ABSTRACT
In budding yeast, macroautophagy/autophagy is required for cells to enter into the meiotic divisions. Our recent publication showed that autophagy is also required for meiotic exit. Inhibition of autophagy as cells enter into the meiotic divisions results in additional rounds of spindle formation, spindle elongation, and aberrant chromosome segregation leading to cell death. Under these conditions, the meiosis II-specific cyclin Clb3 is absent, and two substrates of the anaphase-promoting complex/cyclosome (APC/C) persist into the additional divisions instead of being degraded after meiosis II. We found that the translational repressor Rim4 is a substrate of autophagy, which could explain these observations through its known role in repressing synthesis of Clb3 and the meiosis-specific co-activator of the APC/C, Ama1. Combined, these results provide new mechanistic insight into the control of meiotic exit through timed autophagic degradation of a master regulator of gene expression.
Unicellular organisms, such as budding yeast, adapt to fluctuating environmental conditions for survival. In diploid yeast, nitrogen limitation in the presence of a non-fermentable carbon source induces the sporulation program, which involves meiosis coupled to packaging of the four haploid nuclei into spores. The spores can survive the nutrient-poor conditions and then germinate when nutrients return. Mutations in core autophagy genes have been known to result in a severe sporulation defect ever since 1993 when they were originally characterized by the pioneering work of the Ohsumi lab.
In addition to its likely role in maintaining amino acid levels for ongoing protein synthesis that is necessary to enter into the first meiotic division, we wondered if autophagy also had an important role during the meiotic divisions and spore formation. Using ribosome profiling, Brar and colleagues showed that translation of ATG8, a core autophagy component involved in phagophore expansion, sharply increases during the meiotic divisions. We found that autophagy activity measured by the flux of GFP-Atg8 to the vacuole also increases during the meiotic divisions [Citation1]. To determine if autophagy was in fact required for the meiotic divisions, we used an analog-sensitive (as) version of Atg1, a master kinase required for phagophore assembly. The Atg1-as protein is inherently functional but can be inactivated by the bulky ATP analog NM-PP1 because of its enlarged ATP-binding pocket. In addition, we built a strain that can be released from prophase I arrest, allowing the treatment with NM-PP1 to occur on a synchronized population of cells entering meiotic divisions.
The inhibition of Atg1 during the meiotic divisions gave a surprising phenotype. Normally, once cells are released from prophase I, they undergo the two meiotic divisions and then assemble four spores around each haploid nucleus. With inhibition of Atg1, cells undergo meiosis I and meiosis II normally, albeit with a delayed meiosis II. However, cells fail to exit meiosis and instead undergo additional rounds of spindle pole body formation (the yeast equivalent of the centrosome), spindle formation, and spindle elongation. Overall, the cells accumulate between 5–9 spindle pole bodies, undergo up to 17 rounds of spindle elongation and disassembly, and fail to form spores. Interestingly, these spindles are able to segregate chromosomes, resulting in small chromatin masses throughout the cell. Similarly, cells in which we forced degradation of Atg14, a subunit of the autophagy-specific phosphatidylinositol 3-kinase complex, also display the same phenotypes. These results suggest that autophagy prevents additional rounds of chromosome segregation after meiosis II.
The highly abnormal phenotype of additional rounds of chromosome segregation led to the question of whether the meiotic cell cycle was misregulated in the absence of autophagy. To address this question, we analyzed several cell cycle regulators. First, we monitored polo kinase, Cdc5, which binds spindle pole bodies and is targeted for proteasomal degradation prior to meiosis exit by the APC/C. With Atg1 inhibition, Cdc5 localization is normal through meiosis II. However, at the end of meiosis II, Cdc5 is not degraded and instead binds to the additional spindle pole bodies that form after meiosis II. Next we monitored the levels of meiotic cyclins Clb1 and Clb3, which are tightly controlled during meiosis to regulate cyclin-dependent kinase (CDK) activity. Normally, Clb1 is present throughout meiotic divisions, whereas Clb3 is only present during meiosis II. Both Clb1 and Clb3 are targeted for degradation by the APC/C as cells exit meiosis. We found that with autophagy inhibited, Clb1 persists through the additional meiotic divisions and Clb3 is absent throughout. These results suggested that with Atg1 inhibition, meiosis II is misregulated and cells failed to undergo meiotic exit.
What mechanism could account for the lack of Clb3 and the persistence of Cdc5 and Clb1 caused by Atg1 inhibition? We found several clues implicating Rim4 in previous work from the Amon and Berchowitz labs. First, they found that CLB3 mRNA is translationally repressed by its association with Rim4. Second, they identified AMA1 among other mRNA substrates of Rim4, which is a meiosis-specific co-activator of the APC/C that targets Cdc5 and Clb1 for degradation. Last, in meiosis II, Rim4 is phosphorylated and degraded, releasing its mRNAs for translation. Thus, we hypothesized that autophagy degrades Rim4, which would parsimoniously account for the protein level changes in Clb3, Cdc5, and Clb1 that we observed when we inhibit Atg1.
We therefore looked for the appearance of any free GFP derived from Rim4-GFP that would result from its vacuolar delivery by autophagy. Indeed, we found that accumulation of free GFP during meiosis II is blocked by Atg1 inhibition. In further support of our model that autophagy regulates meiotic exit by degradation of Rim4, we found that expression of either CLB3 or AMA1 in meiosis II suppresses the additional meiotic divisions when autophagy is inhibited, albeit the defect in spore formation is not rescued.
Our results indicate that autophagy of Rim4 is a key regulatory step in meiosis II needed to stave off the latent threat of runaway meiotic divisions to meiotic exit (). An important future direction is understanding how Rim4 is selected as an autophagy substrate and how that degradation occurs specifically in meiosis II. Rim4 is heavily phosphorylated by a meiosis-specific kinase called Ime2, and the phosphorylation can lead to its destabilization. We hypothesize that accumulative phosphorylation renders its timely autophagic degradation, likely through an autophagy receptor that binds to phosphorylated Rim4. Next, we will need to understand how Rim4-associated mRNAs are spared from degradation by presumably being released prior to phagophore engulfment. We suspect that Atg1 inhibition does not disrupt Rim4’s phosphorylation by Ime2, suggesting that actual degradation of Rim4 is needed to drive translational derepression of its mRNA targets. Finally, other Rim4-suppressed mRNA targets or autophagy substrates need to be identified to elucidate the role of autophagy in spore formation. Overall, our results demonstrate that we have uncovered a regulatory pathway controlling meiotic exit through the timed autophagic degradation of a translational repressor.
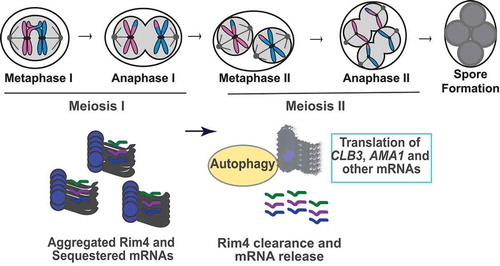
Figure 1. Model of meiotic chromosome segregation and the temporal regulation of Rim4’s autophagic degradation and release of mRNAs. Aggregated Rim4 binds CLB3, AMA1, and other mRNAs until meiosis II, when Rim4 is degraded by autophagy. The mRNAs are then translated for meiotic exit and spore formation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Wang F, Zhang R, Feng W, et al. Autophagy of an amyloid-like translational repressor regulates meiotic exit. Dev Cell. 2020;52(2):141–51 e5.
