after infection. A control siRNA (UUCUCCGAACGUGUCACGU) was maintained in our laboratory and used as a negative control. The silencing efficiency was measured by immunoblotting.
Immunoprecipitation and GST affinity-isolation assays
EECs were transfected for 48 h and incubated on ice with immunoprecipitation lysis buffer (Beyotime, P0013). For each sample, 500 μL of lysate was incubated with 2 μg of antibody and 800 μL of protein A/G plus agarose (Santa Cruz Biotechnology, sc-2003) overnight. The agarose beads were washed 4 times with 1 mL of lysis buffer containing 1% NP-40 (Beyotime, ST366). The precipitates were detected by SDS-PAGE and immunoblotting. For the GST affinity-isolation assays, GST or GST-HSPA1A protein was bound to glutathione agarose (Thermo Scientific, 21516) according to the manufacturer’s instructions, and the beads were washed four times. The beads were incubated with the target proteins harvested from the transfected EECs in affinity-isolation lysis buffer for 2 h at 4°C. The eluted proteins were detected by SDS-PAGE and immunoblotting.
Immunoblotting analysis
Protein samples were prepared from the harvested cells after increasing time intervals following infection and subjected to immunoblotting using primary antibodies that recognized different forms of the cellular proteins. At the indicated time points, cell lysates were generated by adding 5× loading buffer to the collected cells. The samples were boiled for 10 min, separated by SDS-PAGE, and then transferred onto 0.22-µm polyvinylidene difluoride membranes (Millipore, ISEQ00010). The membranes were blocked with 5% non-fat milk and incubated with primary antibodies followed by HRP-conjugated secondary antibodies. The bound antibodies were detected with ECL immunoblotting detection reagents (Millipore, WBKLS0500). Images were obtained with a CanoScan LiDE 100 scanner (Canon), and the intensity of the target protein blots was analyzed with ImageJ software (NIH). All target proteins and internal loading controls were detected and verified within the same linear range.
Transmission electron microscopy (TEM)
Ultra-thin sections (70 nm) of cells were prepared and examined under a Hitachi HT-7700 transmission electron microscope (Hitachi, Tokyo, Japan) as described previously [Citation37]. Briefly, EECs were mock-infected or infected with UV-irradiated PPRV or PPRV at an MOI of 3 for 1.5 h or 12 h. Then, the cells were washed three times with PBS and collected by centrifugation at 800 × g for 5 min. One drop of 2% preheated agarose was added to the cell pellet and uniformly mixed. After solidification, the agar was cut into 1 mm3 blocks and placed into a 2.5% glutaraldehyde/0.1 M phosphate buffer solution (Na2HPO4 · 12H2O, NaH2PO4 · 2 H2O, pH 7.4) at 4°C for fixation; the sections were then washed 3 times with phosphate buffer solution for 10 min and post-fixed in 1% osmium tetroxide at 4°C for 1 h. Following dehydration with a graded series of ethanol solutions, the cells were embedded in a mixture of Epoxy 812 and warmed at 35°C, 45°C, and 60°C for 12 h, 12 h, and 48 h, respectively. Ultra-thin sections were prepared, stained with 4% uranyl acetate and observed under a transmission electron microscope.
Confocal immunofluorescence microscopy
Following the indicated treatments, GFP-LC3 EECs [Citation79,Citation80], pCDNA3.1-N EECs and pCDNA3.1-C EECs were washed 4 times with PBS and fixed in 4% paraformaldehyde. The cells were washed again 4 times with PBS and treated with 0.1% Triton X-100 (Sangon Biotech, A110694) for 15 min. Then, the cells were incubated with 1% bovine serum albumin (BSA; Sigma-Aldrich, A7906) and the appropriate primary antibodies for 1 h at 37°C before being washed and incubated simultaneously with FITC- or TRITC-conjugated secondary antibodies. Finally, the cells were treated with a Hoechst 33342 (Sigma-Aldrich, B2261) solution for 5 min and analyzed under a confocal microscope (CLSM; Leica SP8, Germany).
Statistical analysis
The data are expressed as the mean ± standard deviation (SD). The significance of the variability between the different treatment groups was calculated with two-way ANOVA followed by Tukey’s multiple comparisons test using GraphPad Prism 6.0 software (Graph Pad Software Inc., San Diego, CA, USA). A P value < 0.05 was considered to indicate statistical significance.
