ABSTRACT
Autophagy, in part, is controlled by the repression and activation of autophagy-related (ATG) genes. Here, we describe a new selective autophagy pathway that targets functional transcriptional regulators to control their activity. This pathway is activated in response to nitrogen starvation and recycles transcriptional activators (Msn2 and Rim15) and a repressor (Ssn2/Med13) of ATG expression. Further analysis of Ssn2/Med13 vacuolar proteolysis revealed that this pathway utilizes the core autophagic machinery. However, it is independent of known nucleophagy mechanisms, receptor proteins, and the scaffold protein Atg11. Instead, Ssn2/Med13 exits the nucleus through the nuclear pore complex (NPC) and associates with the cytoplasmic nucleoporin Gle1, a member of the RNA remodeling complex. Dbp5 and Nup159, that act in concert with Gle1, are also required for Ssn2/Med13 clearance. Ssn2/Med13 is retrieved from the nuclear periphery and degraded by Atg17-initiated phagophores anchored to the vacuole. Efficient transfer to phagophores depends on the sorting nexin heterodimer Snx4/Atg24-Atg20, which binds to Atg17, and relocates to the perinucleus following nitrogen starvation. To conclude, this pathway defines a previously undescribed autophagy mechanism that targets select transcriptional regulators for rapid vacuolar proteolysis, utilizing the RNA remodeling complex, the sorting nexin heterodimer Snx4-Atg20, Atg17, and the core autophagic machinery. It is physiologically relevant as this Snx4-assisted vacuolar targeting pathway permits cells to fine-tune the autophagic response by controlling the turnover of both positive and negative regulators of ATG transcription.
Abbreviations: AIM: Atg8 interacting motif; ATG: autophagy-related; CKM: CDK8 kinase module; IDR: intrinsically disordered region; IP6: phosphoinositide inositol hexaphosphate; NPC: nuclear pore complex; PAS: phagophore assembly site; UPS: ubiquitin-proteasomal system
Introduction
Macroautophagy (hereafter autophagy) is a controlled catabolic process critical for maintaining homeostasis and cellular survival during adverse conditions such as starvation or cytotoxic stress. Initially, identified as a mechanism to recycle superfluous cytosolic proteins [Citation1], it is apparent that autophagy is also a critical regulator of specific cargos, including misfolded proteins and aggregates, damaged organelles, lipids, and pathogens [Citation2]. As such, autophagy pathways have been reclassified as being either nonselective or selective autophagy [Citation3]. In budding yeast, nonselective autophagy is upregulated in response to starvation or cytotoxic stress and triggers the formation of autophagosomes that capture superfluous cytosolic proteins [Citation4,Citation5]. It is initiated by TORC1 (target of rapamycin kinase complex) inhibition [Citation6] that triggers a cascade of events, resulting in the pentameric Atg1 kinase complex being recruited to the phagophore assembly site (PAS). Atg1 activates the Atg17 scaffold complex [Citation7] to capture Atg9 vesicles independent of cargo which are needed to nucleate phagophores anchored to the vacuole by Vac8 [Citation8]. Upon the formation of the double-membraned phagophores, cytosolic cargos are randomly sequestered into these structures, and following closure, the autophagosome cargos are subsequently degraded by vacuolar proteolysis [Citation9,Citation10].
Selective autophagy pathways operate both in normal vegetative conditions to maintain homeostasis [Citation3] and in response to stress such as starvation and cytotoxic stress [Citation2]. They share 19 “core autophagy proteins” with nonselective autophagy and also use receptor proteins to coordinate the tethering of specific cargos, including damaged organelles and protein aggregates to Atg8 at phagophores [Citation11]. Receptor proteins also bind to the selective autophagy scaffold protein Atg11 [Citation12] resulting in the nucleation of phagophores in the direct vicinity of the cargo [Citation12,Citation13]. The relationship between Atg11, Atg17, and Atg9 changes between nutrient-rich and deplete conditions [Citation12]. In nutrient-rich conditions, receptor proteins activate and dimerize Atg11, which allows it to outcompete Atg17 for Atg9 vesicles. Furthermore, Atg17 is maintained in an inactive state by forming a constitutive Atg17–Atg31–Atg29 subcomplex [Citation8]. Upon nitrogen starvation, Atg11 is degraded by the ubiquitin-proteasome system (UPS) and the Atg1 kinase activates Atg17. These two events permit Atg17 to sequester Atg9 vesicles to nucleate phagophores independent of cargo.
Nuclear autophagy or nucleophagy is the least well understood of the selective autophagy mechanisms. Underscoring its importance, various pathologies namely cancer, and neurodegeneration are linked with perturbed nucleophagy [Citation14]. It is best characterized in yeast where macronucleophagy utilizes a receptor protein and involves the sequestration of a portion of the nucleus into autophagosomes [Citation15]. In contrast, micronucleophagy (piecemeal autophagy of the nucleus), is autophagosome independent and forms nuclear-vacuole junctions that pinch off portions of the nucleus directly into the vacuolar lumen [Citation16]. Recently, an autophagic mechanism has been described that removes defective nuclear pore complexes (NPCs) [Citation17,Citation18].
In yeast, 43 unique autophagy-related (ATG) genes have been identified that control this highly coordinated and complex process [Citation19]. Accordingly, these genes are tightly regulated at multiple levels. Recently we have shown that the Ssn8/Cnc1-Ssn3/Cdk8 kinase negatively regulates ATG8 expression within the Ume6-Rpd3 HDAC axis [Citation20]. This kinase, together with Ssn2/Med13 and Srb8/Med12, form the evolutionarily conserved CDK8 kinase module (CKM) of the Mediator complex. In budding yeast, the CKM predominantly represses transcription of a diverse set of meiosis and stress response genes [Citation21,Citation22] by interacting with DNA bound transcription factors and RNA polymerase II [Citation23–25].
Activation of genes controlled by the CKM is achieved by disrupting its association with the Mediator [Citation26]. Studies from our group revealed that this is achieved by CKM disassembly. However, we observed that the mechanisms used to disassemble the CKM is dependent upon environmental cues (outlined in ) [Citation27]. In short, oxidative stress triggers Ssn8/Cnc1/cyclin C translocation to the cytoplasm [Citation28] where it mediates stress-induced mitochondrial fission and regulated cell death (RCD) in both yeast [Citation27] and mammalian cells [Citation29–31]. Its nuclear release is dependent upon Ssn2/Med13’s destruction by the UPS [Citation32,Citation33]. In contrast, following a survival cue (nitrogen starvation), Ssn8/Cnc1 is rapidly destroyed by the UPS before its nuclear release which prevents mitochondrial fission [Citation20].
Figure 1. Ssn2/Med13 is degraded via vacuolar proteolysis following nitrogen starvation. (A) Model outlying how the cyclin-dependent kinase 8 module (CKM) of the mediator complex is disassembled following stresses that mediate cell death or survival pathways. Before stress Ssn8/Cnc1/cyclin C (CC), Ssn3/Cdk8, Ssn2/Med13 and Srb8/Med12 form the CKM that predominantly represses stress response genes (SRGs) [Citation21,Citation22]. Repression is relieved after oxidative stress and nitrogen depletion by CKM disassembly, mediated by different mechanisms. After oxidative stress, Ssn2/Med13 is destroyed by the UPS [Citation32,Citation33], which allows Ssn8/Cnc1 localization to the mitochondria where it triggers stress-induced mitochondrial fission and promotes cell death [Citation27]. In contrast, following nitrogen starvation, Ssn8/Cnc1 is rapidly destroyed by the UPS before its nuclear release to prevent mitochondrial fission [Citation20] whereas the fate of Ssn2/Med13 is the subject of this current manuscript. (B) Western blot analysis of extracts prepared from wild-type cells expressing endogenous Ssn2/Med13-9xMYC (RSY2211) resuspended in nitrogen starvation medium (SD-N) or treated with 200 ng/ml rapamycin for the indicated times. (C) As in B except that endogenous Ssn2/Med13 protein levels were monitored in ump1∆ (RSY1961, Ssn2/Med13-13xMYC) and pep4∆ prb1∆.1 (RSY2215) strains. (D) Degradation kinetics and half-life of Ssn2/Med13 protein levels obtained in B and C. Error bars indicate S.D., N = 3 of biologically independent experiments. (E) Wild-type (RSY10) or pep4∆ prb1∆.1 (RSY449) cells expressing Ssn2/Med13-GFP (pSW218) were starved for nitrogen for indicated times. For all cleavage assays, free GFP refers to the protease resistant GFP moiety that accumulates after the full-length fusion protein is degraded via the vacuole. GFP accumulation was monitored by western blot analysis using anti-GFP antibodies. An asterisk indicates a nonspecific proteolytic fragment. (F) As in E except that wild-type cells were resuspended in SD-N or treated with 0.8 mM H2O2. For all blots, Pgk1 levels were used as loading controls
![Figure 1. Ssn2/Med13 is degraded via vacuolar proteolysis following nitrogen starvation. (A) Model outlying how the cyclin-dependent kinase 8 module (CKM) of the mediator complex is disassembled following stresses that mediate cell death or survival pathways. Before stress Ssn8/Cnc1/cyclin C (CC), Ssn3/Cdk8, Ssn2/Med13 and Srb8/Med12 form the CKM that predominantly represses stress response genes (SRGs) [Citation21,Citation22]. Repression is relieved after oxidative stress and nitrogen depletion by CKM disassembly, mediated by different mechanisms. After oxidative stress, Ssn2/Med13 is destroyed by the UPS [Citation32,Citation33], which allows Ssn8/Cnc1 localization to the mitochondria where it triggers stress-induced mitochondrial fission and promotes cell death [Citation27]. In contrast, following nitrogen starvation, Ssn8/Cnc1 is rapidly destroyed by the UPS before its nuclear release to prevent mitochondrial fission [Citation20] whereas the fate of Ssn2/Med13 is the subject of this current manuscript. (B) Western blot analysis of extracts prepared from wild-type cells expressing endogenous Ssn2/Med13-9xMYC (RSY2211) resuspended in nitrogen starvation medium (SD-N) or treated with 200 ng/ml rapamycin for the indicated times. (C) As in B except that endogenous Ssn2/Med13 protein levels were monitored in ump1∆ (RSY1961, Ssn2/Med13-13xMYC) and pep4∆ prb1∆.1 (RSY2215) strains. (D) Degradation kinetics and half-life of Ssn2/Med13 protein levels obtained in B and C. Error bars indicate S.D., N = 3 of biologically independent experiments. (E) Wild-type (RSY10) or pep4∆ prb1∆.1 (RSY449) cells expressing Ssn2/Med13-GFP (pSW218) were starved for nitrogen for indicated times. For all cleavage assays, free GFP refers to the protease resistant GFP moiety that accumulates after the full-length fusion protein is degraded via the vacuole. GFP accumulation was monitored by western blot analysis using anti-GFP antibodies. An asterisk indicates a nonspecific proteolytic fragment. (F) As in E except that wild-type cells were resuspended in SD-N or treated with 0.8 mM H2O2. For all blots, Pgk1 levels were used as loading controls](/cms/asset/074f3161-0a58-4e48-953e-1b5467de46b9/kaup_a_1877934_f0001_oc.jpg)
This study reveals that the transcriptional repressor Ssn2/Med13 is degraded by vacuolar proteolysis via a previously undescribed autophagic pathway. Similar to known selective autophagic pathways, it requires the core autophagic machinery but surprisingly, does not use the known nucleophagy mechanisms or the scaffold protein Atg11. Instead, this pathway utilizes the nucleoporin Gle1, the sorting-nexin heterodimer Snx4-Atg20, and Atg17-initiated autophagosomes. Moreover, two transcriptional activators that regulate ATG expression also utilize this pathway upon nitrogen starvation. This suggests that Snx4-assisted vacuolar targeting of transcriptional regulators defines a new autophagy pathway for controlling ATG expression, that allows for fine-tuning of the autophagic response.
