ABSTRACT
Lipidation of Atg8-family ubiquitin-like proteins (UBLs) plays important roles in macroautophagy/autophagy. This process is catalyzed by an E1-E2-E3 trienzyme cascade, in which an E1 enzyme, Atg7, directs Atg8 to its E2 enzyme, Atg3, forming a thioester bond-linked Atg3~ Atg8 intermediate; then the composite E3, Atg12–Atg5-Atg16, interacts with the Atg3~ Atg8 intermediate and promotes Atg8 transfer from the catalytic cysteine of Atg3 to the head group of phosphatidylethanolamine (PE) lipids. Despite progress that has been made toward understanding the Atg8 lipidation pathway, the molecular mechanism of Atg3 as it orchestrates between the E1 and E3 remains unclear. Here we summarize our recent work reporting an element in Atg3, termed the E1, E2, and E3-interacting region (E123IR), is an allosteric switch: in the absence of other binding partners, the E123IR restrains Atg3′s catalytic loop, while the E1 or E3 enzyme directly binds this region to remove this brace and thereby conformationally activate Atg3 to elicit Atg8 lipidation in vitro and in vivo.
KEYWORDS:
A critical step in most forms of autophagy is conjugation of Atg8-family ubiquitin-like proteins to the lipid phosphatidylethanolamine (PE), which is catalyzed by an autophagy-specific E1–E2–E3 enzymatic cascade (). Besides a core catalytic domain similar to canonical E2s, Atg3 contains unique structures: an N-terminal membrane-binding helix (N); an intrinsically disordered flexible region (FR); a handle region (HR); and a flexible C-terminal tail (C) (). Although it had been reported that in some species the Atg3 FR binds to both E1 and E3, and that for yeast proteins the E3 triggers structural rearrangement of Atg3’s catalytic cysteine, detailed knowledge of if and how these functions are coupled had remained elusive. In our recent study, we discovered an element in Atg3 serving as an allosteric switch that regulates Atg8 lipidation [Citation1].
Figure 1. Allosteric regulation through a switch element in the autophagy E2 Atg3. (A) Atg8 is activated by the E1, Atg7, and then transferred to the E2 enzyme, Atg3. Atg12–Atg5-Atg16 acts as an E3 catalyzing PE linkage to Atg8. (B) Diagram representing Atg3 sequence and highlighting unique elements: N, N-terminal region; Cat, catalytic domain; FR, flexible region; E123IR, E1-, E2-, and E3-interacting region; HR, handle region; and C, extreme C-terminal region. (C) Schematic model for allosteric regulation of Atg3 activity by E123IR. Atg3 is initially autoinhibited by the Atg3 E123IR. E123IR is relocated upon binding to the Atg7 NTD, thereby triggering rearrangement of the Atg3 catalytic core to activate Cys234 for attacking the Atg7~ Atg8 intermediate. Atg8 is transferred from Atg7 to Atg3, producing the Atg3~ Atg8 intermediate. Relieved from Atg7, the Atg3 E123IR protects the Atg3~ Atg8 intermediate from wayward discharge to errant nucleophiles. Atg12–Atg5-Atg16 binds the E123IR and activates the Atg3~ Atg8 intermediate for nucleophilic attack from PE.
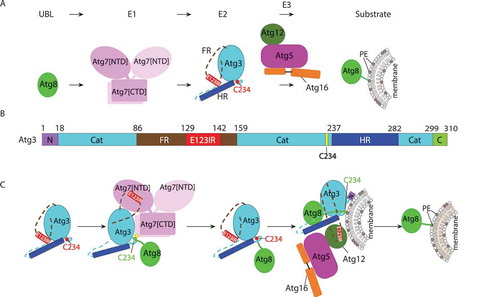
First, we identified the Atg12 binding region in yeast Atg3 FR. Interestingly, this region had previously been shown to bind Atg7’s N-terminal domain (NTD). Furthermore, this E1 and E3 binding region had been shown to contact the Atg3 catalytic (Atg3[cat]) domain in the prior crystal structure of the full-length Atg3 alone. However, the functional importance of these intramolecular interactions was unknown. Our NMR and mutagenesis results show this element as a bona fide Atg3[cat]-binding region in solution. Thus, we termed this Atg3[E123IR], for E1-, E2-, and E3-interacting region.
