ABSTRACT
The small non-coding VTRNA1-1 (vault RNA 1–1) is known to confer resistance to apoptosis in several malignant cell lines and to also modulate the macroautophagic/autophagic flux in hepatocytes, thus highlighting its pro-survival role. Here we describe a new function of VTRNA1-1 in regulating in vitro and in vivo tumor cell proliferation, tumorigenesis and chemoresistance. Knockout (KO) of VTRNA1-1 in human hepatocellular carcinoma cells reduced nuclear localization of TFEB (transcription factor EB), leading to a downregulation of the coordinated lysosomal expression and regulation (CLEAR) network genes and lysosomal compartment dysfunction. We demonstrate further that impaired lysosome function due to loss of VTRNA1-1 potentiates the anticancer effect of conventional chemotherapeutic drugs. Finally, loss of VTRNA1-1 reduced drug lysosomotropism allowing higher intracellular compound availability and thereby significantly reducing tumor cell proliferation in vitro and in vivo. These findings reveal a so far unknown role of VTRNA1-1 in the intracellular catabolic compartment and describe its contribution to lysosome-mediated chemotherapy resistance.
Abbreviations: ATP6V0D2: ATPase H+ transporting V0 subunit d2; BafA: bafilomycin A1; CLEAR: coordinated lysosomal expression and regulation; CQ: chloroquine; DMSO: dimethyl sulfoxide; GST-BHMT: glutathionine S-transferase N-terminal to betaine–homocysteine S-methyltransferase; HCC: hepatocellular carcinoma; LAMP1: lysosomal associated membrane protein 1; LLOMe: L-leucyl-L-leucine methyl ester; MAP1LC3B/LC3: microtubule associated protein 1 light chain 3 beta; MAPK: mitogen-activated protein kinase; MITF: melanocyte inducing transcription factor; MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; ncRNA: non-coding RNA; RNP: ribonucleoprotein; SF: sorafenib; SQSTM1/p62: sequestosome 1; STS: staurosporine; tdRs: tRNA-derived RNAs; TFE3: transcription factor binding to IGHM enhancer 3; TFEB: transcription factor EB; vtRNA: vault RNA transcript.
Introduction
A new class of small non-coding RNA (ncRNA), named vault RNA (vtRNA), was discovered in 1986 in the Rome lab [Citation1]. Vault RNAs are integral components of the large 13-MDa vault particle, a hollow barrel-shaped ribonucleoprotein (RNP) complex, consisting of multiple copies of three proteins: MVP (major vault protein), PARP4/vPARP (poly(ADP-ribose) polymerase family member 4) and TEP1 (telomerase associated protein 1) [Citation2]. Notably, only a minor fraction of their cellular transcript is associated with the vault particles [Citation3,Citation4] hinting to complex-independent functions. Vault RNAs are short POLR3 (RNA polymerase III) transcripts with a length varying between about 80 and 150 nucleotides (nt). In humans, four vtRNA paralogs have been identified, VTRNA1-1, VTRNA1-2, VTRNA1-3 and VTRNA2-1 [Citation4,Citation5]. vtRNAs have been suggested to be involved in a multitude of functions including cell proliferation [Citation6,Citation7], apoptosis [Citation8], autophagy [Citation9], serving as microRNA precursors [Citation10] and have also been linked to chemotherapy resistance [Citation11,Citation12]. Many of these findings are compatible with the hypothesis that altered vault RNA expression is associated with tumorigenesis. Recently, we demonstrated that the VTRNA1-1 paralog protects various human cancer cell lines from undergoing apoptosis in a vault particle-independent manner [Citation8,Citation13]. Yet, the exact function and mechanism of action of vault RNAs remains undeciphered in molecular terms.