Results
Ssn2/Med13 is actively degraded following nitrogen starvation
We started this investigation by addressing if Ssn2/Med13 was destroyed following nitrogen starvation. Wild-type cells expressing endogenous Ssn2/Med13-9xMYC were starved for nitrogen (SD-N), and western blot analysis showed that Ssn2/Med13 protein levels decreased with a half-life of 2.6 h (). Similarly, Ssn2/Med13 was rapidly degraded in replete medium containing rapamycin, a drug that mimics nitrogen starvation by inhibiting TORC1 [Citation34] ( and S1A). As the half-life of Ssn2/Med13 is >6 h in unstressed cultures [Citation32], and SSN2/MED13 mRNA increased following 4 h in SD-N (Fig. S1B), these results indicate that Ssn2/Med13 is actively degraded following TORC1 inhibition.
Ssn2/Med13 degradation following nitrogen starvation is mediated by the vacuole
Ssn2/Med13 levels were next monitored in ump1∆, a mutant deficient in 20S proteasome assembly [Citation35], and no change in degradation kinetics was observed (). In contrast, in pep4∆ prb1∆.1, a vacuolar protease mutant [Citation1,Citation36] Ssn2/Med13 was stable in SD-N () with a half-life >15 h. This indicates that Ssn2/Med13 degradation requires vacuolar proteolysis. Confirming this, the same results were obtained in wild type, ump1∆ and pep4∆ prb1∆.1 cells harboring a low copy, functional Ssn2/Med13-3xHA plasmid [Citation33] (Fig. S1C and D). Moreover, we found that after nitrogen starvation GFP accumulated in Ssn2/Med13-GFP cleavage assays. This indicates that Ssn2/Med13-GFP is degraded in the vacuole as the compact fold of GFP renders it resistant to vacuolar hydrolases [Citation37]. Accordingly, repeating these cleavage assays in pep4∆ prb1∆.1 cells abolished the formation of free GFP and stabilized full-length Ssn2/Med13-GFP (, quantified in Fig. S1E). As anticipated from our previous studies [Citation32], Ssn2/Med13-GFP was destroyed following 0.8 mM H2O2, and no GFP accumulation was seen ( and S1E). These results show that the proteolysis machinery employed to degrade Ssn2/Med13 is dependent upon environmental cues.
To visualize Ssn2/Med13 vacuolar degradation we used live-cell imaging of endogenous Ssn2/Med13-mNeongreen in pep4∆ prb1∆.1. In SD media, Ssn2/Med13-mNeongreen is nuclear () but after 4 h in SD-N, Ssn2/Med13 accumulated in the vacuole (). The deconvolved collapsed images in also captured Ssn2/Med13-mNeongreen transitioning between these organelles. After 24 h in SD-N, Ssn2/Med13-mNeongreen is predominantly vacuolar (Fig. S2A), and similar results were obtained when endogenous Ssn2/Med13-YFP was expressed in wild-type cells treated with PMSF that blocks the activity of vacuolar serine proteases (Fig. S2B) [Citation1]. We also monitored the transition of Ssn2/Med13-mNeongreen to the vacuole in pep4∆ prb1∆.1 over time (). After 1 h Ssn2/Med13 foci are seen (yellow arrow) localizing to the nuclear membrane. After 3 h Ssn2/Med13 is visualized in the vacuole (pink arrows), with the vacuolar pool being easier to capture at later timepoints (orange arrow, and see ). These results are consistent with a model in which nitrogen starvation triggers Ssn2/Med13 vacuolar proteolysis.
Figure 2. Ssn2/Med13 translocates from the nucleus to the vacuole in nitrogen starvation. (A) Endogenous Ssn2/Med13-mNeongreen localization was monitored in pep4∆ prb1∆.1 cells (RSY2305) expressing Nab2-mCherry (a nuclear marker) before (growing in SD) and after 4 h in SD-N. Representative single plane images are shown. (B) As in A, except that cells expressed a vacuolar marker (Vph1-mCherry). (C) As in B, except that slices were taken through the whole cell which were then collapsed and deconvolved. Representative deconvolved images are shown. Scale bar: 5 µm. (D) Fluorescence microscopy of Ssn2/Med13-mNeonGreen for the timepoints indicated
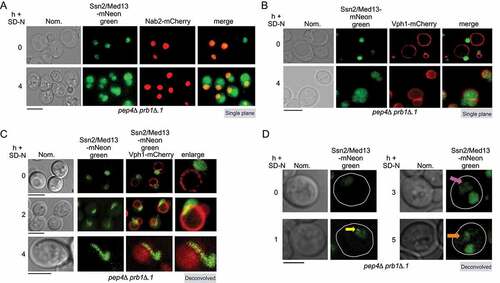
Figure 3. Ssn2/Med13 requires the core autophagy machinery for vacuolar degradation. (A) Schematic depicting the five stages of autophagy and the major autophagy protein complexes associated with each stage. (B) Western blot analysis of extracts prepared from wild-type (RSY2211), atg1∆ (RSY2214), and atg8∆ (RSY2231) cells expressing endogenous Ssn2/Med13-9xMYC resuspended in SD-N media for the indicated times. (C) Degradation kinetics and half-life of Ssn2/Med13 protein levels obtained in B. Error bars indicate S.D., N = 3 of biologically independent experiments. (D) The indicated mutants expressing Ssn2/Med13-GFP (pSW218) were starved for nitrogen for the indicated times and accumulation of free GFP monitored by western blot analysis using anti-GFP antibodies. For all experiments, Pgk1 protein levels were used as a loading control. (E and F) Endogenous Ssn2/Med13-mNeongreen localization was monitored in atg8∆ cells (RSY2307) following SD-N. Vacuoles were visualized with either CMAC (E) or using a vacuolar marker, Vph1-mCherry (F). Representative deconvolved or single-plane images are shown. Scale bar: 5 µm
Ssn2/Med13 degradation requires core autophagy machinery
Next, we investigated if Ssn2/Med13 degradation was dependent upon proteins required for autophagosome biogenesis (outlined in ). Ssn2/Med13 was significantly stabilized following nitrogen starvation in mutants defective in induction (atg1∆) and phagophore assembly (atg8∆) [Citation38] (half-lives >15 h, ). Likewise, GFP accumulation from Ssn2/Med13-GFP was abolished in autophagy mutants (vps34∆, atg9∆, atg4∆ and vam3∆) required for different stages of the pathway ( and S1E). We did observe some turnover of full-length Ssn2/Med13-GFP which we attribute to a combination of Ssn2/Med13 being expressed from the ADH1 promotor rather than the endogenous locus used for degradation assays, and UPS activity. Consistent with this, Ssn2/Med13-GFP was more stable in atg1∆ ump1∆ than atg1∆ cells (Fig. S2C). Therefore, both the vacuole and core autophagy proteins are needed for Ssn2/Med13 degradation.
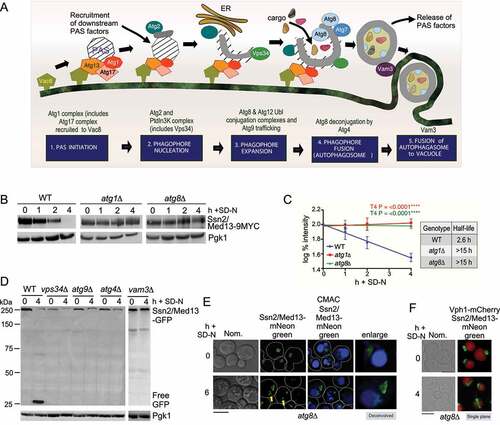
Ssn2/Med13 colocalizes with the autophagy machinery after nitrogen starvation
If the degradation of Ssn2/Med13 requires the autophagic machinery, we would expect Ssn2/Med13 to accumulate outside of the vacuole in mutants defective in autophagic cargo delivery. Consistent with this model, we observed Ssn2/Med13 accumulating on the surface of the vacuole in atg8∆ mutants (yellow arrow, ). Here it colocalizes with Atg2-mCherry and Atg7-mCherry, two components of the phagophore ( and S2D). In vam3∆, a fusion deficient t-SNARE mutant [Citation39,Citation40], Ssn2/Med13 colocalizes with Atg8-mCherry which conjugates to inner autophagosome membranes [Citation41] (). Using this series of colocalization studies in null mutants we were able to demonstrate the sequential movement of Ssn2/Med13 from the nucleus (Nab2), to the PAS (Atg2 and Atg7), followed by sequestration within autophagosomes (Atg8), and ultimately degraded within the vacuole (Vph1). Despite this, Ssn2/Med13 and Atg8 do not interact in a two-hybrid assay (Fig. S2E, and see Fig. S2F for Y2H control of Atg8 interaction with Atg7). Furthermore, co-immunoprecipitation experiments of endogenously tagged proteins suggest that Ssn2/Med13 and Atg8 do not interact in pep4∆ prb1∆.1 cells. In these experiments, the GFP-Atg8 affinity isolation of Ssn2/Med13-HA was not significantly above background (Fig. S2G). This result was surprising as Ssn2/Med13 contains 33 potential Atg8-interacting motifs (AIMs) including 13 located within its large 700 amino acid intrinsically disordered region (IDR) (Fig. S2H). However, while AIMs are well defined, potential motifs require experimental confirmation [Citation42–44]. Given the large number found in Ssn2/Med13, coupled with the negative Y2H and co-immunoprecipitation results, we felt that this approach was outside of the scope of this report. Moreover, the results are more consistent with a model in which another mechanism targets Ssn2/Med13 to Atg8.
Figure 4. Ssn2/Med13 colocalizes with autophagic machinery following nitrogen starvation. (A) Localization of endogenous Ssn2/Med13-mNeongreen and Atg2-mCherry were monitored in atg8∆ cells (RSY2547) following 24 h of SD-N. (B) Same as in A except endogenous Atg7-mCherry colocalization with Ssn2/Med13-mNeongreen is monitored in atg8∆ cells (RSY2545). A 3D image is shown in the (bottom panel). (C) To visualize Ssn2/Med13 within autophagosomes, Ssn2/Med13-mNeongreen Atg8-mCherry (CUP-mCherry-Atg8) colocalization was monitored in vam3∆ cells (RSY2535). Representative deconvolved or single-plane images are shown. Scale bar: 5 µm
Ssn2/Med13 degradation does not use known selective autophagy pathways
Selective autophagy relies on cargo receptors that bind to both Atg8 and cargos [Citation2,Citation45]. Deletions of known receptor proteins (Cue5, Atg19, Atg34, Atg36, Atg32), including the newly defined Ubx5 [Citation46] and the redundant Atg19 Atg34 receptors [Citation47] did not influence the autophagic degradation of Ssn2/Med13 (Fig. S3A, B, and Fig. S1E). Moreover, the receptors required for micro-and macro- nucleophagy (Nvj1, Atg39, and Atg40, ) [Citation15,Citation48] do not mediate Ssn2/Med13 degradation (). We also did not observe Ssn2/Med13 forming large protein aggregates after nitrogen starvation although very occasionally small foci were observed at early time points (see ). Consistent with this, deletion of Hsp42 or Hsp104, two chaperones known for sequestration of protein aggregates [Citation49–51] did not affect Ssn2/Med13 degradation following nitrogen starvation (Fig. S3C). Taken together, these results show that Ssn2/Med13 vacuolar degradation is independent of the known receptor proteins.