Supplemental Material
Download Zip (896.3 KB)Acknowledgments
We are grateful to Yaping Jin (Northwest A&F University Yangling, Shaanxi, China) for the caprine endometrial epithelial cells. We thank the China Animal Health and Epidemiology Centre (Qingdao, China) for the antibodies used in these studies. We thank the Life Science Research Core Services of Northwest A&F University for providing the confocal microscope. We also thank Yanqing Wang for guidance in the use of the confocal microscope.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Abubakar M, Mahapatra M, Muniraju M, et al. Serological detection of antibodies to peste des petits ruminants virus in large ruminants. Transbound Emerg Dis. 2017 Apr;64(2):513–519. PubMed PMID: 26200233; PubMed Central PMCID: PMCPMC5347956.
- Khalafalla AI, Saeed IK, Ali YH, et al. An outbreak of peste des petits ruminants (PPR) in camels in the Sudan. Acta Trop. 2010 Nov;116(2):161–165. PubMed PMID: 20707980.
- Zakian A, Nouri M, Kahroba H, et al. The first report of peste des petits ruminants (PPR) in camels (Camelus dromedarius) in Iran. Trop Anim Health Prod. 2016 Aug;48(6):1215–1219. PubMed PMID: 27155951.
- Liu F, Li J, Li L, et al. Peste des petits ruminants in China since its first outbreak in 2007: A 10-year review. Transbound Emerg Dis. 2018 Jun;65(3):638–648. PubMed PMID: 29322642.
- Kumar N, Maherchandani S, Kashyap SK, et al. Peste des petits ruminants virus infection of small ruminants: a comprehensive review. Viruses. 2014 Jun 6;6(6):2287–2327. PubMed PMID: 24915458; PubMed Central PMCID: PMCPMC4074929.
- Jagtap SP, Rajak KK, Garg UK, et al. Effect of immunosuppression on pathogenesis of peste des petits ruminants (PPR) virus infection in goats. Microb Pathog. 2012 Apr;52(4):217–226. PubMed PMID: 22248720.
- Truong T, Boshra H, Embury-Hyatt C, et al. Peste des petits ruminants virus tissue tropism and pathogenesis in sheep and goats following experimental infection. PloS One. 2014;9(1):e87145. PubMed PMID: 24498032; PubMed Central PMCID: PMCPMC3907444.
- Abubakar M, Ali Q, Khan HA. Prevalence and mortality rate of peste des petitis ruminant (PPR): possible association with abortion in goat. Trop Anim Health Prod. 2008 Jun;40(5):317–321. PubMed PMID: 18509938.
- Borel N, Sachse K, Rassbach A, et al. Ovine enzootic abortion (OEA): antibody response in vaccinated sheep compared to naturally infected sheep. Vet Res Commun. 2005 Mar;29(Suppl 1):151–156. PubMed PMID: 15943073.
- Taylor W. The global eradication of peste des petits ruminants (PPR) within 15 years–is this a pipe dream? Trop Anim Health Prod. 2016 Mar;48(3):559–567. PubMed PMID: 26851956.
- Maan S, Kumar A, Gupta AK, et al. Concurrent infection of Bluetongue and Peste-des-petits-ruminants virus in small ruminants in Haryana State of India. Transbound Emerg Dis. 2018 Feb;65(1):235–239. PubMed PMID: 28116836.
- Sen A, Saravanan P, Balamurugan V, et al. Vaccines against peste des petits ruminants virus. Expert Rev Vaccines. 2010 Jul;9(7):785–796. PubMed PMID: 20624051.
- Saravanan P, Sen A, Balamurugan V, et al. Comparative efficacy of peste des petits ruminants (PPR) vaccines. Biologicals. 2010 Jul;38(4):479–485. PubMed PMID: 20199873.
- Caufour P, Rufael T, Lamien CE, et al. Protective efficacy of a single immunization with capripoxvirus-vectored recombinant peste des petits ruminants vaccines in presence of pre-existing immunity. Vaccine. 2014 Jun 24;32(30):3772–3779. PubMed PMID: 24837763.
- Guirimand T, Delmotte S, Navratil V. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res. 2015 Jan;43(Database issue):D583–D587. PubMed PMID: 25392406; PubMed Central PMCID: PMCPMC4383936.
- Gregoire IP, Richetta C, Meyniel-Schicklin L, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011 Dec;7(12):e1002422. PubMed PMID: 22174682; PubMed Central PMCID: PMCPMC3234227.
- Joubert PE, Meiffren G, Gregoire IP, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009 Oct 22;6(4):354–366. PubMed PMID: 19837375.
- Hu B, Zhang Y, Jia L, et al. Binding of the pathogen receptor HSP90AA1 to avibirnavirus VP2 induces autophagy by inactivating the AKT-MTOR pathway. Autophagy. 2015;11(3):503–515. PubMed PMID: 25714412; PubMed Central PMCID: PMCPMC4502722.
- Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009 Nov;19(6):359–378. PubMed PMID: 19750559; PubMed Central PMCID: PMCPMC2852112.
- Hammouchi M, Loutfi C, Sebbar G, et al. Experimental infection of alpine goats with a Moroccan strain of peste des petits ruminants virus (PPRV). Vet Microbiol. 2012 Nov 9;160(1–2):240–244. PubMed PMID: 22633480.
- Birch J, Juleff N, Heaton MP, et al. Characterization of ovine Nectin-4, a novel peste des petits ruminants virus receptor. J Virol. 2013 Apr;87(8):4756–4761. PubMed PMID: 23388720; PubMed Central PMCID: PMCPMC3624396.
- Muhlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011 Nov 2;480(7378):530–533. PubMed PMID: 22048310; PubMed Central PMCID: PMCPMC3245798.
- Noyce RS, Richardson CD. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 2012 Sep;20(9):429–439. PubMed PMID: 22721863.
- Pratakpiriya W, Ping Teh AP, Radtanakatikanon A, et al. Expression of canine distemper virus receptor nectin-4 in the central nervous system of dogs. Sci Rep. 2017 Mar 23;7(1):349. PubMed PMID: 28336928; PubMed Central PMCID: PMCPMC5428276.
- Melia MM, Earle JP, Abdullah H, et al. Use of SLAM and PVRL4 and identification of pro-HB-EGF as cell entry receptors for wild type phocine distemper virus. PloS One. 2014;9(8):e106281. PubMed PMID: 25171206; PubMed Central PMCID: PMCPMC4149546.
- Viret C, Rozieres A, Faure M. Autophagy during early virus-host cell interactions. J Mol Biol. 2018 Jun 8;430(12):1696–1713. PubMed PMID: 29698649.
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. PubMed PMID: 19653858; PubMed Central PMCID: PMCPMC2831538.
- Chang H, Li X, Cai Q, et al. The PI3K/Akt/mTOR pathway is involved in CVB3-induced autophagy of HeLa cells. Int J Mol Med. 2017 Jul;40(1):182–192. PubMed PMID: 28560385; PubMed Central PMCID: PMCPMC5466389.
- Sun P, Zhang S, Qin X, et al. Foot-and-mouth disease virus capsid protein VP2 activates the cellular EIF2S1-ATF4 pathway and induces autophagy via HSPB1. Autophagy. 2018;14(2):336–346. PubMed PMID: 29166823; PubMed Central PMCID: PMCPMC5902195.
- Fan X, Han S, Yan D, et al. Foot-and-mouth disease virus infection suppresses autophagy and NF-small ka, CyrillicB antiviral responses via degradation of ATG5-ATG12 by 3C(pro). Cell Death Dis. 2017 Jan 19;8(1):e2561. PubMed PMID: 28102839; PubMed Central PMCID: PMCPMC5386389.
- Ma J, Sun Q, Mi R, et al. Avian influenza A virus H5N1 causes autophagy-mediated cell death through suppression of mTOR signaling. J Genet Genomics. 2011 Nov 20;38(11):533–537. PubMed PMID: 22133684.
- Huang WR, Chiu HC, Liao TL, et al. Avian reovirus protein p17 functions as a nucleoporin Tpr suppressor leading to activation of p53, p21 and PTEN and Inactivation of PI3K/AKT/mTOR and ERK signaling pathways. PloS One. 2015;10(8):e0133699. PubMed PMID: 26244501; PubMed Central PMCID: PMCPMC4526660.
- Richetta C, Gregoire IP, Verlhac P, et al. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 2013;9(9):e1003599. PubMed PMID: 24086130; PubMed Central PMCID: PMCPMC3784470.
- Xia M, Gonzalez P, Li C, et al. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J Virol. 2014 May;88(9):5152–5164. PubMed PMID: 24574393; PubMed Central PMCID: PMCPMC3993837.
- Kabak YB, Sozmen M, Yarim M, et al. Immunohistochemical detection of autophagy-related microtubule-associated protein 1 light chain 3 (LC3) in the cerebellums of dogs naturally infected with canine distemper virus. Biotech Histochem. 2015;90(8):601–607. PubMed PMID: 26179070.
- Zhang Y, Wu S, Lv J, et al. Peste des petits ruminants virus exploits cellular autophagy machinery for replication. Virology. 2013 Mar 1;437(1):28–38. PubMed PMID: 23318276.
- Yang B, Xue Q, Qi X, et al. Autophagy enhances the replication of Peste des petits ruminants virus and inhibits caspase-dependent apoptosis in vitro. Virulence. 2018;9(1):1176–1194. PubMed PMID: 30067475; PubMed Central PMCID: PMCPMC6086290.
