ABSTRACT
How energy deprivation induces macroautophagy/autophagy is not fully understood. Here, we show that Atg11, a receptor protein for cargo recognition in selective autophagy, is required for the initiation of glucose starvation-induced autophagy. Upon glucose starvation, Atg11 facilitates the interaction between Snf1 and Atg1, thus is required for Snf1-dependent Atg1 activation. Phagophore assembly site (PAS) formation requires Atg11 via its control of the association of Atg17 with Atg29-Atg31. The binding of Atg11 with Atg9 is crucial for recruiting Atg9 vesicles to the PAS and, thus, glucose starvation-induced autophagy. We propose Atg11 as a key initiation factor controlling multiple key steps in energy deprivation-induced autophagy.
Abbreviations: AMPK: AMP-activated protein kinase; Ams1: α-mannosidase; Ape1: aminopeptidase I; Cvt: cytoplasm-to-vacuole targeting; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; GFP: green fluorescent protein; MBP: myelin basic protein; MMS: methanesulfonate; PAS: phagophore assembly site; PNBM: p-nitrobenzyl mesylate; SD-G: glucose starvation medium; SD-N: nitrogen starvation medium; ULK1, unc-51 like autophagy activating kinase 1; WT: wild type
Introduction
Macroautophagy/autophagy is a highly conserved protein degradation pathway occurring in taxa from yeast to humans, which delivers required degradable substances into the vacuole/lysosome through the autophagosome [Citation1]. It is known to be critical in maintaining cell homeostasis [Citation2,Citation3]. Autophagy is a multi-step process, including initiation, membrane growth and extension, autophagosome fusion with vacuole/lysosome, and vacuole/lysosome degradation. Each step has specific proteins governing different stages [Citation4]. Recently, studies in yeast and human genetics have greatly aided our understanding of the molecular mechanisms and physiological functions of autophagy [Citation5–7]. More than 40 autophagy-related (ATG) genes have now been identified to be involved in various types of autophagy [Citation8].
While great progress has been made in understanding nutrient starvation-induced autophagy, little is known about the molecular mechanism and functions of energy deprivation-induced autophagy. In yeast cells, our previous study revealed mitochondria as a hub essential for glucose starvation-induced autophagy [Citation9]. Activation of the AMP-activated protein kinase (AMPK) homolog Snf1 occurs upon glucose starvation. Activated Snf1 then maintains mitochondrial respiration by phosphorylating Mec1, thus promoting the binding of Atg1 with Atg13 to initiate autophagy. In mammals, most studies on glucose starvation-induced autophagy have their focus on identifying the substrate proteins of AMPK. AMPK, as an intracellular energy sensor, is crucial for glucose starvation-induced autophagy by phosphorylating ULK1 (unc-51 like autophagy activating kinase 1), the PIK3C3/VPS34-containing complex and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) [Citation10–13].
Atg1 encodes a Ser/Thr kinase essential for autophagy, thus, linking its activity to autophagy initiation [Citation14]. Atg13, as a key autophagy-regulating protein, is highly phosphorylated by the target of rapamycin complex 1 (TORC1) kinase under normal growth conditions. Treatment of cells with rapamycin or nitrogen starvation downregulates the activity of TORC1, resulting in rapid dephosphorylation of Atg13 to enhance the association of Atg13 with Atg1 [Citation15]. Atg17, Atg29, and Atg31 proteins constitutively form a stable complex independent of nutrient conditions [Citation16]. Upon nitrogen starvation, Atg1 and Atg13 form a polymer complex with Atg17-Atg31-Atg29, which activates Atg1 to initiate autophagy. Genetic analysis has shown that the Atg1-Atg13-Atg17-Atg31-Atg29 complex serves as a platform for the recruitment of other Atg proteins [Citation17]. However, how Atg1 activates under glucose starvation is still not fully understood.
Atg11 is a receptor protein that directly binds to autophagy receptors and to Atg1 to deliver specific cargoes into the autophagosome. Although Atg11 is essential for most types of selective autophagy, the activity of Atg1 or autophagy under nitrogen starvation conditions does not require Atg11 [Citation18,Citation19]. Atg11 has four domains, namely, CC1, CC2, CC3, and CC4. Under normal conditions, the association of Atg11 with the receptor protein Atg19 and the cytoplasm-to-vacuole targeting (Cvt) pathway require the CC4 domain. Recently, it was found that the region where the Atg11 homolog RB1CC1/FIP200 in mammals binds SQSTM1/p62 is a globular domain [Citation20]. The CC2 domain is essential for the binding of Atg11-Atg20 and the formation of homo-oligomers of the Atg11 at the phagophore assembly site (PAS). Also, the interactions between Atg1 and Atg11 require the CC2 and CC3 domains [Citation18]. Nevertheless, the role of Atg11 in glucose starvation-induced autophagy remains unclear.
In this study, we show that glucose starvation-induced autophagy requires Atg11. By revealing a strong link between Atg11 and the kinase activity of Atg1, the formation of the PAS, and the recruitment of Atg9 vesicles to the PAS, we propose Atg11 as a key initiation factor controlling multiple key steps in glucose starvation-induced autophagy.
Results
Atg11 is required for glucose starvation-induced autophagy
To discover the required autophagy genes for glucose starvation-induced autophagy, we performed a GFP-Atg8 processing assay of cultured wild-type, and atg KO strains in glucose starvation medium (SD-G) and nitrogen starvation medium (SD-N). As shown in Figure S1A–D, knockout of the core genes of autophagy completely blocked GFP-Atg8 cleavage in SD-G, indicating that the core machinery of autophagy was also indispensable for glucose starvation-induced autophagy. Furthermore, we found that the Atg17-Atg31-Atg29 complex, specifically required for nitrogen starvation-induced autophagy [Citation16], is also involved in response to glucose starvation conditions. ATG17 deletion completely blocked autophagy, while the knockout of ATG29 and ATG31 dramatically inhibited autophagy. These results indicate that the Atg17-Atg31-Atg29 complex plays an important role in glucose starvation-induced autophagy (). In addition, we found that the absence of autophagy receptors did not affect GFP-Atg8 cleavage (Figure S1E and F), suggesting that these known autophagy receptor proteins are not involved in glucose starvation-induced autophagy.