Other classes of small ncRNAs, such as microRNAs (miRNA) and tRNA-derived RNAs (tdRs) have also been linked to tumorigenesis. miRNA levels were found to be altered in all cancer types studied [Citation14] and have been suggested to function as tumor suppressors or oncogenes (oncomiRs) [Citation15]. miRNAs seem to exert multifaceted functions on tumor progression, modulating tumor growth, metastatic potential, chemoresistance and regulation of metabolism [Citation16,Citation17]. Similarly, tdRs have been identified to be significantly upregulated under various stress conditions and therefore have garnered much attention in the recent years [Citation18–21]. This newly discovered class of ncRNAs has been shown to influence cancer development by modulating global translation via regulating the expression of genes coding for ribosomal components [Citation22], interacting with RNA binding proteins [Citation23], by regulating kinase activity [Citation24] and promoting cell proliferation [Citation25]. It is evident that small ncRNAs are important regulatory molecules orchestrating gene expression that can either drive or prevent oncogenic processes and therefore possess potential as prognostic biomarkers or putative drug targets for cancer treatment.
Autophagy is among the biological processes that have been recently connected to vault RNA levels [Citation9]. Autophagy is an evolutionarily conserved degradation pathway activated by cellular stress [Citation26]. Basal autophagy is essential for the degradation and the recycling of proteins and organelles. Especially during stress conditions an effective autophagy machinery is beneficial for cell survival [Citation26]. Damaged organelles and proteins are taken up by autophagosomes which fuse with lysosomes for degradation [Citation27]. Lysosomes are a key entity of the cellular digestive system and their function requires the concerted action of hydrolases, the acidification machinery and lumenal protective membrane proteins [Citation28]. Lysosome biogenesis and function are closely coordinated by TFEB (transcription factor EB). TFEB orchestrates the coordinated lysosomal expression and regulation (CLEAR) network genes, involved in lysosomal biogenesis, lysosome-to-nucleus signaling and lipid catabolism [Citation28]. The acidic environment of the lysosomes is a double-edged sword for chemotherapy. Numerous anti-tumor drugs, due to their formulation as a weak-base and hydrophobicity, can be sequestered into the lysosomal lumen, a phenomenon known as lysosomotropism, thus reducing drug-sensitivity and inducing chemoresistance [Citation29].
Here, we describe a novel pro-survival function of VTRNA1-1/vtRNA1-1 in regulating in vitro and in vivo tumor cell proliferation, tumorigenesis and chemoresistance, via its newly disclosed role in supporting the function of lysosomes. We showed that VTRNA1-1 knockout (KO) in human hepatocellular carcinoma (HCC) cells, leads to lysosomal compartment dysfunction by inhibiting TFEB nuclear translocation thus resulting in a downregulation of the CLEAR network genes. We observed that VTRNA1-1 depletion resulted in an increased activation of the MAPK (mitogen-activated protein kinase) cascade, MAPK1/ERK2-MAPK3/ERK1, responsible for TFEB inactivation and cytoplasmic retention. Finally, we demonstrated that lack of VTRNA1-1 reduces drug lysosomotropism and significantly inhibits tumor cell proliferation in vitro and in an in vivo mouse model. These findings reveal a role of VTRNA1-1 in supporting the function of the intracellular catabolic compartment in human hepatocellular carcinoma cells and highlight its importance in lysosome-mediated chemotherapy resistance.
Results
VTRNA1-1 plays a crucial role in tumor cell proliferation and tumorigenesis
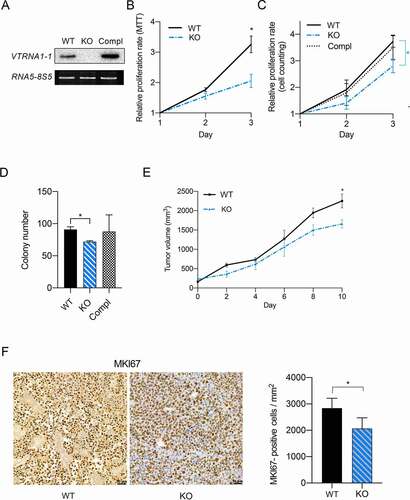
The anti-apoptotic effect of VTRNA1-1/vtRNA1-1 has been the subject of intensive studies in our laboratory [Citation8,Citation13] and lead us to hypothesize about its involvement in tumorigenesis and cancer progression. To test the influence of VTRNA1-1 expression on tumor cell proliferation and colony formation in vitro, we used the human hepatocellular carcinoma cell lines Huh-7 (WT), and Huh-7 VTRNA1-1 KO (). We found that proliferation and colony formation were significantly reduced in Huh-7 VTRNA1-1 KO cells (). Re-introducing the VTRNA1-1 gene could fully rescue cell proliferation in these complementation cells (, C). Likewise, the colony formation assay showed a similar trend upon VTRNA1-1 complementation (). The lack of VTRNA1-1 seems to suggest an important role of this ncRNA in tumor cell viability and proliferation. To further assess the effects of vtRNA1-1 on the tumor growth using an in vivo subcutaneous xenograft mouse model, we injected Huh-7 cells into mice and allowed to develop measurable tumors. Consistent with our in vitro results, the growth was significantly reduced in the VTRNA1-1 KO-transplanted tumors compared to the WT ().