- Petkova DS, Verlhac P, Rozieres A, et al. Distinct contributions of autophagy receptors in measles virus replication. Viruses. 2017 May 22;9(5). PubMed PMID: 28531150; PubMed Central PMCID: PMCPMC5454435. DOI:10.3390/v9050123
- Gregoire IP, Rabourdin-Combe C, Faure M. Autophagy and RNA virus interactomes reveal IRGM as a common target. Autophagy. 2012 Jul 1;8(7):1136–1137. PubMed PMID: 22722598; PubMed Central PMCID: PMCPMC3429549.
- Oglesbee MJ, Pratt M, Carsillo T. Role for heat shock proteins in the immune response to measles virus infection. Viral Immunol. 2002;15(3):399–416. PubMed PMID: 12479391.
- Oglesbee M, Krakowka S. Cellular stress response induces selective intranuclear trafficking and accumulation of morbillivirus major core protein. Lab Invest. 1993 Jan;68(1):109–117. PubMed PMID: 8423670.
- Dokladny K, Zuhl MN, Mandell M, et al. Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J Biol Chem. 2013 May 24;288(21):14959–14972. PubMed PMID: 23576438; PubMed Central PMCID: PMCPMC3663517.
- Dokladny K, Myers OB, Moseley PL. Heat shock response and autophagy–cooperation and control. Autophagy. 2015;11(2):200–213. PubMed PMID: 25714619; PubMed Central PMCID: PMCPMC4502786.
- Yang Y, Fiskus W, Yong B, et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):6841–6846. PubMed PMID: 23569248; PubMed Central PMCID: PMCPMC3637746.
- Yang B, Qi X, Guo H, et al. Peste des petits ruminants virus enters caprine endometrial epithelial cells via the caveolae-mediated endocytosis pathway. Front Microbiol. 2018;9:210. PubMed PMID: 29497407; PubMed Central PMCID: PMCPMC5818419.
- Yang B, Qi X, Chen Z, et al. Binding and entry of peste des petits ruminants virus into caprine endometrial epithelial cells profoundly affect early cellular gene expression. Vet Res. 2018 Jan 24;49(1):8. PubMed PMID: 29368634; PubMed Central PMCID: PMCPMC5784595.
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. PubMed PMID: 26799652; PubMed Central PMCID: PMCPMC4835977.
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013 Oct;13(10):722–737. PubMed PMID: 24064518; PubMed Central PMCID: PMCPMC5340150.
- Sarbassov DD, Guertin DA, Ali SM, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005 Feb 18;307(5712):1098–1101. PubMed PMID: 15718470.
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009 Oct 15;122(Pt 20):3589–3594. PubMed PMID: 19812304; PubMed Central PMCID: PMCPMC2758797.
- Petkova DS, Viret C, Faure M. IRGM in autophagy and viral infections. Front Immunol. 2012;3:426. PubMed PMID: 23335927; PubMed Central PMCID: PMCPMC3547297.
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004 Nov 5;306(5698):990–995. PubMed PMID: 15528435; PubMed Central PMCID: PMCPMC1705980.
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008 Jan 11;132(1):27–42. PubMed PMID: 18191218; PubMed Central PMCID: PMCPMC2696814.
- Talloczy Z, Virgin H, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006 Jan–Mar;2(1):24–29. PubMed PMID: 16874088.
- Chaumorcel M, Souquere S, Pierron G, et al. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008 Jan;4(1):46–53. PubMed PMID: 18340111.
- Delpeut S, Rudd PA, Labonte P, et al. Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J Virol. 2012 Aug;86(16):8527–8535. PubMed PMID: 22647692; PubMed Central PMCID: PMCPMC3421762.
- Pokharel SM, Shil NK, Bose S. Autophagy, TGF-beta, and SMAD-2/3 signaling regulates interferon-beta response in respiratory syncytial virus infected macrophages. Front Cell Infect Microbiol. 2016;6:174. PubMed PMID: 28018859; PubMed Central PMCID: PMCPMC5149518.
- Kang Y, Yuan R, Xiang B, et al. Newcastle disease virus-induced autophagy mediates antiapoptotic signaling responses in vitro and in vivo. Oncotarget. 2017 Sep 26;8(43):73981–73993. PubMed PMID: 29088762; PubMed Central PMCID: PMCPMC5650317.
- Charest A, Lane K, McMahon K, et al. Association of a novel PDZ domain-containing peripheral Golgi protein with the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor) protein syntaxin 6. J Biol Chem. 2001 Aug 3;276(31):29456–29465. PubMed PMID: 11384996.