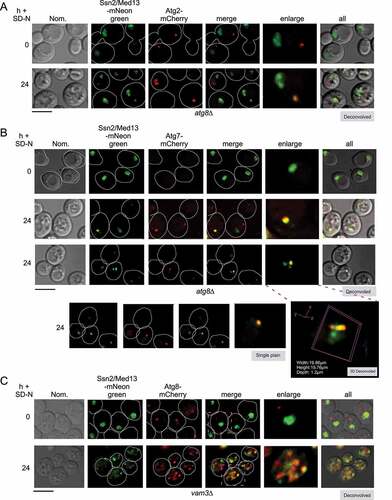
Figure 5. The autophagic degradation of Ssn2/Med13 requires the nucleoporin Gle1 and is independent of known nucleophagy pathways. (A) Western blot analysis of Ssn2/Med13-GFP cleavage assays after 4 h nitrogen depletion in micro-nucleophagy (nvj1∆, RSY2106) and macro-nucleophagy (atg39∆ atg40∆, RSY2123) mutants. (B) Same as A except Ssn2/Med13-GFP cleavage assays were performed in atg11∆ (RSY2248), atg17∆ (RSY2104), and vac8∆ cells (RSY2097). (C) Co-immunoprecipitation analysis of endogenous Gle1-GFP and Ssn2/Med13-3 HA. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 cells expressing endogenous Gle1-GFP (RSY2423) and Ssn2/Med13-3 HA (pKC801, lanes 1 and 4) or a vector control (lane 2). pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 was immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown. Western blot analysis of Gle1-GFP proteins in the whole cell lysates for the three conditions tested is shown (input – bottom panel). (D) Map of Ssn2/Med13 depicting different structural regions and known interacting proteins (upper panel). Different colors represent different regions of the protein and structural regions are denoted by amino acid positions. Ssn2/Med13-Gle1 Y2H analysis. Y2H Gold cells (RSY2000) harboring Gal4-BD-Gle1 and the indicated Gal4-AD-Ssn2/Med13 subclone or empty vector control were streaked on medium selecting for plasmid maintenance (left) or induction of the ADE2 and HIS3 reporter genes (right) by Y2H interaction (lower panel). See fig. S5A, B for western blot analysis of the different constructs. (E) Fluorescence microscopy of endogenous Ssn2/Med13-mNeongreen and Gle1-RedStar (RSY2450) starved for nitrogen for 2 h in pep4∆ prb1∆.1. The yellow arrow is pointing to Ssn2/Med13 in the vacuole whereas the blue arrow shows colocalization. OE represents an over-exposed image to better show the colocalization of Gle1 and Ssn2/Med13 (pink arrows) Scale: 5 μm. (F) Fluorescence microscopy of Ssn2/Med13-mNeongreen localization in the Gle1 auxin-inducible degron (Gle1-AID) strain (RSY2473) expressing Nab2-mCherry before and after SD-N. An 8 h time point was used as the strain background is BY4741 which is not as sensitive to environmental stress as W303a. Scale: 5 µm. (G) Ssn2/Med13-GFP cleavage assays in the auxin-inducible deletion strains indicated. Cells expressing Ssn2/Med13-GFP (pSW320) were treated with 250 μM auxin for 30 m before proceeding with autophagic cleavage assays in SD-N (left panel). The GLE1 cis alleles were expressed in the GLE1::AID strain (RSY2456) harboring either pSW3345, or pSW2571 (IP6 and Dbp5 cis alleles respectively). For all blots, Pgk1 levels were used as loading controls
![Figure 5. The autophagic degradation of Ssn2/Med13 requires the nucleoporin Gle1 and is independent of known nucleophagy pathways. (A) Western blot analysis of Ssn2/Med13-GFP cleavage assays after 4 h nitrogen depletion in micro-nucleophagy (nvj1∆, RSY2106) and macro-nucleophagy (atg39∆ atg40∆, RSY2123) mutants. (B) Same as A except Ssn2/Med13-GFP cleavage assays were performed in atg11∆ (RSY2248), atg17∆ (RSY2104), and vac8∆ cells (RSY2097). (C) Co-immunoprecipitation analysis of endogenous Gle1-GFP and Ssn2/Med13-3 HA. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 cells expressing endogenous Gle1-GFP (RSY2423) and Ssn2/Med13-3 HA (pKC801, lanes 1 and 4) or a vector control (lane 2). pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 was immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown. Western blot analysis of Gle1-GFP proteins in the whole cell lysates for the three conditions tested is shown (input – bottom panel). (D) Map of Ssn2/Med13 depicting different structural regions and known interacting proteins (upper panel). Different colors represent different regions of the protein and structural regions are denoted by amino acid positions. Ssn2/Med13-Gle1 Y2H analysis. Y2H Gold cells (RSY2000) harboring Gal4-BD-Gle1 and the indicated Gal4-AD-Ssn2/Med13 subclone or empty vector control were streaked on medium selecting for plasmid maintenance (left) or induction of the ADE2 and HIS3 reporter genes (right) by Y2H interaction (lower panel). See fig. S5A, B for western blot analysis of the different constructs. (E) Fluorescence microscopy of endogenous Ssn2/Med13-mNeongreen and Gle1-RedStar (RSY2450) starved for nitrogen for 2 h in pep4∆ prb1∆.1. The yellow arrow is pointing to Ssn2/Med13 in the vacuole whereas the blue arrow shows colocalization. OE represents an over-exposed image to better show the colocalization of Gle1 and Ssn2/Med13 (pink arrows) Scale: 5 μm. (F) Fluorescence microscopy of Ssn2/Med13-mNeongreen localization in the Gle1 auxin-inducible degron (Gle1-AID) strain (RSY2473) expressing Nab2-mCherry before and after SD-N. An 8 h time point was used as the strain background is BY4741 which is not as sensitive to environmental stress as W303a. Scale: 5 µm. (G) Ssn2/Med13-GFP cleavage assays in the auxin-inducible deletion strains indicated. Cells expressing Ssn2/Med13-GFP (pSW320) were treated with 250 μM auxin for 30 m before proceeding with autophagic cleavage assays in SD-N (left panel). The GLE1 cis alleles were expressed in the GLE1::AID strain (RSY2456) harboring either pSW3345, or pSW2571 (IP6 and Dbp5 cis alleles respectively). For all blots, Pgk1 levels were used as loading controls](/cms/asset/b886c2ce-3d9a-4d73-9ee6-220ca22dd39e/kaup_a_1877934_f0005_oc.jpg)
Ssn2/Med13 degradation requires the Atg17 scaffold complex
Selective autophagy of excess or damaged cellular components in physiological conditions requires the scaffold protein Atg11 to build phagophores at the site of the cargo [Citation52]. During nitrogen starvation, Atg11 is destroyed by the UPS [Citation12] and Atg17 tethers Atg1 and other Atg proteins to the PAS [Citation53] located at the vacuole [Citation9]. Analysis of the autophagic degradation of Ssn2/Med13-GFP ( and S1E) and the degradation of endogenous Ssn2/Med13-9xMYC (Fig. S3D and E) revealed that Atg17 but not Atg11 is required for Ssn2/Med13 destruction. These results suggest that the autophagic degradation of Ssn2/Med13 is mediated by phagophores tethered to the vacuole. Consistent with this, we observed a significant reduction in the appearance of GFP in vac8∆ cells ( and S1E). As one role of Vac8 is to tether the Atg17-initiated PAS complex to the vacuole [Citation9], these results suggest that Ssn2/Med13 is transported to Atg17-built phagophores located on the vacuole membrane for autophagic destruction.
The cytoplasmic nucleoporin Gle1 associates with Ssn2/Med13 after nitrogen starvation
To further define components of the Ssn2/Med13 degradative pathway, a pull-down approach of Ssn2/Med13-3xHA followed by mass-spectroscopy was used (Fig. S4A and B and Table SI for a list of candidate interactors). The conserved, essential nucleoporin Gle1, that localizes to the cytoplasmic filament of the nuclear pore complex (NPC) was identified [Citation54]. Co-immunoprecipitation analysis revealed that Ssn2/Med13-3xHA and endogenous Gle1-GFP interacted following 1 h SD-N, confirming this association (). Y2H analysis showed that the large central IDR of Ssn2/Med13, which provides a flexible interaction hub for multiple partners [Citation33,Citation55], interacts with Gle1 ( and S5). Live-cell imaging of endogenous Ssn2/Med13-mNeongreen and Gle1-RedStar show Ssn2/Med13 moves from being diffuse nuclear to associating with the punctate Gle1-RedStar that surrounds the nucleus after nitrogen starvation ().
Gle1, Dbp5, and Nup159 are required for Ssn2/Med13 nuclear export
Gle1 is an essential protein [Citation56], therefore, the auxin-inducible degron (AID) system was used to reduce Gle1 protein levels (Fig. S4C). Deletion of Gle1 does not affect autophagy (monitored using GFP-Atg8 cleavage assays) and Gle1 does not associate with Atg8 using Y2H analysis (Fig. S4D, E, and S5). Significantly, in the auxin-treated GLE1::AID degron Ssn2/Med13 was retained in the nucleus () and the autophagic degradation of Ssn2/Med13-GFP was significantly decreased following nitrogen starvation ( – left panel).
Gle1, the DEAD-box protein Dbp5, and phosphoinositide inositol hexaphosphate (IP6) are members of the RNA remodeling complex [Citation57–61]. By binding to Nup159, this complex is positioned toward the NPC’s central channel, permitting it to efficiently capture proteins as they exit the NPC [Citation62]. Mutation of the cis-acting alleles IP6 and Dbp5 on Gle1 [Citation63] revealed that only Dbp5 is required for efficient autophagic cleavage degradation of Ssn2/Med13 ( middle panel). Also, autophagic degradation of Ssn2/Med13 was significantly decreased in the NUP159::AID strain (-right panel). As the RNA remodeling complex plays additional Nup159-independent cytoplasmic roles in RNA translation [Citation64–66], these results support a model in which Nup159-dependent positioning of Gle1 and Dbp5 in the central channel is important for the final stages of Ssn2/Med13 nuclear export. Intriguingly, the known β karyopherins, (Crm1, Msn5, Cse1 and Los1 [Citation67]) are not required for Ssn2/Med13 transport across the NPC (Fig. S4 F-I). Dbp5 has also been found in the nucleus [Citation68], suggesting the possibility that it might play an additional role in enabling Ssn2/Med13 nuclear export. One caveat to this model, is that these factors possess overlapping activities, which has been demonstrated for other cargos [Citation69].