Figure 1. Atg11 is required for glucose starvation-induced autophagy. GFP-Atg8 was expressed in the yeast strains listed from (A–D). Cells were grown to the log-growth phase, then subjected to nitrogen starvation or glucose starvation for 4 h. Autophagic activity was assessed by western blot analysis of GFP-Atg8 cleavage
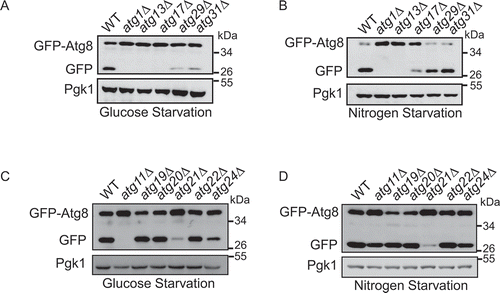
The Cvt pathway is essential for the delivery of Ams1 (α-mannosidase) and Ape1 (aminopeptidase I) to the vacuole. The completion of this process requires the participation of additional ATG proteins such as Atg11, Atg19, Atg20, Atg22, and Atg24 [Citation21]. Unexpectedly, Atg11 appeared essential for glucose starvation-induced autophagy but not for the nitrogen starvation-induced autophagy (). Together, these data indicate that glucose starvation-induced autophagy shares core machinery with other types of autophagy, except for Atg11.
Atg11 is essential for activating Atg1 in glucose starvation conditions
Next, we tested whether Atg1 activation under glucose starvation conditions require Atg11. First, we tested whether glucose starvation can activate Atg1. The bio-orthogonal ATPγS analog and p-nitrobenzyl mesylate (PNBM) system can measure the activity of kinase and identify kinase substrates [Citation22]. We used this system to examine the kinase activity of Atg1. As a positive control, we treated cells with rapamycin to activate Atg1 [Citation23]. We found that the phosphorylation level of substrate MBP (myelin basic protein) increased in both SD-G and rapamycin-treated cells. These results suggested that glucose starvation activated Atg1 (Figure S2A). We then attempted to explore whether the kinase activity of Atg1 is essential for glucose starvation-induced autophagy. We constructed two kinase-dead mutant plasmids, Atg1D211A and Atg1T226A, which were then separately transformed into atg1∆ yeast strains [Citation23]. We found that Atg1 was activated by glucose starvation in the wild type (WT), but not in the Atg1D211A and Atg1T226A mutants (Figure S2B). Under either glucose- or nitrogen-starvation conditions, re-introduction of the Atg1D211A or Atg1T226A mutants into atg1∆ yeast failed to rescue the autophagic activity (Figure S2C-E). Subsequently, we purified the Atg1 complex from wild type, atg11∆, atg13∆, and atg17∆, each in either full medium, SD-N, or SD-G. An in vitro Atg1 kinase assay revealed that Atg13 and Atg17 were essential for the activity of Atg1 in both SD-N and SD-G. Interestingly, the activity of Atg1 under glucose selectively required Atg11, but not nitrogen starvation conditions ().
Figure 2. Atg11 is essential for activating Atg1 in glucose starvation conditions. (A) Atg1-3XFLAG was expressed in wild type, atg11∆, atg13∆, atg17∆ yeast strains. Cells were subjected to nitrogen starvation (SD-N) or glucose starvation medium (SD-G) for 1 h. Cell lysates were immunoprecipitated with anti-FLAG magnetic beads. An In vitro phosphorylation assay was carried out with MBP protein as substrate and immunoprecipitated-Atg1 as kinase for analyzing Atg1 kinase activity. S.E and L.E stands for short exposure and long exposure, respectively. (B) Cells expressing HA-Snf1 were cultured in full medium, SD-N, SD-G, or treated with rapamycin for 1 h. Cell lysates were then immunoblotted with anti-p-Thr172 PRKAA/AMPKα antibody or anti-HA antibody. (C) His6-tagged trigger factor (TF)-Atg1 and TF-Mec1-1300-2000 aa protein was purified with a Ni2+ column from E. Coli; Snf1-WT and Snf1-KD protein were purified with TAP tag from glucose starvation-treated snf1∆ yeast cells expressing Snf1-3XFLAG-TEV-ZZ or Snf1-KD-3XFLAG-TEV-ZZ. Purified Snf1-WT or Snf1-KD were incubated with TF-Atg1 or TF-Mec1-1300-2000aa protein and ATP-γ-S in Snf1 kinase buffer at 30°C for 30 min, and followed by PNBM treatment for another hour. The phosphorylation of Atg1 was detected using an anti-thiophosphate ester (anti-thioP) antibody. Snf1 kinase activity was monitored by immunoblotting with an anti-p-PRKAA/AMPKα (Thr172) antibody. (D) The Atg1-3XFLAG protein was purified using anti-FLAG magnetic beads from BY4741 and snf1∆ yeast strain under full medium and SD-G. An in vitro Atg1 phosphorylation assay was carried out with MBP protein as a substrate and immunoprecipitated-Atg1 as a kinase. (E) HA-Snf1 and Atg1-3XFLAG were co-expressed in wild type and atg11∆ yeast strains. Cells were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG magnetic beads and analyzed by western blot with the indicated antibodies. IC: mouse IgG1 isotype control
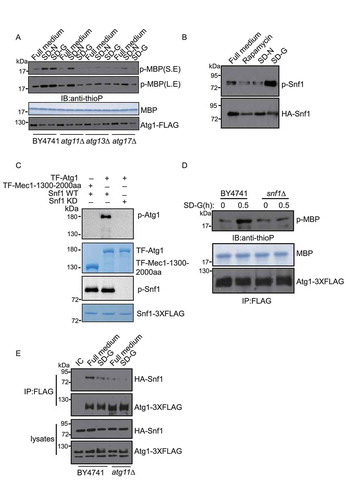