- Cheng J, Wang H, Guggino WB. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J Biol Chem. 2004 Jan 16;279(3):1892–1898. PubMed PMID: 14570915.
- Delpeut S, Noyce RS, Richardson CD. The tumor-associated marker, PVRL4 (nectin-4), is the epithelial receptor for morbilliviruses. Viruses. 2014 Jun 2;6(6):2268–2286. PubMed PMID: 24892636; PubMed Central PMCID: PMCPMC4074928.
- Noyce RS, Bondre DG, Ha MN, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011 Aug;7(8):e1002240. PubMed PMID: 21901103; PubMed Central PMCID: PMCPMC3161989.
- Reymond N, Fabre S, Lecocq E, et al. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J Biol Chem. 2001 Nov 16;276(46):43205–43215. PubMed PMID: 11544254.
- Frenzke M, Sawatsky B, Wong XX, et al. Nectin-4-dependent measles virus spread to the cynomolgus monkey tracheal epithelium: role of infected immune cells infiltrating the lamina propria. J Virol. 2013 Mar;87(5):2526–2534. PubMed PMID: 23255790; PubMed Central PMCID: PMCPMC3571369.
- Zhang X, Lu G, Qi J, et al. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol. 2013 Jan;20(1):67–72. PubMed PMID: 23202587.
- Cobbold SP. The mTOR pathway and integrating immune regulation. Immunology. 2013 Dec;140(4):391–398. PubMed PMID: 23952610; PubMed Central PMCID: PMCPMC3839643.
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011 Jan;12(1):21–35. PubMed PMID: 21157483; PubMed Central PMCID: PMCPMC3390257.
- Zhou Y, Geng P, Liu Y, et al. Rotavirus-encoded virus-like small RNA triggers autophagy by targeting IGF1R via the PI3K/Akt/mTOR pathway. Biochim Biophys Acta Mol Basis Dis. 2018 Jan;1864(1):60–68. PubMed PMID: 29017894.
- Hansen MD, Johnsen IB, Stiberg KA, et al. Hepatitis C virus triggers Golgi fragmentation and autophagy through the immunity-related GTPase M. Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):E3462–E3471. PubMed PMID: 28389568; PubMed Central PMCID: PMCPMC5410803.
- Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015 May 7;58(3):507–521. PubMed PMID: 25891078; PubMed Central PMCID: PMCPMC4427528.
- Kim MY, Shu Y, Carsillo T, et al. hsp70 and a novel axis of type I interferon-dependent antiviral immunity in the measles virus-infected brain. J Virol. 2013 Jan;87(2):998–1009. PubMed PMID: 23135720; PubMed Central PMCID: PMCPMC3554074.
- Desai S, Liu Z, Yao J, et al. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J Biol Chem. 2013 Mar 29;288(13):9165–9176. PubMed PMID: 23386620; PubMed Central PMCID: PMCPMC3610989.
- Gruet A, Dosnon M, Blocquel D, et al. Fuzzy regions in an intrinsically disordered protein impair protein-protein interactions. Febs J. 2016 Feb;283(4):576–594. PubMed PMID: 26684000.
- Oglesbee M, Ringler S, Krakowka S. Interaction of canine distemper virus nucleocapsid variants with 70K heat-shock proteins. J Gen Virol. 1990 Jul;71(Pt 7):1585–1590. PubMed PMID: 2374009.
- Lei Y, Wen H, Yu Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012 Jun 29;36(6):933–946. PubMed PMID: 22749352; PubMed Central PMCID: PMCPMC3397828.
- Yasukawa K, Oshiumi H, Takeda M, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009 Aug 18;2(84):ra47. PubMed PMID: 19690333.
- Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010 May 14;141(4):656–667. PubMed PMID: 20478256; PubMed Central PMCID: PMCPMC3059894.
- Pei J, Zhao M, Ye Z, et al. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy. 2014 Jan;10(1):93–110. PubMed PMID: 24262968; PubMed Central PMCID: PMCPMC4389882.
- Qi X, Qu Y, Nan Z, et al. Caprine endometrial stromal cells modulate the effects of steroid hormones on cytokine secretion by endometrial epithelial cells in vitro. Reprod Biol. 2012 Nov;12(3):309–315. PubMed PMID: 23153702.
- Qi XF, Nan ZC, Jin YP, et al. Stromal-epithelial interactions modulate the effect of ovarian steroids on goat uterine epithelial cell interleukin-18 release. Domest Anim Endocrinol. 2012 May;42(4):210–219. PubMed PMID: 22226936.
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27(3):493–497.