The sorting nexin heterodimer Snx4-Atg20 is required for efficient autophagic degradation of Ssn2/Med13
To further define components of this new autophagy mechanism, null alleles of candidate proteins from the mass spectroscopy were screened using Ssn2/Med13-GFP cleavage assays. In cells deleted for the conserved sorting nexin Snx4 (Atg24), we found that free GFP was significantly reduced ( and S1E). Snx4 forms distinct heterodimer complexes, with either Snx41 or Atg20 (Snx42), which mediate retrograde trafficking of cargo from the vacuole and endosomes to the Golgi. This role maintains homeostasis and is dispensable for nonselective autophagy [Citation70–73]. In addition, Snx4-Atg20 promotes selective autophagy of mitochondria and pexoxisomes and is required for other selective autophagy pathways including the cytoplasm to vacuole pathway (CVT), ribophagy, and proteaphagy [Citation74–78].
Figure 6. The sorting nexin heterodimer, Snx4-Atg20 is required for efficient autophagic degradation of Ssn2/Med13. (A) Western blot analysis of Ssn2/Med13-GFP cleavage assays after 4 h nitrogen starvation in wild-type and snx4∆ (RSY2272). (B) Co-immunoprecipitation analysis of GFP-Snx4 and Ssn2/Med13-3 HA. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 cells expressing GFP-Snx4 (RSY2299) and Ssn2/Med13-3 HA (pKC801, lanes 1 and 2) or a vector control (lane 4). Pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 or Snx4 was immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown (input – bottom panel). (C) Western blot analysis of extracts prepared from wild-type (RSY2211), snx4∆ (RSY2276), atg20∆ (RSY2277), and snx41∆ (RSY2394) expressing endogenous Ssn2/Med13-9MYC resuspended in SD-N for the indicated times. (D) Degradation kinetics and half-lives of Ssn2/Med13 protein levels obtained in C. Error bars indicate S.D., N = 3 of biologically independent experiments. (E) Fluorescence microscopy of endogenous Ssn2/Med13-mNeongreen localization in snx4∆ pep4∆ prb1∆.1 (RSY2324) expressing the vacuole marker Vph1-mCherry. Cells were visualized before (SD) and after 2 h of SD-N treatment and representative single plane images are shown. Scale: 5 µm. (F) As in E except that endogenous Ssn2/Med13-mNeongreen localization was followed in nitrogen-starved pep4∆ prb1∆.1 cells. Representative single plane images of the results are shown. Bar: 5 µm. (G) Quantification of Ssn2/Med13-mNeongreen accumulation in vacuoles obtained from results in D and E. 100 cells counted per sample. N = 3 biological samples. **** P = >0.0001. For all blots, Pgk1 levels were used as loading controls
![Figure 6. The sorting nexin heterodimer, Snx4-Atg20 is required for efficient autophagic degradation of Ssn2/Med13. (A) Western blot analysis of Ssn2/Med13-GFP cleavage assays after 4 h nitrogen starvation in wild-type and snx4∆ (RSY2272). (B) Co-immunoprecipitation analysis of GFP-Snx4 and Ssn2/Med13-3 HA. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 cells expressing GFP-Snx4 (RSY2299) and Ssn2/Med13-3 HA (pKC801, lanes 1 and 2) or a vector control (lane 4). Pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 or Snx4 was immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown (input – bottom panel). (C) Western blot analysis of extracts prepared from wild-type (RSY2211), snx4∆ (RSY2276), atg20∆ (RSY2277), and snx41∆ (RSY2394) expressing endogenous Ssn2/Med13-9MYC resuspended in SD-N for the indicated times. (D) Degradation kinetics and half-lives of Ssn2/Med13 protein levels obtained in C. Error bars indicate S.D., N = 3 of biologically independent experiments. (E) Fluorescence microscopy of endogenous Ssn2/Med13-mNeongreen localization in snx4∆ pep4∆ prb1∆.1 (RSY2324) expressing the vacuole marker Vph1-mCherry. Cells were visualized before (SD) and after 2 h of SD-N treatment and representative single plane images are shown. Scale: 5 µm. (F) As in E except that endogenous Ssn2/Med13-mNeongreen localization was followed in nitrogen-starved pep4∆ prb1∆.1 cells. Representative single plane images of the results are shown. Bar: 5 µm. (G) Quantification of Ssn2/Med13-mNeongreen accumulation in vacuoles obtained from results in D and E. 100 cells counted per sample. N = 3 biological samples. **** P = >0.0001. For all blots, Pgk1 levels were used as loading controls](/cms/asset/4ef4ea61-3c91-4f49-a6a9-8bb99487c956/kaup_a_1877934_f0006_oc.jpg)
Co-immunoprecipitation analysis confirmed the interaction between Ssn2/Med13 and Snx4 in SD-N (). Ssn2/Med13 degradation assays revealed that Snx4 and Atg20, but not Snx41, mediate Ssn2/Med13 autophagic degradation (). The half-life of Ssn2/Med13 in these mutants was 6.5 and 7.0 h, respectively, compared to >15 h seen in core autophagic mutants. Cytosolic and vacuolar Ssn2/Med13-mNeongreen levels were drastically reduced in snx4∆ pep4∆ prb1∆.1 mutants compared to the pep4∆ prb1∆.1 control (). These data demonstrate that the Snx4-Atg20 heterodimer is required for maximal efficient Ssn2/Med13 autophagic degradation, but in its absence, limited autophagic degradation of Ssn2/Med13 still can occur.
Snx4 localizes to the nuclear periphery to transport Ssn2/Med13 to autophagosomes
Snx4 binds to the scaffold protein Atg17 [Citation74], whose major autophagic role is binding the Atg1 kinase to the PAS [Citation10]. Here we report that both Snx4 and Atg17 mediate Ssn2/Med13 degradation ( and ). This suggests a model in which upon nuclear export Snx4 recognizes Ssn2/Med13 and delivers it to the growing phagophore by Snx4-Atg17 association. Consistent with this, Ssn2/Med13-3xHA and endogenous Atg17-GFP co-immunoprecipitated following 2 h in SD-N. This interaction was drastically decreased in the absence of Snx4 (). Also, endogenous Atg17-RedStar colocalized with Ssn2/Med13-mNeongreen after nitrogen starvation (Fig. S3F). This suggests that Snx4-Atg17 interaction promotes the efficient recruitment of Ssn2/Med13 to autophagosomes. Quantitative colocalization analysis of GFP-Snx4 with the nuclear marker Nab2-mCherry, showed that nitrogen depletion triggers a ~ 10-fold increase in perinuclear GFP-Snx4 foci (, S6A and B). Moreover, Ssn2/Med13-mNeongreen colocalized with both perinuclear and cytosolic Snx4 foci in SD-N ( and S6C). Together, this suggests that Snx4 localizes to the nuclear periphery to retrieve and transport Ssn2/Med13 to phagophores via Atg17 association.
Figure 7. Snx4 localizes to the nuclear periphery to retrieve Ssn2/Med13. (A) Co-immunoprecipitation analysis of endogenous Atg17-GFP and Ssn2/Med13-3 HA in the presence and absence of Snx4. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 (RSY2395) or snx4∆ pep4∆ prb1∆.1 cells (RSY2396) expressing endogenous Atg17-GFP and Ssn2/Med13-3 HA (pKC801, lanes 1, 2, and 6) or a vector control (lanes 4 & 5). Pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 and Atg17 were immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown (input – bottom panel). (B) Representative images showing perinuclear and perivacuolar localization of GFP-Snx4 in wild-type expressing Nab2-mCherry (nuclear marker) before and after nitrogen starvation. Bar: 5 μm. The number of perinuclear foci was counted (N = 2) before and after nitrogen starvation. At least 100 cells were counted per sample. Data are the percentage of perinuclear foci among the total number of foci. Scale: 5 µm. (C) Fluorescence microscopy of pep4∆ prb1∆.1 cells expressing endogenous Ssn2/Med13-mNeongreen and mCherry-Snx4 (RSY2424) before and after nitrogen depletion. Hoechst staining was used to visualize the nucleus. Representative deconvolved images are shown. Bar: 5 μm. (D) GFP-Snx4 and endogenous Gle1-RedStar colocalize in wild-type cells (RSY2451) following nitrogen starvation. Representative images are shown. Bar: 5 μm
![Figure 7. Snx4 localizes to the nuclear periphery to retrieve Ssn2/Med13. (A) Co-immunoprecipitation analysis of endogenous Atg17-GFP and Ssn2/Med13-3 HA in the presence and absence of Snx4. Whole cell lysates were immunoprecipitated with the antibodies shown from nitrogen-starved pep4∆ prb1∆.1 (RSY2395) or snx4∆ pep4∆ prb1∆.1 cells (RSY2396) expressing endogenous Atg17-GFP and Ssn2/Med13-3 HA (pKC801, lanes 1, 2, and 6) or a vector control (lanes 4 & 5). Pep4∆ prb1∆.1 cells expressing Ssn2/Med13-3 HA alone (lane 3) was included as a control. [] represents no antibody control. For input controls Ssn2/Med13 and Atg17 were immunoprecipitated from whole cell lysates with the indicated antibodies for the three conditions tested is shown (input – bottom panel). (B) Representative images showing perinuclear and perivacuolar localization of GFP-Snx4 in wild-type expressing Nab2-mCherry (nuclear marker) before and after nitrogen starvation. Bar: 5 μm. The number of perinuclear foci was counted (N = 2) before and after nitrogen starvation. At least 100 cells were counted per sample. Data are the percentage of perinuclear foci among the total number of foci. Scale: 5 µm. (C) Fluorescence microscopy of pep4∆ prb1∆.1 cells expressing endogenous Ssn2/Med13-mNeongreen and mCherry-Snx4 (RSY2424) before and after nitrogen depletion. Hoechst staining was used to visualize the nucleus. Representative deconvolved images are shown. Bar: 5 μm. (D) GFP-Snx4 and endogenous Gle1-RedStar colocalize in wild-type cells (RSY2451) following nitrogen starvation. Representative images are shown. Bar: 5 μm](/cms/asset/5df5f8ba-a50d-41b2-a165-883c0d9a7fb2/kaup_a_1877934_f0007_oc.jpg)
The above model predicts that Snx4 and Gle1 interact whilst “handing-off” Ssn2/Med13 from the NPC to the sorting nexin complex. Live-cell imaging showed that perinuclear Snx4 foci are adjacent to, or colocalize with, Gle1 in unstressed cultures. After nitrogen starvation, an increased number of perinuclear Snx4 foci colocalize with Gle1 ( and S6D). This is consistent with the model that Snx4 localizes to the NPC to retrieve Ssn2/Med13.
Snx4 specifically targets Ssn2/Med13 for autophagic degradation
To further understand the sequential stages of this autophagy pathway, we next asked if Ssn2/Med13 nuclear localization was required for retrieval and transport by Snx4. To address this, we fused Ssn2/Med13-GFP to the N terminus of Crn1, a protein associated with actin rafts, which relocalized Ssn2/Med13 to the plasma membrane [Citation79] (). Autophagic degradation assays with Crn1-Ssn2/Med13-GFP showed that GFP accumulated in wild-type cells, and this was mostly dependent on Snx4 (). This suggests that Ssn2/Med13 can be targeted for Snx4-assisted autophagic destruction even when located outside the nucleus. To confirm specificity, we revealed that free GFP accumulation in Pgk1-GFP cleavage assays, a well known nonselective autophagic substrate [Citation80], is Snx4 independent (Fig. S7A). This further illustrates that Snx4 is dispensable for nonselective autophagy but mediates many forms of selective autophagy.