In mammals, glucose starvation activates AMPK, which then phosphorylates and activates ULK1 to initiate autophagy [Citation23]. In yeast, Snf1 is the homolog of AMPK, and the activity of Snf1 can be detected by p-Thr172 PRKAA/AMPKα antibodies [Citation24]. We used this antibody to test the activity of Snf1 under glucose starvation conditions. As shown in , the phosphorylation level of Snf1 dramatically increased upon glucose starvation, but not upon nitrogen starvation or rapamycin treatment, suggesting that glucose starvation conditions resulted in activated Snf1 in yeast. An in vitro kinase assay showed that Snf1 can directly phosphorylate Atg1 and that Snf1-phosphorylated Atg1 could phosphorylate MBP. These results suggested that in yeast, Snf1 activates Atg1 under glucose starvation conditions (). It has also been reported that AMPK associates with ULK1 (Atg1 in yeast) to phosphorylate and activate ULK1 in mammals [Citation11]. To test whether Snf1-mediated Atg1 activation requires Atg11 in yeast, we examined the association of Snf1 with Atg1 in wild type and atg11∆ yeast strains. Immunoprecipitation assays showed that the interaction of Snf1 and Atg1 decreased significantly in atg11∆ yeast strains, indicating that the association of Snf1 with Atg1 requires Atg11 (). Furthermore, we tested the binding of Snf1 and Atg1 in atg13∆, atg17∆, and atg29∆ yeast strains. Co-immunoprecipitation results showed that the binding of Snf1 and Atg1 remains unaltered in atg13∆ and atg29∆ yeast strains, while ATG17 depletion led to the binding levels of Snf1 and Atg1 in full medium to decrease to similar levels observed under glucose starvation conditions. These data indicated that the binding of Snf1 to Atg1 partially involved Atg17, but the binding of Snf1 to Atg1 was mainly controlled by Atg11 (Figure S2F). Thus, we concluded that Atg11 regulates the kinase activity of Atg1 by controlling the association of Snf1 with Atg1 in glucose starvation conditions.
Atg11 is required for the formation of the Atg17-Atg31-Atg29 complex in glucose starvation conditions
Atg17, Atg29, and Atg31 proteins constitutively form an Atg17-Atg31-Atg29 ternary complex, thereby recruiting other autophagic proteins such as Atg1 and Atg13 to the PAS. In this way, the Atg17-Atg31-Atg29 ternary complex forms a scaffold complex essential for the PAS organization [Citation16]. Our imaging data showed that the localization of Atg17, Atg29, and Atg31 proteins dispersed well in the atg11∆ yeast strain upon glucose starvation. In contrast, under nitrogen starvation conditions, Atg17, Atg29, and Atg31 proteins retained puncta formation despite atg11 knockout. Consistent with the result that atg11 knockout did not affect nitrogen starvation-induced autophagy ( and S3A), suggesting that Atg11 is essential for PAS organization under glucose starvation conditions. Using the same approach, we also found that the localization of Atg1 and Atg13 proteins diffused in atg11∆ yeast cells upon glucose starvation (Figure S3B-D). Similarly, after glucose starvation, the localization of Atg11 dispersed in atg17∆, atg29∆, and atg31∆ yeast strains, while Atg11 formed puncta in atg1∆ and atg13∆ yeast strains (Figure S4A and B). These results indicate that the localization of Atg11 and Atg17-Atg31-Atg29 is interdependent upon glucose starvation.
Figure 3. Atg11 is required for the formation of the Atg17-Atg31-Atg29 complex in glucose starvation conditions. (A-C) Atg17-2XGFP, Atg29-2XGFP and Atg31-2XGFP were expressed in the wild type and atg11∆ strains. Cells were cultured in full medium, SD-N or SD-G for 1 h, then analyzed by microscopy. Scale bar, 2 µm. (D) Atg17-6XHA, Atg29-TAP and Atg31-2XGFP were co-expressed in wild type and atg11∆ yeast strains. Cells were grown to the log-growth phase and then subjected to SD-N or SD-G for 1 h. Cell lysates were immunoprecipitated with anti-HA magnetic beads and then analyzed with the indicated antibodies. IC: mouse IgG1 isotype control. (E) Cell co-expressing Atg31-2XGFP and Atg29-TAP in the wild type or atg11∆ strains were then subjected to SD-N and SD-G for 1 h. Cell lysates were immunoprecipitated with rabbit IgG agarose beads and then analyzed by western blot using the indicated antibody. (F) Gel filtration chromatography analysis by Superose 6 10/300 GL column for proteins lysed from wild type (WT) and atg11∆ yeast after cells were deprived from glucose for 1 h. Each fraction was detected by immunoblotting using anti-HA, anti-GFP and anti-Protein A antibodies. Positions of molecular weight (MW) between fractions are shown
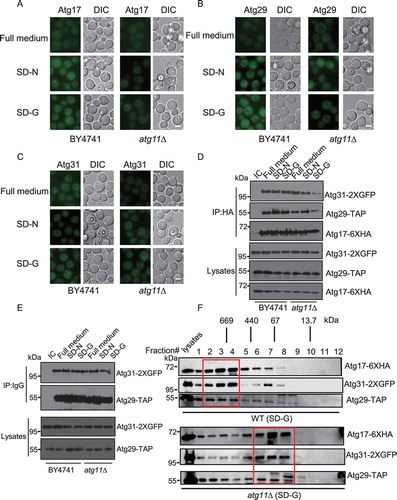
Next, we examined interactions within the Atg17-Atg31-Atg29 complex in the atg11∆ strain. Our data showed that upon glucose starvation, but not nitrogen starvation, the deletion of Atg11 led to significantly decreased binding of Atg29 and Atg31 to Atg17, while the interaction between Atg29 and Atg31 remains unaltered ().