To understand effects of the Atg3[E123IR] element binding to Atg3[cat], we determined a crystal structure of Atg3 lacking the entire FR and N-terminal residues (Atg3[∆NFR]). The catalytic Cys loop and adjacent regions of Atg3[∆NFR] show substantial differences from the prior structure of full-length Atg3. These results suggest that by removing the Atg3[E123IR] element, the catalytic Cys is rearranged to approach key catalytic residues in Atg3[cat]. This is in-line with prior studies of cysteine accessibility in yeast Atg3, and with prior structures of full-length Atg3 bound to Atg7. Further NMR experiments showed that the Atg3[E123IR] element also binds to a stable proxy for the Atg3[∆FR]~Atg8 intermediate, indicating a potential for the Atg3[E123IR] to play an inhibitory role in the context of the Atg3~ Atg8 thioester-bonded intermediate. Notably, such interactions might have different effects in this context wherein Atg3’s catalytic Cys would necessarily be engaged in distinctive contacts with Atg8’s C terminus. Strong support for the Atg3[E123IR]-Atg3[cat] interaction came from mutants designed to disrupt this, which show increased E3-independent activity of the Atg3~ Atg8 intermediate and Atg8 lipidation compared to the wild-type counterpart.
A combination of mutagenesis and NMR studies, analyzed in the context of the existing structures presenting canonical E2~UBL intermediates, support a model whereby the active Atg3~Atg8 intermediate forms a “closed” conformation. A major fraction of the surface residues in prior structures represent closed conformations of canonical E2~UBL complexes, where the E2 and its covalently-linked UBL interact through extensive noncovalent interactions. The closed conformation is thought to indirectly contribute to immobilization of the thioester bond, thereby increasing susceptibility to nucleophilic attack.
Besides residues at the predicted Atg8-Atg3[cat] interface, 3 regions show importance for activity: the AIM/LIR binding site in Atg8, Atg3 residues corresponding to E3-binding regions of canonical E2s, and the extreme C terminus of Atg3. These regions are not conserved in canonical E2s, and the latter region is not fully visible in Atg3 crystal structures. Nonetheless, these autophagy-specific regions are consistent with roles in allosteric regulation, and may suggest E3 or substrate binding sites or potential other mechanisms.
In summary, our recent publication suggests that the Atg3 catalytic site is braced by an element within its FR; this brace underlies regulation by interacting with E1 and E3; and that a key function of Atg12–Atg5-Atg16 is to bind this brace to activate the intrinsic reactivity of the Atg3~ Atg8 intermediate (). For many RING E3s in the ubiquitin pathway, they interact with both the E2 and ubiquitin to stabilize the closed conformation, while another part of the E3 recruits substrate to facilitate site-specific ubiquitination. For the autophagy E3, does it bind Atg3 and Atg8 simultaneously or are there other means of stabilizing the closed conformation? Alanine scanning mutagenesis of Atg8 revealed a crucial surface around the AIM/LIR-binding region, raising the possibility that an AIM/LIR-like sequence in Atg3 binds to Atg8 in the closed form. One potential candidate is the “MEGW” motif at the extreme C terminus of Atg3, as it is spatially close to the binding site, and mutations at this motif show a significant defect in Atg3 activity. Such interactions would conceptually parallel those observed in a recent crystal structure of an activated ubiquitin intermediate, in which residues C-terminal of the catalytic domain of the canonical E2 enzyme Cdc34 bind ubiquitin to anchor the closed conformation. Notably, another crucial surface discovered by alanine scanning mutagenesis of Atg3 corresponds to E3-binding regions of canonical E2s. Indeed, it seems that the E3 plays other roles than just releasing constraint from the E123IR, given that removal of the E123IR brace by Atg7 is insufficient to activate the Atg3~ Atg8 intermediate, and that the Atg3 mutations designed to displace the E123IR are much less efficient than Atg12–Atg5-Atg16. Thus, we anticipate additional roles of the E3 activating Atg3~ Atg8 to be visualized in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Zheng Y, Qiu Y, Grace CRR, et al. A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade. Nat Commun. 2019;10:3600.