Figure 8. The BAR domain of Snx4 interacts with the C-terminal region of Ssn2/Med13. (A) Fluorescence microscopy of Crn1-Ssn2/Med13-GFP (pSW288) in wild-type cells growing in SD. Hoechst staining was used to visualize the nucleus. Bar: 5 μm. (B) Western blot analysis of Ssn2/Med13-GFP (pSW218) or Crn1-Ssn2/Med13-GFP (pSW288) cleavage assays performed in wild-type or snx4∆ cells following nitrogen starvation. (C) Western blot analysis of endogenous Snx4-6 HA (pSH30) degradation assays performed in wild-type cells following nitrogen starvation. (D) Western blot analysis of endogenous Gle1-FLAG (RSY2456) degradation assays following nitrogen starvation. (E) Western blot analysis of Ssn2/Med13-GFP (pSW218) cleavage assays performed in wild-type cells. (F) Y2H Gold cells harboring Gal4-BD-Snx4 and the indicated Gal4-AD-Ssn2/Med13 construct or vector control were plated on medium selecting for plasmid maintenance (-LEU, -TRP) (left) or interaction by induction of the ADE2 and HIS3 reporter genes (right). See Figure S5 for western blot analysis of the different constructs. (G) Predicted structural analysis of the Ssn2/Med13 tail region using Phyre2 plot analysis of this region [Citation81]. Y2H analysis of full-length Gal4-BD-Snx4 (pSH8) and Gal4-AD-Ssn2/Med13 subclones containing either the first (pSH18) or second (pSH19) domain region of the Ssn2/Med13 tail. Cells were streaked on medium selecting for plasmid maintenance (left) or induction of reporter genes (right) by Y2H interaction. (H) Map of Snx4 depicting known domains (left panel). Y2H analysis of Snx4 PX (pSH16) and BAR binding domain constructs (pSH17) with the Ssn2/Med13907−1163AD construct and empty vector. (I) Cells were streaked on medium selecting for plasmid maintenance (left) or induction of reporter genes (right) by Y2H interaction (right panel). (J) Same as in F expect Y2H Gold cells harboring Gal4-BD-Atg20 (pSH13) and the indicated Gal4-AD-Ssn2/Med13 construct or vector control were plated on medium selecting for plasmid maintenance (-LEU, -TRP) (left) or interaction by induction of the ADE2 and HIS3 reporter genes (right). (K) Map of Ssn2/Med13 structure depicting interactive regions between indicated proteins. For all blots, Pgk1 levels were used as loading controls
![Figure 8. The BAR domain of Snx4 interacts with the C-terminal region of Ssn2/Med13. (A) Fluorescence microscopy of Crn1-Ssn2/Med13-GFP (pSW288) in wild-type cells growing in SD. Hoechst staining was used to visualize the nucleus. Bar: 5 μm. (B) Western blot analysis of Ssn2/Med13-GFP (pSW218) or Crn1-Ssn2/Med13-GFP (pSW288) cleavage assays performed in wild-type or snx4∆ cells following nitrogen starvation. (C) Western blot analysis of endogenous Snx4-6 HA (pSH30) degradation assays performed in wild-type cells following nitrogen starvation. (D) Western blot analysis of endogenous Gle1-FLAG (RSY2456) degradation assays following nitrogen starvation. (E) Western blot analysis of Ssn2/Med13-GFP (pSW218) cleavage assays performed in wild-type cells. (F) Y2H Gold cells harboring Gal4-BD-Snx4 and the indicated Gal4-AD-Ssn2/Med13 construct or vector control were plated on medium selecting for plasmid maintenance (-LEU, -TRP) (left) or interaction by induction of the ADE2 and HIS3 reporter genes (right). See Figure S5 for western blot analysis of the different constructs. (G) Predicted structural analysis of the Ssn2/Med13 tail region using Phyre2 plot analysis of this region [Citation81]. Y2H analysis of full-length Gal4-BD-Snx4 (pSH8) and Gal4-AD-Ssn2/Med13 subclones containing either the first (pSH18) or second (pSH19) domain region of the Ssn2/Med13 tail. Cells were streaked on medium selecting for plasmid maintenance (left) or induction of reporter genes (right) by Y2H interaction. (H) Map of Snx4 depicting known domains (left panel). Y2H analysis of Snx4 PX (pSH16) and BAR binding domain constructs (pSH17) with the Ssn2/Med13907−1163AD construct and empty vector. (I) Cells were streaked on medium selecting for plasmid maintenance (left) or induction of reporter genes (right) by Y2H interaction (right panel). (J) Same as in F expect Y2H Gold cells harboring Gal4-BD-Atg20 (pSH13) and the indicated Gal4-AD-Ssn2/Med13 construct or vector control were plated on medium selecting for plasmid maintenance (-LEU, -TRP) (left) or interaction by induction of the ADE2 and HIS3 reporter genes (right). (K) Map of Ssn2/Med13 structure depicting interactive regions between indicated proteins. For all blots, Pgk1 levels were used as loading controls](/cms/asset/27f5bbe9-c52b-4a6e-ab16-2303aa1471ce/kaup_a_1877934_f0008_oc.jpg)
We next addressed whether Gle1 and Snx4 remain part of the Ssn2/Med13 complex delivered to the vacuole. Endogenous Snx4-6HA () and Gle1-FLAG () were not degraded following nitrogen starvation, suggesting that neither protein is incorporated into autophagosomes or degraded by the vacuole. Importantly, by monitoring the timing of GFP accumulation in Ssn2/Med13-GFP cleavage assays, we observed that the earliest detection of GFP occurred after 2 h of nitrogen depletion when both Gle1 and Snx4 are present (). These data support the model that Gle1 and Snx4 mediate Ssn2/Med13 delivery to the autophagosome, but these proteins themselves are not vacuolar substrates.
Snx4 BAR domains interact with the C-terminal domain of Ssn2/Med13
To further understand the interaction between Gle1, Snx4, and Ssn2/Med13, we used Y2H analysis to ask if Snx4 and Gle1 associate with the same or different domains of Ssn2/Med13. The results show that Snx4 interacts with the structured C-terminal tail domain of Ssn2/Med13 ( and S5). Phyre2 plot analysis of this region [Citation81] revealed two potential domains of which only one, Ssn2/Med13[907–1163] interacted with Snx4 (). Therefore Snx4 interacts with a previously undescribed region of Ssn2/Med13 that lies adjacent to the Gle1 interaction domain. Moreover, as Snx4 is not a nuclear protein, this interaction may be direct and define a new role for Snx4 in transporting nuclear proteins.
Snx4 is a conserved member of the SNX-BAR subfamily of sorting nexin proteins [Citation82]. Common to all SNX family members it contains a phosphoinositide-binding phox homology (PX) domain, which bind phosphatidylinositol 3-phosphate enriched endosomal membranes. It also contains two BAR (Bin/|Amphiphysin/Rvs) domains that bind to curved membranes upon dimerization [Citation78,Citation83] (). Y2H interaction analyses between these Snx4 domains and Ssn2/Med13[907–1163] indicated that the Ssn2/Med13-Snx4 interaction occurs through the BAR domain region (). These results indicate that the Snx4 BAR domains recognize cargos and, dimerization partners.
Atg20 interacts with a different region of Ssn2/Med13
We also used Y2H analysis to ask if Ssn2/Med13 interacts with Atg20, the heterodimer partner of Snx4 that is required for efficient degradation of Ssn2/Med13 (, D). The results show that Atg20 also interacts with a different region of Ssn2/Med13, overlapping with the region that binds to Gle1, lying adjacent to the Snx4 binding region (, and S5D). Taken together, these data support our model that Ssn2/Med13 first exits the nucleus via Gle1 and then is released from NPC to the Snx4-Atg20 heterodimer for rapid delivery to the autophagic machinery. These Y2H results demonstrate a sequential hand-off mechanism from Gle1 to the Snx4-Atg20 heterodimer as Gle1 and Atg20 bind to the same region of Ssn2/Med13.
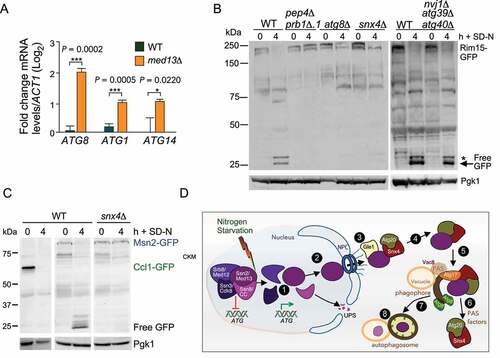
Ssn2/Med13 negatively regulates the transcription of a subset of ATG genes
We next explored if this newly described autophagy pathway affects viability during nitrogen starvation. Consistent with previous studies [Citation84], efficient survival in prolonged nitrogen starvation conditions requires Snx4 (Fig. S7B). Snx4 is dispensable for nonselective autophagy but is required for many selective autophagy pathways [Citation71]. This suggests that the role of Snx4 is essential for cellular adaptation and survival in prolonged starvation conditions. Survival during periods of nutrient depletion requires the upregulation of ATG genes. As Ssn2/Med13 destruction following H2O2 relieves repression on stress-responsive genes [Citation32], we asked if a similar strategy was used following nitrogen starvation. RT-qPCR analysis of ATG mRNA levels in unstressed wild-type and Ssn2/Med13∆ cells showed a ~ fourfold increase in ATG8 mRNA levels () that was mirrored by Atg8 protein levels (Fig. S7C). Furthermore, ATG1 and ATG14 mRNA levels were also elevated (), indicating Ssn2/Med13 represses transcription of a subset of ATG genes. Moreover, these results are consistent with the model that the destruction of two CKM members following nitrogen starvation, Ssn8/Cnc1 by the UPS [Citation20] (see ) and Ssn2/Med13 by autophagy are mechanisms used to relieve this repression. Snx4 assists in this autophagy program that by targeting a transcriptional repressor provides a positive feedback loop for the autophagic response.
Figure 9. Transcriptional regulators controlling ATG genes are autophagy substrates. (A) RT-qPCR analysis probing for ATG8, ATG1, and ATG14 mRNA expression in wild-type and ssn2/med13∆(RSY2444) cells in unstressed conditions. ∆∆Ct results for relative fold change (log2) values using wild-type unstressed cells as a control. Transcript levels are given relative to the internal ACT1 mRNA control. (B) Western blot analysis of Rim15-GFP (pFD846) cleavage assays in indicated mutants after nitrogen starvation. The asterisk denotes a background band. (C) Western blot analysis of Ccl1-GFP (pSW230) and Msn2-GFP (pSW217) cleavage assays in wild-type or snx4∆ cells. For all blots, Pgk1 levels were used as loading controls. (D) Summary model of the Snx4-assisted autophagy pathway. In unstressed cells, the CKM represses a subset of ATG genes. Following nitrogen starvation, an unknown signal triggers CKM disassembly releasing Ssn2/Med13. Chromatin-free Ssn2/Med13 is transported by unknown mechanisms through the NPC to the cytoplasmic nucleoporin Gle1. Gle1 releases Ssn2/Med13 to the Snx4-Atg20 heterodimer which transports it to Atg17-initiated phagophores, anchored to the vacuole. These sequester Ssn2/Med13 and the autophagosomes fuse with the vacuole for proteolysis
Snx4-assisted autophagy degrades other transcriptional regulators following nitrogen starvation
Next, we wanted to explore the idea that Snx4-assisted autophagy is a mechanism used by the cell to fine-tune the autophagic response at the level of transcription. We first examined the transcriptional activator Rim15 that also regulates ATG genes. During nitrogen starvation, Rim15 enters the nucleus to directly phosphorylate and inhibit the activity of the transcriptional repressors Ume6 [Citation85] and Rph1 [Citation86]. Surprisingly, Rim15-GFP cleavage assays revealed Rim15 is degraded via Snx4-assisted vacuolar degradation ().