To further explore the effect of Atg11 on the formation of the Atg17-Atg31-Atg29 ternary complex under glucose starvation conditions, we subjected the lysates of wild-type BY4741 and atg11∆ yeast cells to gel filtration analysis using a Superose 6 10/300 GL column. As shown in , in response to glucose starvation, Atg17, Atg29, and Atg31 proteins of wild-type yeast cells form a complex around 670 kDa, and their co-elution peaks appear in fractions #2-4. However, Atg17, Atg29, and Atg31 proteins of co-elution peaks appear in fractions #6-8 in atg11∆ yeast strains, indicating that the Atg17-Atg31-Atg29 complex dissociates in the atg11∆ mutant under glucose starvation conditions. Furthermore, we found that atg11 knockout did not affect the integrity of the Atg6-Atg14-Vps34 protein complex under glucose starvation conditions (Figure S4C). Thus, we concluded that Atg11 is necessary for the formation of the PAS by regulating the association of Atg17 with Atg29-Atg31 upon glucose deprivation.
The recruitment of Atg9 vesicles to the PAS by Atg11 is required for glucose starvation-induced autophagy
One key step of autophagosome formation is the recruitment of Atg9 vesicles to the PAS. Atg9 is the only integral membrane protein of the autophagic machinery, which is sorted into vesicles [Citation25]. In response to nitrogen starvation, PAS recruits Atg9 vesicles to provide part of the autophagosomal membrane [Citation26]. We tested whether the recruitment of Atg9 vesicles to the PAS upon glucose starvation requires Atg11. Previous reports showed that the CC2 domain of Atg11 regulates the association of Atg11 with Atg9 under normal medium conditions (). However, disruption of Atg9-Atg11 binding only impaired the Cvt pathway but did not affect nitrogen starvation-induced autophagy [Citation27]. To identify the required sites within the CC2 domain of the Atg11 protein essential for Atg9-Atg11 interaction, and to test whether glucose starvation-induced autophagy requires this binding site, we constructed various truncated and site-mutated Atg11 plasmids for yeast two-hybrid analysis. We found a crucial Isoleucine 569 (I569) residue of Atg11 for the association of Atg11 with Atg9 ( and S4D). Also, an immunoprecipitation assay showed that the Atg11I569E mutant decreased the interaction between Atg9 and Atg11 in the full medium, and their interaction almost disappeared under glucose starvation conditions. This observation indicated that the I569 site of Atg11 mediates the Atg9-Atg11 interaction (). Next, we wanted to know whether the CC2 domain of Atg11, if alone, was sufficient for binding to Atg9. Since the CC2 domain has only 41 amino acids, we added a GST tag to the N-terminal of the CC2 domain. As shown in Figure S5A, the CC2 domain of Atg11 could bind to Atg9, and the I569E mutant significantly reduces the association of Atg11 CC2 with Atg9. The co-localization of PAS marker Ape1 with Atg9 also dramatically decreased in the Atg11I569E mutant strain in response to glucose starvation (). This result suggests that the Atg11(I569) site regulates the recruitment of Atg9 vesicles to the PAS under glucose starvation conditions. We further investigated whether glucose starvation-induced autophagy requires the Atg11 (I569) site. Both GFP-Atg8 localization and processing assays confirmed that the I569 site is crucial for vacuole localization and the subsequent cleavage of GFP-Atg8 (). These results imply that the recruitment of Atg9 vesicles to the PAS is important for glucose starvation-induced autophagy.
Figure 4. The recruitment of Atg9 vesicles to the PAS by Atg11 is required for glucose starvation-induced autophagy. (A) The scheme of the Atg11 domains and the deletions. CC1∆ involves the deletion of amino acids at the 272–321 position of the Atg11 protein, CC2∆ involves the deletion of amino acids at the 536–576 position, CC3∆ involves the deletion of amino acids at the 627–859 position, CC4∆ involves the deletion of amino acids at the 859–1178 position. Modified from [Citation18]. (B) Yeast two-hybrid analysis of the Atg11 and Atg9 interaction. The yeast strain AH109 was transformed with plasmids expressing AD-fused with Atg11, Atg11 CC2∆, Atg11 566-576aa∆, or Atg11I569E and plasmids expressing a BD-fused with the N terminus of Atg9. These strains were grown on SD-Leu-Trp (+Ade) and SD-Leu-Trp-Ade (−Ade) agar plates at 30°C for 3 d. (C) Cells co-expressing an empty vector, HA-Atg11 WT, HA-Atg11-CC2∆, or HA-Atg11I569E with Atg9-TAP in the atg11∆ strain were starved with glucose for 0 h and 1 h. Cell lysates were immunoprecipitated with rabbit IgG agarose beads and then analyzed by western blot using the anti-HA antibody. (D) Images of cells expressing Atg9-2XGFP and RFP-Ape1, which were starved in SD-G for 1 h. Scale bar, 2 µm. (E) Cells from (D) were quantified for the number of cells with Atg9-2XGFP dots co-localized with RFP-Ape1 dots. n = 300 cells pooled from three independent experiments. Data are presented as means ± SD. (F) Cells expressing GFP-Atg8 and Vph1-Cherry in atg11∆, HA-Atg11-WT, HA-Atg11-CC2∆, and HA-Atg11I569E strains were cultured in SD-N or SD-G medium for 4 h. Autophagic activity was assessed by the quantification of the translocation of GFP-Atg8 into vacuoles. Scale bar, 2 µm. (G) Cells from (F) were analyzed for the translocation of GFP-Atg8 into vacuoles. n = 300 cells pooled from three independent experiments. Data are presented as means ± SD. (H) Cells from (F) were analyzed by western blot for GFP-Atg8 cleavage
![Figure 4. The recruitment of Atg9 vesicles to the PAS by Atg11 is required for glucose starvation-induced autophagy. (A) The scheme of the Atg11 domains and the deletions. CC1∆ involves the deletion of amino acids at the 272–321 position of the Atg11 protein, CC2∆ involves the deletion of amino acids at the 536–576 position, CC3∆ involves the deletion of amino acids at the 627–859 position, CC4∆ involves the deletion of amino acids at the 859–1178 position. Modified from [Citation18]. (B) Yeast two-hybrid analysis of the Atg11 and Atg9 interaction. The yeast strain AH109 was transformed with plasmids expressing AD-fused with Atg11, Atg11 CC2∆, Atg11 566-576aa∆, or Atg11I569E and plasmids expressing a BD-fused with the N terminus of Atg9. These strains were grown on SD-Leu-Trp (+Ade) and SD-Leu-Trp-Ade (−Ade) agar plates at 30°C for 3 d. (C) Cells co-expressing an empty vector, HA-Atg11 WT, HA-Atg11-CC2∆, or HA-Atg11I569E with Atg9-TAP in the atg11∆ strain were starved with glucose for 0 h and 1 h. Cell lysates were immunoprecipitated with rabbit IgG agarose beads and then analyzed by western blot using the anti-HA antibody. (D) Images of cells expressing Atg9-2XGFP and RFP-Ape1, which were starved in SD-G for 1 h. Scale bar, 2 µm. (E) Cells from (D) were quantified for the number of cells with Atg9-2XGFP dots co-localized with RFP-Ape1 dots. n = 300 cells pooled from three independent experiments. Data are presented as means ± SD. (F) Cells expressing GFP-Atg8 and Vph1-Cherry in atg11∆, HA-Atg11-WT, HA-Atg11-CC2∆, and HA-Atg11I569E strains were cultured in SD-N or SD-G medium for 4 h. Autophagic activity was assessed by the quantification of the translocation of GFP-Atg8 into vacuoles. Scale bar, 2 µm. (G) Cells from (F) were analyzed for the translocation of GFP-Atg8 into vacuoles. n = 300 cells pooled from three independent experiments. Data are presented as means ± SD. (H) Cells from (F) were analyzed by western blot for GFP-Atg8 cleavage](/cms/asset/68577456-fe80-42d7-a81c-dde261227b99/kaup_a_1719724_f0004_oc.jpg)
The CC1, CC2, CC3, and CC4 domains of Atg11 are required for glucose starvation-induced autophagy
To identify the domain of Atg11 required for glucose starvation-induced autophagy, we constructed CC1, CC2, CC3, and CC4 domain-deletion mutants of Atg11 and performed rescue experiments by examining GFP-Atg8 distribution and cleavage under conditions, as indicated. The deletion of the CC1, CC2, CC3, and CC4 domains of Atg11 prevented the localization of GFP-Atg8 to the vacuole and accordingly reduced the generation of cleaved GFP ( and S5B). These data indicated that glucose starvation-induced autophagy requires CC1, CC2, CC3, and CC4 domains.
Figure 5. The CC1, CC2, CC3, and CC4 domains of Atg11 are required for glucose starvation-induced autophagy. (A) Cells expressing GFP-Atg8 and Vph1-Cherry in atg11∆, HA-Atg11 WT, HA-Atg11-CC1∆, HA-Atg11-CC2∆, HA-Atg11-CC3∆, and HA-Atg11-CC4∆ strains were cultured in SD-N or SD-G medium for 4 h. Autophagic activity was assessed for the translocation of GFP-Atg8 into vacuoles. n = 300 cells pooled from three independent experiments. Data are presented as means ± SD. (B) Autophagic activity from (A) was assessed using western blot analysis of GFP-Atg8 and Ape1 cleavage. (C-E) Cells co-expressing an empty vector, HA-Atg11 WT, HA-Atg11-CC1∆, HA-Atg11-CC2∆, HA-Atg11-CC3∆, or HA-Atg11-CC4∆ with Atg1-3XFLAG, Atg29-3XFLAG, or Atg31-3XGLAG in the atg11∆ strain were grown to the log phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG magnetic beads and then analyzed by western blot using the indicated antibody. (F) Cells co-expressing an empty vector, HA-Atg11 WT, HA-Atg11-CC1∆, HA-Atg11-CC2∆, HA-Atg11-CC3∆, or HA-Atg11-CC4∆ with Snf1-3XFLAG in the atg11∆ strain were grown to the log phase. Cell lysates were immunoprecipitated with anti-FLAG magnetic beads and then analyzed by western blot using the indicated antibody
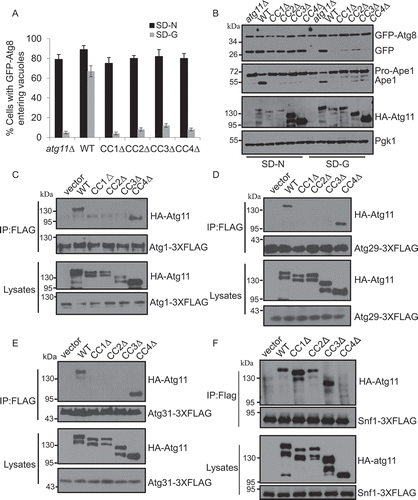
We further explored which proteins within the Atg1-Atg13-Atg17-Atg31-Atg29 complex associated with Atg11. Yeast two-hybrid assays showed that Atg11 bound to Atg1, Atg29, and Atg31, instead of Atg13 and Atg17 (Figure S5C). Next, we tested each domain of the Atg11 protein necessary for its associations with Atg1, Atg29, and Atg31. Immunoprecipitation experiments showed that under glucose starvation conditions, the CC1, CC2, and CC3, but not the CC4 domains of Atg11 were essential for the association of Atg11 with Atg1, Atg29, and Atg31 (). Next, to explore whether the association of Atg11 with Atg1, Atg29, and Atg31 depend on Atg17 or Atg29 under glucose starvation conditions, we knocked out either ATG17 or ATG29. Immunoprecipitation results showed that the association of Atg11 with Atg1 required Atg17 and Atg29, and the combination of Atg11 and Atg29 is Atg17-dependent. However, the combination with Atg31 does not require Atg17 or Atg29 (Figure S5D-F). Since Atg11 mediates the interaction between Atg1 and Snf1, we then tested which domain of Atg11 was crucial for the binding of Snf1-Atg11. As shown in , the absence of CC4 domain dramatically decreased the association of Atg11 with Snf1. This result suggests that the CC4 domain mediates the association of Atg11 with Snf1. This result was consistent with the finding that the CC1, CC2, CC3, and CC4 domain deletions of Atg11 blocked autophagy induced by glucose starvation. Together, our data show that glucose starvation-induced autophagy through their binding to Atg1, Atg9, Atg29, Atg31, and Snf1 requires the CC1, CC2, CC3, and CC4 domains of Atg11.