To address if Snx4-assisted autophagy may have a more global role in degrading transcription factors that control ATG expression, the transcriptional activators Msn2 and Ccl1, as well as the repressor Rph1 were tested [Citation86–88]. Free GFP accumulated from Msn2-GFP and Rph1-GFP in wild-type cells following nitrogen starvation, illustrating that these transcription factors are degraded via the vacuole. However, only Msn2 degradation requires Snx4 ( and S7D). These results support the conclusion that Snx4-assisted autophagy of transcriptional regulators targets a unique subset of nuclear proteins. These findings also provide the first evidence that the autophagic pathway directly targets regulatory proteins that control its own processes.
Discussion
Here, by following Ssn2/Med13’s fate after nitrogen starvation, we have uncovered a previously undescribed autophagy pathway in which both negative and positive transcriptional regulators of ATG genes are degraded following nitrogen starvation. For clarity, a list of the proteins tested and their requirements are provided in . Our findings, together with previous studies, suggest a two-step pathway involving the NPC and Snx4 for Ssn2/Med13 translocation from the nucleus to the autophagic machinery (outlined in ). First, Ssn2/Med13 disassociates from the CKM, shuttles through the NPC, and associates with the cytoplasmic nucleoporin Gle1. In the second step, Ssn2/Med13 is handed-off from Gle1 to the sorting nexin heterodimer Snx4-Atg20. This heterodimer transports Ssn2/Med13 to Atg17-initiated phagophores located on the vacuole, by localizing to Atg17. Lastly, Snx4-Atg20 is recycled back to the cytosol, and Ssn2/Med13 is degraded by vacuolar proteolysis. This pathway is also utilized to degrade Rim15 and Msn2, but not Rph1 following nitrogen starvation suggesting that the cell degrades specific transcription factors following nitrogen starvation.
Table 1. Proteins tested in this report and their requirement for Snx4-assisted autophagy of Med13
Snx4-assisted vacuolar degradation of transcriptional regulators is distinct from previously identified selective autophagy mechanisms for several reasons. Firstly, different from nucleophagy pathways, it does not use a blebbing mechanism to export cargos [Citation89]. Instead, cargos are rapidly transported through the NPC to the awaiting RNA remodeling complex, with Snx4-Atg20 enabling their final passage to the vacuole. Secondly, to our knowledge, Snx4-assisted autophagy of transcriptional regulators is not used to maintain homeostasis. Instead, it rapidly responds to environmental cues such as nitrogen starvation and targets functional proteins for autophagic degradation. Also other selective autophagy pathways that utilize Snx4 heterodimers [Citation74,Citation84,Citation90–92], predominantly operate to maintain homeostasis, by removing damaged and surplus proteins and organelles. Proteaphagy, the autophagic removal of nuclear 19S and 20S subunits most resembles Snx4-assisted degradation of transcription factors [Citation84]. However, there are significant differences between the pathways, with proteaphagy being dependent upon the ß-karyopherin Crm1 for passage through the NPC and utilizing either Snx4-Atg41 or Atg20 for transport to autophagosomes [Citation84]. Similarities include both pathways utilizing the Atg17-scaffold complex. The exception to this is the degradation of Rpn1 (19S proteasome subunit) and RNA that utilizes both scaffolds [Citation93–95]. The requirement for Atg17 to degrade Ssn2/Med13 following nitrogen starvation also is consistent with the recent discovery that Atg11 is rapidly degraded by the UPS in nitrogen starvation [Citation12]. Lastly, although nonselective autophagy of cytosolic proteins is upregulated upon nitrogen starvation, we did not observe a requirement for Snx4, in this mechanism (see Fig. S7A). However, Snx4 does recognize plasma membrane-tethered Ssn2/Med13 (), further illustrating the specificity of this pathway. Thus, to delineate this new pathway from other Snx4 pathways, we propose to call it Snx4-assisted autophagy of transcription factors.
How does Ssn2/Med13 find its way to the phagophore? Ssn2/Med13 is a large, 160-kDa protein that must require an active process to transit from the nucleus to the cytoplasm. Its unusual structure, (see and discussed in [Citation33]) suggests it functions both as a scaffold protein and a communication/interaction hub [Citation33,Citation96]. Consistent with this, the IDR of Ssn2/Med13 interacts with Gle1, Atg20, Ssn8/Cnc1, and Ssn3/Cdk8 and is the target of additional regulatory protein kinases including Slt2 and Snf1 [Citation33,Citation97]. The established role of the Gle1-Dbp5 complex in releasing RNPs (ribonucleoproteins) from mRNPs (messenger ribonucleoprotein particles) [Citation98] sets a precedent for Gle1 shuttling proteins through the NPC. Moreover, recent structural data places the Gle1-Dbp5 remodeling complex right over the NPC’s central channel, allowing it to efficiently capture proteins as they reach the cytoplasmic side of the NPC [Citation62]. Although speculative, a similar capture and release mechanism may be used in the Ssn2/Med13 transition between Gle1 to Snx4. In support of this model, both Gle1 and Dbp5 are required for the autophagic degradation of Ssn2/Med13 and Gle1 and Snx4 colocalize at the NPC (). Furthermore, Nup159, the nucleoporin that tethers Gle1 and Dbp5 to the NPC is also required for Ssn2/Med13 degradation (). Recently, Nup159 has been suggested to act as a receptor protein for NPC-phagy [Citation17,Citation99,Citation100]. This is an Atg11 dependent process and therefore unlikely to regulate Ssn2/Med13 autophagic degradation. Furthermore, mass spectrometry analysis of the Atg8 interactome did not identify Gle1 as an interacting nucleoporin [Citation17] and we show here that Gle1 does not interact with Atg8 by Y2H analysis (Fig. S4E). Lastly, the vacuolar degradation kinetics of Nup159 is significantly delayed (24 h) compared to the rapid degradation of Ssn2/Med13-GFP where free GFP can be seen within 2 h (). Taken together, the role of Nup159 in Ssn2/Med13 autophagic degradation is most likely to be connected to its requirement for tethering the remodeling complex to the NPC.
The best-characterized cargo of Snx4-Atg20 is Snc1, a plasma membrane-directed v-SNARE [Citation70,Citation101]. Similar to our results with Ssn2/Med13, Snc1 associates with the BAR domain of Snx4 using both co-immunoprecipitation analysis as well as Y2H assays [Citation102]. Likewise, SNX-BARs also directly recognize many retrograde cargos transiting the endosome in mammals [Citation103,Citation104] suggesting that this may be a common function of BAR domains. The mechanism by which Snx4 delivers Ssn2/Med13 to phagophores currently remains unknown. In selective autophagy, receptor proteins bind to the cargo as well as with Atg8. This is important as the phagophore in this case has to be anchored to the cargo [Citation10]. In Snx4-assisted autophagy, the phagophores are anchored to the vacuole and Snx4 transports the cargo to the phagophore which may negate the need for a “conventional” receptor protein. Supporting this, Snx4 colocalizes with Ssn2/Med13 at both perinuclear and cytoplasmic locations. Our finding that efficient Ssn2/Med13 and Atg17 co-immunoprecipitation requires Snx4 suggests that this sorting nexin relays Ssn2/Med13 from the NPC to the autophagic machinery. In addition, Snx4 mediates Ssn2/Med13 vacuolar degradation when this nuclear protein is relocalized to the plasma membrane, illustrating that Snx4 specifically targets and transports Ssn2/Med13 to growing autophagosomes. Defining the Snx4 interaction motif on Ssn2/Med13, as well as Msn2 and Rim15, may result in a common “Snx4 recognition motif” that could be used to identify additional transcriptional regulators that are degraded by this mechanism. This would be useful because, despite the recent discovery of additional Snx4 cargos [Citation101], no consensus Snx4-dependent sorting signal has been identified [Citation73]. This is important as Snx4 is evolutionarily conserved [Citation105], with recent discoveries that Snx4 dysregulation is now associated with the etiology of many diseases, including, cancer, Parkinson disease, and Alzheimer disease [Citation106].
The most unexpected finding from this research was that the complex process of autophagy is used to degrade transcriptional regulators. More unexpectedly, was that two transcriptional activators of ATG genes are also regulated by this pathway. The molecular details of how these transcriptional regulators are initially targeted and then transported through the NPC are unknown. These details will define the nuclear components required for this pathway. This work also provides developing information on how autophagy is regulated at the level of transcription. The process is selective as Rph1, which is also degraded by vacuolar proteolysis following nitrogen starvation, does not require the autophagic machinery (Fig. S7D). Likewise, the zinc-deficiency triggered vacuolar degradation of RNA polymerase I is autophagy-independent [Citation107]. Taken together, Snx4-assisted autophagy may shed light on how other nuclear proteins are regulated following stress. This is important as nucleophagy is the least well defined of all the autophagy pathways and in recent years links between deficiencies and human disease are starting to emerge [Citation89]. For example, a mutation in human Gle1 has recently been linked to amyotrophic lateral sclerosis [Citation108]. Given the highly conserved nature of both Ssn2/Med13 and Snx4 [Citation105,Citation109], these studies are likely to be relevant to mammalian systems.
Materials and methods
Yeast strains and plasmids
Experiments were primarily performed with endogenously labeled proteins in the S. cerevisiae W303 background [Citation110] and are listed in Table S2. All strains were constructed using replacement methodology [Citation111]. Other strains used were from the Research Genetics yeast knockout collection and are derived from BY4741 strain background. The Y2H assays were performed in the Y2H Gold strain (Takara 630,489, PT4084-1; Matchmaker Gold Yeast Two-Hybrid System). In accordance with the Saccharomyces Genome Database members of the CDK8 module, SSN8/CNC1/UME3/SRB11, SSN3/CDK8/UME5/SRB10, SRB8/MED12/SSN5 and SSN2/MED13/UME2/SRB9, will use SSN8/CNC1, SSN3/CDK8, SRB8/MED12, and SSN2/MED13 gene designations, respectively.
Plasmids used in this study are listed in Table S3. The wild-type epitope-tagged plasmids Vph1-mCherry, Nab2-mCherry, Ssn2/Med13-HA, and SSN2/MED13 Y2H GAL4 activating-domain plasmids have been previously described [Citation20,Citation33]. The GAL4-BD-SNX4 fusion plasmids were constructed by amplifying SNX4 alleles from wild-type genomic DNA with oligonucleotides containing SalI flanking sites and cloning into the SalI site of the GAL4 binding domain plasmid pAS2 [Citation112]. The Crn1-GFP-Ssn2/Med13 plasmid was created by amplifying the first 400 amino acids of Crn1 from genomic DNA and cloning it in-frame to the N terminus of GFP-Ssn2/Med13 (pSW218). Plasmid construction details are available upon request. All constructs were verified by sequencing.