Discussion
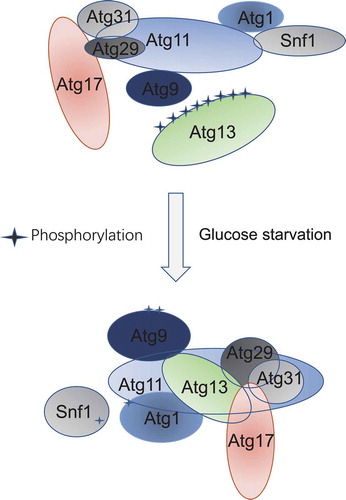
Our findings strongly suggest that Atg11 is essential for glucose starvation-induced autophagy. ATG11 deletion significantly decreased the association of Snf1 with Atg1, the formation of the Atg17-Atg31-Atg29 complex, and the recruitment of Atg9 vesicles to the PAS (). These results provide strong evidence that Atg11 is a multifunctional initiator for energy deprivation-induced autophagy and that it participates in the activation of Atg1, the assembly of the PAS, and the recruitment of Atg9 vesicles to the PAS under energy-deficient conditions.
Figure 6. Model of Atg11 participating in initiation of glucose starvation-induced autophagy. In response to glucose starvation, Atg11 participates in the assembly of the PAS by regulating the association of Atg17 with Atg29-Atg31. Simultaneously, Atg1 is activated by Atg11 mediating binding of Atg1 with Snf1, and Atg9 vesicles are recruited to the PAS by Atg11 to initiate glucose starvation-induced autophagy
It is well known that autophagy requires the Atg1 kinase activity. Many studies demonstrated that the kinase activity of Atg1 regulates autophagy by phosphorylating autophagic proteins such as Atg4, Atg9, Atg14, and Vps30/BECN1 [Citation28–31]. Therefore, exploring the molecular mechanism of Atg1 activation is very important for us to understand the regulation of autophagy better. Atg13 and Atg17 proteins are necessary for Atg1 activation under nitrogen starvation conditions [Citation15]. Recently, it has also been revealed that the activation of Atg1 in the process of autophagy induced by either the DNA damage agent methyl methanesulfonate (MMS) or by phosphate starvation requires Atg11 [Citation32,Citation33]. However, the mechanism of Atg1 activation remained elusive. In this study, we found that Atg11 is essential for Atg1 activation under glucose starvation. As in mammalian cells, glucose starvation immediately activates Snf1 and subsequently phosphorylates and activates Atg1 in yeast. Since both Atg11 and Snf1 bind to Atg1 and are involved in Atg1 activation in glucose starvation conditions, we detected the association of Snf1 with Atg1 in the atg11∆ yeast strain and found that the binding of Snf1 and Atg1 decreased significantly upon atg11∆ deletion. These data showed that Atg11 regulates the kinase activity of Atg1 by affecting the association of Snf1-Atg1 under glucose starvation conditions.
In S. cerevisiae, either rapamycin- or nitrogen starvation-induced autophagy both require Atg17, which serves as the primary scaffold protein for the PAS. However, selective autophagy requires Atg11 [Citation34]. Our data revealed that glucose starvation-induced autophagy requires both Atg11 and Atg17. The absence of ATG11 resulted in the dissociation of the Atg17-Atg31-Atg29 ternary complex, which prevented the recruitment of other autophagic proteins such as Atg1 and Atg13 to the PAS. Simultaneously, the absence of any protein in the Atg17-Atg31-Atg29 complex led to the cytosolic distribution of Atg11. These results suggest that under glucose starvation, Atg11, and the Atg17-Atg31-Atg29 complex function together to recruit other autophagic proteins to the PAS.
Atg11 is essential for the Cvt pathway and most types of selective autophagy. We found that the CC1, CC2, and CC3 domains of Atg11 are crucial for the association of Atg11 with Atg1, Atg29, and Atg31. Previous studies found that the CC4 domain of Atg11 is required for its association with Atg19, but not with Atg1 [Citation7,Citation18]. Our results showed that the interaction between Snf1 and Atg11 also requires the CC4 domain. Reports show that Atg11 can bind directly to Atg9 through the CC2 domain. However, the association of Atg9 with Atg11 only affected the Cvt pathway but did not affect autophagy induced by nitrogen starvation. We further identified that the Atg11 (I569) site is crucial for the recruitment of Atg9 vesicles to the PAS and autophagy during glucose starvation. Together, these results suggested Atg11, a key initiation factor, recruits other autophagic proteins and Atg9 vesicles into the PAS to induce autophagy under glucose starvation conditions.
Despite the above results, the question as to how the ATG11 deletion affects the association of Atg17 with Atg29 and Atg31 during glucose starvation, rather than nitrogen starvation, remains unsolved. To answer this, we performed a yeast two-hybrid assay, and our results showed that Atg11 could bind to Atg29 and Atg31, which provides a structural basis for Atg11’s regulation of the formation of Atg17-Atg31-Atg29. Secondly, we speculated that there might be extra proteins that bind the Atg17-Atg31-Atg29 complex in response to glucose starvation, or that potential post-translational modifications of any member of this complex could lead to conformational changes of the whole complex that may also require the involvement of Atg11. We then went on to further elucidate how Atg11 participates in the formation of the Atg17-Atg31-Atg29 complex by comparing the binding proteins and analyzing the post-translational modifications of the Atg17-Atg31-Atg29 complex under energy-deficient conditions.
In summary, we have elucidated the role of Atg11 in the process of autophagy induced by glucose starvation. Our results suggest that Atg11 is a key initiation factor in Atg1 activation, PAS assembly, and Atg9 vesicle recruitment to the PAS. Further studies on the protein machinery, molecular mechanism, and physiological function of energy deficiency-induced autophagy will be forthcoming.