Cell growth
Yeast cells were grown in either rich, nonselective medium (YPDA: 2% [w:v] glucose, 2% w:v Bacto peptone, 1% w:v yeast extract, 0.001% w:v adenine sulfate) or synthetic minimal dextrose medium (SD: 0.17% w:v yeast nitrogen base without amino acids and ammonium sulfate, 0.5% w:v ammonium sulfate, 1x supplement mixture of amino acids, 2% w:v glucose) allowing plasmid selection as previously described [Citation22]. For nitrogen-starvation experiments, cells were grown to mid-log in SD medium spun down, washed in 2x volume of water, and resuspended in SD-N media for indicated time points [Citation113]. The GLE1- and NUP159-Auxin-inducible depletion strains () were a gift from K. Cunningham (John Hopkins University) [Citation114]. Live cells were treated with 250 μM auxin (Indole-3-acetic acid; GoldBio, I-110) dissolved in ethanol 30 min before and during nitrogen starvation. The doxycycline-inducible Crm1 N-end rule degron strain (Fig. S4) was generated by integrating pMK632 [Citation115] into the CRM1 locus in the presence of the pCM188 TET activator plasmid to create RSY2348 (Ubi-Leu-3 HA-CRM1::NATMX) as described in detail in [Citation20]. Cells were treated with 2 μg/mL of doxycycline (Simga Aldrich, D9891) for 1 h before and during nitrogen starvation.
Cellular assays
RT-PCR analysis was executed as previously described in [Citation20,Citation28]. Oligonucleotides used during these studies are available upon request. All RT-PCR studies were conducted with three biological samples in technical triplicates. For other assays the standard deviation from three replicate reactions is indicated in the figures. P values were determined using the unpaired Students t-test. The six-day nitrogen starvation viability assays were executed exactly as described [Citation20] with 30,000 cells counted per timepoint using FACS and the studies were conducted in biological duplicates. Data are mean ± standard deviation
Western blot assays
Protein extracts for western blot studies were prepared using a NaOH lysis procedure exactly as described in [Citation97]. In short, protein extracts were prepared from 25 ml per timepoint except for Ssn2/Med13-9xMYC cultures in which 50 ml was needed to visualize the protein. Proteins were separated on 6–10% SDS polyacrylamide gels depending upon their size using the Bio-Rad Mini-Trans Blot cell. To detect proteins, 1:5000 dilutions of anti-MYC (UpState/EMD Millipore Corp.,05–724), anti-HA (Abcam, ab9110), or anti-Pgk1 (Invitrogen, 459,250) antibodies were used. Western blot signals were detected using 1:5000 dilutions of either goat anti-mouse (Abcam, ab97027) or goat anti-rabbit (Abcam, ab97061) secondary antibodies conjugated to alkaline phosphatase and CDP-Star chemiluminescence kit (Invitrogen, T2307). Signals were quantified relative to Pgk1 or Tubulin (Developmental Studies Hybridoma Bank, University of Iowa) controls using CCD camera imaging (Kodak Inc.) All degradation assays were performed in triplicate. Standard deviation and significance were calculated from the mean ± standard deviation using GraphPad Prism 7.
Cleavage assays
Strains harboring GFP-fusion proteins were grown to mid-log in SD, washed, and resuspended in SD-N. Protein extracts were prepared using NaOH as described above (25 mL/timepoint). Proteins were separated using Invitrogen BlotTM 4–12% Bis-Tris Plus gradient gels with 1X MOPS SDS running buffer (NW04122BOX). Proteins were transferred to PVDF membranes in 1X BlotTM transfer buffer for 1 h 30 min (BT00061). GFP-tagged proteins were detected using 1:5000 dilution of anti-GFP (FUJIFILM Wako Pure Chemical Corp., 012–20,461) antibodies and goat anti-mouse secondary antibodies conjugated to alkaline phosphatase Quantification of free GFP was calculated exactly as previously described [Citation17].
Co-Immunoprecipitation
For co-immunoprecipitation experiments, 1 L of cells were grown to mid-log, washed, and resuspended in SD-N media (250 mL/timepoint). Protein extracts were prepared using a glass bead lysis method exactly as described in [Citation33], except protein A beads (GoldBio, P-400-5) were pre-washed with IP wash solution (500 mM NaCl, 25 mM Tris, pH 7.4). One mg of total protein was immunoprecipitated per timepoint. Anti-GFP antibodies (Invitrogen,A11122) or anti-HA antibodies were used for immunoprecipitations. Co-immunoprecipitation blot was probed with antibodies against HA or T7 (Novagen,69,522) epitope. Due to the drastic difference in size between Ssn2/Med13-3xHA and GFP tagged proteins (Snx4 and Atg17) input controls were run on separate gels. For all input controls 500 μg of protein was immunoprecipitated from whole-cell lysates and separated on either 6% (Ssn2/Med13-3 HA) or 10% (GFP-Snx4 and Atg17-GFP) SDS polyacrylamide gels. All co-immunoprecipitation experiments were performed in pep4∆ prb1∆.1 strains. Endogenous Gle1-GFP was not able to be analyzed via immunoprecipitation using glass bead lysis, therefore western blot analysis following NaOH lysis methodology was used as input controls.
Fluorescence microscopy
For all microscopy experiments, cells were grown to mid-log phase, washed, and resuspended in SD-N for the time points indicated. Deconvolved images were obtained using a Nikon microscope (Model E800) with a 100x objective with 1.2x camera magnification (Plan Fluor Oil, NA 1.3) and a CCD camera (Hamamatsu Model C4742). Data were collected using NIS software and processed using Image Pro software. All images of individual cells were optically sectioned (0.2-μM slices at 0.3 μM spacing) and deconvolved, and slices were collapsed to visualize the entire fluorescent signal within the cell. 3D imaging was obtained using the Image Pro software. The nuclei were visualized in live cells using Hoechst staining (Cayman Chemical, 15,547). Hoechst (5 μM), dissolved in water, was added to cells growing in either SD or SD-N (5 μM) 30 min before they were visualized by microscopy. The vacuole was visualized in live cells in (Fig S2A) using FM4-64 (Invitrogen, T3166) and phenylmethane-sulfonyl-fluoride (PMSF; Sigma, P7626) treatment of cells was executed exactly as described [Citation116]. CMAC staining was executed exactly as described [Citation117]. In order to optimize the visualization of the lowly expressed endogenous Ssn2/Med13-mNeongreen, the Keyence microscope was used. This scope has a high sensitivity CCD and high-speed autofocus, low photobleaching mode that aids in monitoring Ssn2/Med13 localization in live cells. Deconvolution and processing capabilities are very limited with the compatible analyzer software. Single plane images were obtained using a Keyence BZ-X710 fluorescence microscope with a 100x objective with 1.0x camera magnification (PlanApoλ Oil, NA 1.45) and a CCD camera. Data were collected using BZ-X Analyzer software. Quantification of Ssn2/Med13-mNeongreen fluorescence within the vacuole was obtained using the Hybrid cell count function within the analyzer software (300 cells were counted per sample). For analysis single extraction settings were used. Red (vacuole, Vph1-mCherry) was set as the target area and green (Ssn2/Med13-mNeongreen) was set as the extraction area. The percentage of cells with vacuolar Ssn2/Med13-mNeongreen was calculated using Area ratio (1st) (ratio of the total area of the extracted areas to the target area) and cell count values. Percentages represent a ratio of extraction area to the target area.
Statistical analysis
All representative results included at least two independent biological experiments. P values were generated from Prism-GraphPad using unpaired Student’s t-tests; NS P ≥ 0.05; *P ≤ 0.05, **P ≤ 0.005; ***P ≤ 0.001; ****P ≤ 0.0001. All error bars indicate mean ± SD. For quantification of Ssn2/Med13-9MYC degradation kinetics band intensities of each time point was first divided by unstressed, T = 0 band intensity. These values were then divided by Pgk1 loading band intensity values which were also normalized to their T = 0 intensities. P-values shown are relative to wild-type T = 4 timepoints.
Acknowledgments
We thank C. De Virgilio, P. Herman, K. Cunningham, S. Wente, K. Weis, and D. Kilonsky for strains and plasmids. We especially thank the members of T. Nazarko, A. Melendez, R. Strich, and the Cooper laboratory for critical reading of this manuscript.
Declaration of interest
The authors declare no competing or financial interests.
Supplemental Material
Download Zip (55.3 MB)Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. Oct.
- Farre JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17(9):537–552. Sep.
- Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20(1):43–48. Jan.
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280(36):31582–31586. Sep 9.
- Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5(5):427–433.
- Noda T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules. 2017;7(4):52. Jul 7.
- Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16(5):2544–2553. May.
- Rao Y, Perna MG, Hofmann B, et al. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat Commun. 2016;7(1):10338. Jan 12.
- Hollenstein DM, Gomez-Sanchez R, Ciftci A, et al. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J Cell Sci. 2019;132(22): Nov 14. doi:10.1242/jcs.235002.
- Hollenstein DM, Kraft C. Autophagosomes are formed at a distinct cellular structure. Curr Opin Cell Biol. 2020;65:50–57. [ Mar 20].
- Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23. Jan.
- Matscheko N, Mayrhofer P, Rao Y, et al. Atg11 tethers Atg9 vesicles to initiate selective autophagy. PLoS Biol. 2019;17(7):e3000377. Jul.
- Zientara-Rytter K, Subramani S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and Autophagy. Cells. 2019;8(1):40. Jan 10.
- Fu N, Yang X, Chen L. Nucleophagy plays a major role in human diseases. Curr Drug Targets. 2018;19(15):1767–1773.
- Mochida K, Oikawa Y, Kimura Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522(7556):359–362. Jun 18.
- Roberts P, Moshitch-Moshkovitz S, Kvam E, et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(1):129–141. Jan.
- Lee CW, Wilfling F, Ronchi P, et al. Selective autophagy degrades nuclear pore complexes. Nat Cell Bio. 2020;22(2):159–166. Feb.
- Vishnoi N, Dhanasekeran K, Chalfant M, et al. Differential turnover of Nup188 controls its levels at centrosomes and role in centriole duplication. J Cell Biol. 2020;219(3): Mar 2. 10.1083/jcb.201906031.
- Delorme-Axford E, Klionsky DJ. Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem. 2018;293(15):5396–5403. Jan 25.
- Willis SD, Hanley SE, Beishke T, et al. Ubiquitin-proteasome-mediated cyclin C degradation promotes cell survival following nitrogen starvation. Mol Biol Cell. 2020;31(10):1015–1031. May 1.
- Cooper KF, Mallory MJ, Strich R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol. 1999;19(5):3338–3348. May.
- Cooper KF, Mallory MJ, Smith JB, et al. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). Embo J. 1997;16(15):4665–4675. Aug 1.
- Nemet J, Jelicic B, Rubelj I, et al. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie. Feb 2014;97: 22–27.
- Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. Sep 7 Sep 7. 2000;407(6800):102–106.
- Jeronimo C, Langelier MF, Bataille AR, et al. Tail and Kinase modules differently regulate core mediator recruitment and function in Vivo. Mol Cell. 2016;64(3):455–466. Nov 3.
- Jeronimo C, Robert F. The mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol. 2017;27(10):765–783. Oct.
- Cooper KF, Khakhina S, Kim SK, et al. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev Cell. 2014;28(2):161–173. Jan 27.
- Cooper KF, Scarnati MS, Krasley E, et al. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for not4-dependent cyclin C destruction. J Cell Sci. 2012;125(Pt 4):1015–1026. Feb 15.
- Wang K, Yan R, Cooper KF, et al. Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Mol Biol Cell. 2015;26(6):1030–1043. Mar 15.
- Ganesan V, Willis SD, Chang KT, et al. Cyclin C directly stimulates Drp1 GTP affinity to mediate stress-induced mitochondrial hyperfission. Mol Biol Cell. 2019;30(3):302–311. Feb 1.
- Jezek J, Chang KT, Joshi AM, et al. Mitochondrial translocation of cyclin C stimulates intrinsic apoptosis through Bax recruitment. EMBO Rep. 2019;20(9):e47425. Sep.
- Khakhina S, Cooper KF, Strich R. Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol Biol Cell. 2014;25(18):2807–2816. Sep 15.
- Stieg DC, Willis SD, Ganesan V, et al. A complex molecular switch directs stress-induced cyclin C nuclear release through SCF(Grr1)-mediated degradation of Med13. Mol Biol Cell. 2018;29(3):363–375. Feb 1.
- Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–379. Mar 4.
- Ramos PC, Hockendorff J, Johnson ES, et al. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92(4):489–499.
- Van Den Hazel HB, Kielland-Brandt MC, Winther JR. Review: biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12(1):1–16. Jan.
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279(29):29889–29894. Jul 16.
- Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. Sep 18.
- Xu H, Jun Y, Thompson J, et al. HOPS prevents the disassembly of trans-SNARE complexes by sec17p/sec18p during membrane fusion. Embo J. 2010;29(12):1948–1960. Jun 16.
- D’Agostino M, Risselada HJ, Lurick A, et al. A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature. 2017;551(7682):634–638. Nov 30.
- Martens S, Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6(1):23.
- Xie Q, Tzfadia O, Levy M, et al. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy. 2016;12(5):876–887. May 3.
- Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126(Pt 15):3237–3247. Aug 1.
- Johansen T, Lamark T. Selective Autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432(1):80–103. Jan 3.
- Martens S, Behrends C. Molecular mechanisms of selective Autophagy. J Mol Biol. 2020;432(1):1–2. Jan 3.
- Marshall RS, Hua Z, Mali S, et al. ATG8-binding UIM proteins define a new class of Autophagy adaptors and receptors. Cell. 2019;177(3):766–781 e24. Apr 18.
- Sawa-Makarska J, Abert C, Romanov J, et al. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16(5):425–433. May.
- Millen JI, Krick R, Prick T, et al. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy. 2009;5(1):75–81. Jan.
- Specht S, Miller SB, Mogk A, et al. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195(4):617–629. Nov 14.
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–1095. Aug 28.
- Parsell DA, Kowal AS, Singer MA, et al. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372(6505):475–478. Dec 1.
- Zientara-Rytter K, Subramani S. Mechanistic Insights into the Role of Atg11 in Selective Autophagy. J Mol Biol. 2020;432(1):104–122. Jan 3.
- Mao K, Chew LH, Inoue-Aono Y, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013;110(31):E2875–84. Jul 30.
- Lin DH, Hoelz A. The structure of the nuclear pore complex (an update). Annu Rev Biochem. 2019;88(1):725–783. Jun 20.
- Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40(3):1623–1634. Mar.
- Murphy R, Wente SR, An RN. A-export mediator with an essential nuclear export signal. Nature. 1996;383(6598):357–360. Sep 26.
- Noble KN, Tran EJ, Alcazar-Roman AR, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25(10):1065–1077. May 15.
- Tran EJ, Zhou Y, Corbett AH, et al. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28(5):850–859. Dec 14.
- Aryanpur PP, Regan CA, Collins JM, et al. Gle1 regulates RNA binding of the DEAD-box helicase Ded1 in its complex role in translation initiation. Mol Cell Biol. 2017;37(21): Nov 1. doi:10.1128/MCB.00139-17.
- Alcazar-Roman AR, Tran EJ, Guo S, et al. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8(7):711–716. Jul.
- Weirich CS, Erzberger JP, Flick JS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8(7):668–676. Jul.
- Fernandez-Martinez J, Kim SJ, Shi Y, et al. Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell. 2016;167(5):1215–1228 e25. Nov 17.
- Adams RL, Mason AC, Glass L, et al. Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells. Traffic. 2017;18(12):776–790. Dec.
- Bolger TA, Folkmann AW, Tran EJ, et al. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134(4):624–633. Aug 22.
- Bolger TA, Wente SR. Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. J Biol Chem. 2011;286(46):39750–39759. Nov 18.
- Gross T, Siepmann A, Sturm D, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315(5812):646–649. Feb 2.
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65(4):570–594.
- Adams RL, Wente SR. Dbp5 associates with RNA-bound Mex67 and Nab2 and its localization at the nuclear pore complex is sufficient for mRNP export and cell viability. PLoS Genet. 2020;16(10):e1009033. Oct.
- Chatterjee K, Majumder S, Wan Y, et al. Sharing the load: mex67-Mtr2 cofunctions with Los1 in primary tRNA nuclear export. Genes Dev. 2017;31(21):2186–2198. Nov 1.
- Ma M, Burd CG, Chi RJ. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic. 2017;18(2):134–144. Feb.
- Ma M, Kumar S, Purushothaman L, et al. Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol Biol Cell. 2018;29(18):2190–2200. Sep 1.
- Suzuki SW, Emr SD. Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol. 2018;217(5):1623–1632. May 7.
- Ma M, Burd CG. Retrograde trafficking and plasma membrane recycling pathways of the budding yeast Saccharomyces cerevisiae. Traffic. 2020;21(1):45–59. Jan.
- Nice DC, Sato TK, Stromhaug PE, et al. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277(33):30198–30207. Aug 16.
- Kanki T, Wang K, Cao Y, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. Jul.
- Shpilka T, Welter E, Borovsky N, et al. Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc Natl Acad Sci U S A. 2015;112(5):1434–1439. Feb 3.
- Deng Y, Qu Z, Naqvi NI. The role of snx41-based pexophagy in magnaporthe development. PLoS One. 2013;8(11):e79128.
- Popelka H, Damasio A, Hinshaw JE, et al. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc Natl Acad Sci U S A. 2017;114(47):E10112–E10121. Nov 21.
- Humphries CL, Balcer HI, D’Agostino JL, et al. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159(6):993–1004. Dec 23.
- Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in Scerevisiae using Pgk1-GFP. Autophagy. 2010;6(6):794–797. Aug.
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. Jun.
- Hanley SE, Cooper KF. Sorting Nexins in Protein Homeostasis. Cells. 2020;10(1):17. Dec 24.
- Stanishneva-Konovalova TB, Derkacheva NI, Polevova SV, et al. The role of bar domain proteins in the regulation of membrane dynamics. Acta Naturae. 2016;8(4):60–69. Oct-Dec. .
- Nemec AA, Howell LA, Peterson AK, et al. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J Biol Chem. 2017;292(52):21466–21480. Dec 29.
- Bartholomew CR, Suzuki T, Du Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109(28):11206–11210. Jul 10.
- Bernard A, Jin M, Gonzalez-Rodriguez P, et al. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol. 2015;25(5):546–555. Mar 2.
- Vlahakis A, Lopez Muniozguren N, Powers T. Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy. Autophagy. 2017;13(11):1804–1812.
- Zhu J, Deng S, Lu P, et al. The Ccl1-Kin28 kinase complex regulates autophagy under nitrogen starvation. J Cell Sci. 2016;129(1):135–144. Jan 1.
- Papandreou ME, Tavernarakis N. Nucleophagy: from homeostasis to disease. Cell Death Differ. 2019;26(4):630–639. Mar.
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17(1):87–97. Jul.
- Kanki T, Wang K, Baba M, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20(22):4730–4738. Nov.
- Shpilka T, Weidberg H, Pietrokovski S, et al. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12(7):226. Jul 27.
- Waite KA, De-La Mota-Peynado A, Vontz G, et al. Starvation induces proteasome Autophagy with different pathways for core and regulatory Particles. J Biol Chem. 2016;291(7):3239–3253. Feb 12.
- Huang H, Kawamata T, Horie T, et al. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. Embo J. 2015;34(2):154–168. Jan 13.
- Kageyama T, Suzuki K, Ohsumi Y. Lap3 is a selective target of autophagy in yeast, Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;378(3):551–557. Jan 16.
- Nagulapalli M, Maji S, Dwivedi N, et al. Evolution of disorder in mediator complex and its functional relevance. Nucleic Acids Res. 2016;44(4):1591–1612. Feb 29.
- Willis SD, Stieg DC, Ong KL, et al. Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress. Microb Cell. 2018;5(8):357–370. Aug.
- Folkmann AW, Noble KN, Cole CN, et al. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2(6):540–548. Nov-Dec. .
- Yin Z, Klionsky DJ. NPC-phagy: selective autophagy of the nuclear pore complexes. Autophagy. 2020;16(10):1735–1736. Oct.
- Tomioka Y, Kotani T, Kirisako H, et al. TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J Cell Biol. 2020;219(7): Jul 6. doi:10.1083/jcb.201910063.
- Bean BD, Davey M, Conibear E. Cargo selectivity of yeast sorting nexins. Traffic. 2017;18(2):110–122. Feb.
- Zhang D, Chen T, Ziv I, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36(6):1018–1033. Dec 25.
- Simonetti B, Danson CM, Heesom KJ, et al. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J Cell Biol. 2017;216(11):3695–3712. Nov 6.
- Kvainickas A, Jimenez-Orgaz A, Nagele H, et al. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J Cell Biol. 2017 Nov 6;216(11):3677–3693.
- Zhang H, Huang T, Hong Y, et al. The retromer complex and sorting nexins in neurodegenerative diseases. Front Aging Neurosci. 2018;10:79.
- Hu YB, Dammer EB, Ren RJ, et al. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4(1):18.
- Lee YJ, Lee CY, Grzechnik A, et al. RNA polymerase I stability couples cellular growth to metal availability. Mol Cell. 2013;51(1):105–115. Jul 11.
- Aditi, Glass L, Dawson TR, et al. An amyotrophic lateral sclerosis-linked mutation in GLE1 alters the cellular pool of human Gle1 functional isoforms. Adv Biol Regul. Sep 2016;62: 25–36.
- Tsai KL, Sato S, Tomomori-Sato C, et al. A conserved mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20(5):611–619. May.
- Ronne H, Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci U S A. 1988;85(8):2696–2700. Apr.
- Janke C, Magiera MM, Rathfelder N, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11):947–962. Aug.
- Wang R, Solomon MJ. Identification of She3 as an SCF(Grr1) substrate in budding yeast. PLoS One. 2012;7(10):e48020.
- Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009;284(51):35885–35895. Dec.18.
- Snyder NA, Kim A, Kester L, et al. Auxin-inducible depletion of the essentialome suggests inhibition of TORC1 by auxins and inhibition of Vrg4 by SDZ 90–215, a natural antifungal cyclopeptide. G3 (Bethesda). 2019;9(3):829–840. Mar 7.
- Gnanasundram SV, Kos M. Fast protein-depletion system utilizing tetracycline repressible promoter and N-end rule in yeast. Mol Biol Cell. 2015;26(4):762–768. Feb 15.
- Journo D, Winter G, Abeliovich H. Monitoring autophagy in yeast using FM 4-64 fluorescence. Methods Enzymol. 2008;451:79–88.
- Conibear E, Stevens TH. Studying yeast vacuoles. Methods Enzymol. 2002;351:408–432.