Materials and methods
Yeast strains and constructs
All yeast strains used in this study are listed in Table S1. The wild-type BY4741 and related deletion strains were purchased from Invitrogen (95,401.H2). The related strains were constructed, as described previously, and verified by polymerase chain reaction (PCR) (Vazyme, P505-d1) or western blot analysis with the indicated antibody [Citation35]. All mutant yeast strains in this study were sequenced and detected by western blot.
Antibodies
Antibodies were purchased and used at the indicated dilutions in this study as follows: anti-HA (Abmart, M20003L; 1:3000), anti-FLAG (Sigma, F1804; 1:2500), anti-GFP (Roche, 11814460001; 1:2500), anti-MYC (Roche, 11814460001; 1:2500), anti-protein A antibody (Raybiotech, 168–10967; 1:2500), anti-p-PRKAA/AMPKα (Thr172) (Cell Signaling Technology, 2535S; 1:2000), anti-thiophosphate ester (Abcam, ab133473; 1:10,000), anti-Pgk1 (Nordic Immunology, NE130/7S; 1:10,000), goat anti-mouse IgG1, human ads-HRP (SouthernBiotech, 1070–05; 1:10,000), goat anti-rabbit, human ads-HRP (SouthernBiotech, 4010–05; 1:10,000).
Microscopy, western blots, and immunoprecipitation
Yeast strains with fluorescent tags were grown to OD600 = 0.8 ~ 1.0 in full medium and treated with rapamycin (Selleck, S1039), nitrogen starvation, or glucose starvation for the indicated times. Cells were observed using an inverted fluorescence microscope (IX83; Olympus). The extraction of yeast protein, immunoblotting, and immunoprecipitation experiments were performed in accordance with previously described methods [Citation36].
In vitro Atg1 kinase assay and alkylation protocol
Cells expressing Atg1-3XFLAG in indicated yeast strains were grown to the log phase, then subjected to glucose starvation for 1 h. Yeast cells (50 OD600 units) were harvested and broken up with glass beads in a lysis buffer (50 mM Tris.HCl, pH 7.5, 150 mM NaCl, 1% NP-40 [Sangon Biotech, A600385-0500], 10% glycerol, 10 mM NaF [Sangon Biotech, A500850-0250], protease inhibitor [Bimake, B14001]). Atg1-3XFLAG proteins were immunoprecipitated with anti-FLAG magnetic beads (Bimake, B26102) from cell lysates following centrifugation. Immunoprecipitated Atg1 were incubated with 5 µg MBP (myelin basic protein; Sigma, M2295) and 1.5 µl 10 mM ATP-γ-S (Sigma, A1388) in 30 µl Atg1 kinase buffer (30 mM Hepes.KOH, pH 7.4, 5 mM NaF, 10 mM MgCl2, 1 mM DTT) on a shaker at 30°C for 30 min, added with 2 µl 50 mM p-nitrobenzyl mesylate (PNBM; Abcam, ab138910) and then incubated for another hour at 30°C. Finally, the reactions were halted by boiling in 2X protein loading buffer for 5 min and then resolved using 12% SDS-PAGE gels. The phosphorylation level of MBP was detected by anti-thiophosphate ester (Abcam, ab92570).
Protein purification from E. coli
E. coli BL21 cells (Tsingke,TSV-A09) with pCold-TF-Atg1 (lab stock) and Mec1-1300-2000 aa (lab stock) were grown in 500 ml LB medium containing 50 µg/ul ampicillin (Sangon Biotech, A600064-0025) to OD600 = 0.5 ~ 0.6 at 37°C, then shaken at 16°C for 20 h in the presence of 0.1 mM IPTG (Sangon Biotech, A600168-0025), harvested by centrifugation and the pellets were suspended in 25 ml lysis buffer (50 mM Tris.HCl, pH 7.5, 500 mM NaCl, 0.1% Triton X-100 [Sangon Biotech, A600198-0500], 0.1 mM PMSF [Sangon Biotech, A610425-0025], 1 mM DTT) and disrupted by sonication. Subsequently, the lysates were centrifuged at 13,000 g for 30 min. The supernatants were mixed with 1 ml (bed volume) of a Ni-NTA agarose resin (GE healthcare, 17-5318-06) and rotated at 4°C for 1 h. The resins were washed with 20 ml washing buffer (20 mM Tris.HCl, pH 7.5, 500 mM NaCl, 30 mM Imidazole [Sangon Biotech, A600277-0100], 0.1 mM PMSF) for 3 successive times. The samples were eluted with 10 ml elution buffer (20 mM Tris.HCl, pH 7.5, 500 mM NaCl, 250 mM imidazole, 0.1 mM PMSF). The eluates were concentrated to 1 ~ 2 mg/ml with Amicon Ultra-4 filter units (Millipore, UFC801096), and 50% glycerol was added to a final concentration of 10% at −80°C for storage.
Tandem affinity purification
snf1∆ yeast cells expressing Snf1-3XFLAG-TEV-ZZ or Snf1-KD-3XFLAG-TEV-ZZ was grown to the log phase, then were subjected to glucose starvation for 1 h. The cell pellet was washed with chilled H2O and then broken up with glass beads in lysis buffer (50 mM Tris.HCl, pH7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, 10 mM NaF, protease inhibitor). The supernatant was obtained by centrifugation at 13,000 g for 30 min. Rabbit IgG agarose beads (Sigmsa, A2909) were washed with lysis buffer 3 times, then incubated with the supernatant for 3 h at 4°C. IgG beads were washed 3 times with lysis buffer, and one time with TEV buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 1 mM EDTA), then 80 μl TEV protease (home-made) was added in a TEV cleavage buffer and incubated overnight at 4°C. The supernatant was incubated with anti-FLAG magnetic beads for 3 h by gentle rotation at 4°C. Anti-FLAG beads were washed with a lysis buffer three times. Snf1-3XFLAG or Snf1-KD-3XFLAG was eluted with FLAG peptide. Finally, the protein was resolved by 10% SDS-PAGE gel and visualized using coomassie brilliant blue staining.
In vitro phosphorylation of Atg1 by Snf1
TF-Atg1 and TF-Mec1-1300-2000aa protein were purified from E. coli using a Ni2+ column; Snf1-WT and Snf1-KD protein were purified using a TAP tag from glucose starvation-treated snf1∆ yeast cells expressing Snf1-3XFLAG-TEV-ZZ or Snf1-KD-3XFLAG-TEV-ZZ. Purified Snf1-WT or Snf1-KD was incubated with TF-Atg1 or TF-Mec1-1300-2000aa protein and ATP-γ-S in a Snf1 kinase buffer (30 mM Hepes.KOH, pH 7.4, 5 mM NaF, 10 mM MgCl2, 1 mM DTT) at 30°C for 30 min, and followed by PNBM treatment for another hour. The phosphorylation of Atg1 was detected using an anti-thiophosphate ester antibody.
Construction of Atg11 CC1∆, CC2∆, CC3∆, and CC4∆ mutants
The definition of CC1, CC2, CC3, and CC4 regions were described as previously [Citation18]. The deletion fragments of CC1, CC2, CC3, and CC4 were in positions aa 272–321, aa 536–576, aa 627–859, and aa 859–1178, respectively. Primers for the deletion of CC1∆, CC2∆, CC3∆, and CC4∆ were designed, and PCR-based mutations were carried out with N-HA-Atg11 in pRS316 (Dr. Zhiping Xie’s gift) as a template. These mutations were identified by sequencing, and the expression of related proteins was detected by western blot.
The identification of Atg11 I569E mutant
PCR-based random mutagenesis was performed in the CC2 region of Atg11, and then the loss-of-interaction mutants of Atg11 with Atg9 were screened using a yeast two-hybrid assay. A missense mutation was identified with single isoleucine to glutamic acid substitution at position 569 (I569E).
Yeast two-hybrid assay
A yeast two-hybrid assay was performed as described previously [Citation37]. The AH109 strain was transformed with the indicated combinations of pGADT7-originated and pGBKT7-originated plasmids, and the related yeast strains were spotted onto SC-LW (+Ade) and SC-LWA (−Ade) plates with incubation at 30°C for 2 ~ 4 d. AH109 yeast strain, pGADT7, and pGBKT7 plasmids were generously provided by Dr. Hitoshi Nakatogawa [Citation37].
Statistical analysis
For all quantitative analyses, the mean values are displayed together with the standard deviation (s.d., shown as error bars). The number of independent experiments and the related statistics are indicated in each figure legend.
Supplemental Material
Download MS Word (2.8 MB)Acknowledgments
We are grateful to Prof. Y. Ohsumi, Prof. H. Nakatogawa, and Prof. Zhiping Xie for the plasmids and yeast strains. We thank Prof. Li Yu, Prof. Qiming Sun, Prof. Yixian Cui and Prof. Jingwei Zhao for reading and discussing the manuscript, and Prof. Fan Yang for his technical support in the gel filtration chromatography and Hangjun Wu in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for his technical assistance on Confocal Laser Scanning Microscopy. We also thank for the support by the Protein facility, Core facilities, Zhejiang University School of Medicine. The research was supported by National Natural Science Foundation of China 91754107 and 31771528 to Cong Yi, and Zhejiang Provincial Natural Science Foundation (Grant No:LY17C160001) to Choufei Wu, and the Key Research Development Program of Zhejiang Province (No. 2018C03018), the Traditional Chinese Medicine Scientific Research Fund Project of Zhejiang Province (No. 2017ZA079), and Major Science and Technology Projects of Medicine and Health in Zhejiang Province (WKJ-ZJ-1923) to Liming Wu.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23.
- Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109(22):8670–8675.
- Riahi Y, Wikstrom JD, Bachar-Wikstrom E, et al. Autophagy is a major regulator of beta cell insulin homeostasis. Diabetologia. 2016;59(7):1480–1491.
- Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24(1):58–68.
- Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42.
- Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell. 2016;166(2):288–298.
- Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41.
- Parzych KR, Ariosa A, Mari M, et al. A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2018;29(9):1089–1099.
- Yi C, Tong J, Lu P, et al. Formation of a Snf1-Mec1-Atg1 module on mitochondria governs energy deprivation-induced autophagy by regulating mitochondrial respiration. Dev Cell. 2017;41(1):59–71 e4.
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24(1):42–57.
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141.
- Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1–2):290–303.
- Chang C, Su H, Zhang D, et al. AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol Cell. 2015;60(6):930–940.
- Matsuura A, Tsukada M, Wada Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192(2):245–250.
- Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513.
- Kabeya Y, Noda NN, Fujioka Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389(4):612–615.
- Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553.
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605.
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279(29):29889–29894.
- Turco E, Witt M, Abert C, et al. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol Cell. 2019;74(2):330–46 e11.
- Khalfan WA, Klionsky DJ. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol. 2002;14(4):468–475.
- Allen JJ, Li M, Brinkworth CS, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4(6):511–516.
- Kamada Y, Yoshino K, Kondo C, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058.
- Orlova M, Barrett L, Kuchin S. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast. 2008;25(10):745–754.
- Lang T, Reiche S, Straub M, et al. Autophagy and the cvt pathway both depend on AUT9. J Bacteriol. 2000;182:2125–2133.
- Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233.
- He C, Song H, Yorimitsu T, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175(6):925–935.
- Sanchez-Wandelmer J, Kriegenburg F, Rohringer S, et al. Atg4 proteolytic activity can be inhibited by Atg1 phosphorylation. Nat Commun. 2017;8:295.
- Papinski D, Schuschnig M, Reiter W, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483.
- Park JM, Jung CH, Seo M, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–564.
- Park JM, Seo M, Jung CH, et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy. 2018;14:584–597.
- Eapen VV, Waterman DP, Bernard A, et al. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc Natl Acad Sci U S A. 2017;114(7):E1158–E67.
- Yokota H, Gomi K, Shintani T. Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2017;483:522–527.
- Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218.
- Janke C, Magiera MM, Rathfelder N, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962.
- Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477.
- Suzuki SW, Yamamoto H, Oikawa Y, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A. 2015;112:3350–3355.