Figure 1. VTRNA1-1/vtRNA1-1 plays a crucial role in tumor cell proliferation and tumorigenesis. (A) Total RNA was extracted from Huh-7 cells and analyzed by northern blotting to confirm successful VTRNA1-1 complementation in Huh-7 KO cells. RNA5-8S5 (RNA, 5.8S ribosomal 5) serves as internal loading control. (B) Mean ± SD relative proliferation of Huh-7 WT, KO and complementation cells was measured with the MTT assay. Values were normalized to day 1, n = 3. (C) Three-day cell proliferation (cells/ml) by automated cell counter. Values were normalized to day 1, n = 5 ± SD. (D) Clonogenic assay: data are expressed as the mean values of three independent experiments ± SD. (E) Tumor volume ± SEM of Huh-7 subcutaneous xenografts mouse model (WT n = 4 and KO n = 5). (F) Huh-7 subcutaneous xenografts immunohistochemistry for MKI67. Counterstain: hematoxylin. Scale bar: 50 μm. The graphs show the mean percentage ± SD of MKI67-positive cell/area per tumor/mouse (five random fields were used for the analysis per tumor/mouse). P values < 0.05 were considered statistically significant and are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Interestingly, VTRNA1-1 expression seems to be also modulated in human derived primary hepatocytes (Figure S1A). In culture, primary hepatocytes are unable to proliferate and progressively de-differentiate from their cell-specific functions [Citation30,Citation31]. Interestingly, we observed a significant increase in VTRNA1-1 expression in hepatocyte cultures for up to 14 days (Figure S1A) while hepatocytes still maintained their differentiated state (Figure S1B). Hepatocytes in culture are known for spontaneously activating apoptosis pathways [Citation32,Citation33]. Consistently, we detected PARP1 (poly(ADP-ribose) polymerase 1) cleavage already at day 0 (before seeding) together with an increased expression of BCL2 (BCL2 apoptosis regulator) (Figure S1C), hinting at the already described activation of an anti-apoptotic cellular response [Citation34]. Thus, increased VTRNA1-1 expression might be similarly involved in pro-survival pathways.
These observations reveal that the cellular levels of VTRNA1-1/vtRNA1-1 contribute to proliferation in immortalized cell lines and are modulated in response to stresses in primary hepatocytes. All together our data indicate that vtRNA1-1 plays a key role in tumor cells proliferation and tumorigenesis.
Defective lysosome function impairs autophagy-mediated clearance in VTRNA1-1 KO cells
Because VTRNA1-1 levels have been reported previously to affect apoptosis [Citation8,Citation13] and autophagy [Citation9], we next asked the question how and if these cellular processes are linked via VTRNA1-1 expression in hepatocytes. Huh-7 sensitivity toward apoptosis was not affected by VTRNA1-1 depletion, since upon staurosporine (STS) treatment, a broad-spectrum protein kinase inhibitor inducing apoptosis, the IC50 values were similar for both WT and VTRNA1-1 KO cell lines (). In agreement cleavage of pro-apoptotic PARP1 was not observed or was negligible in these cells (). These results fit with the observation that Huh-7 are remarkably resistant to apoptosis even after treatment with STS [Citation35]. Similar results were obtained using the pro-apoptotic DNA-damaging anticancer drug doxorubicin (Figure S2A) as well as during high stress culture conditions (serum-deprived culture medium) where KO of VTRNA1-1 actually slightly enhances cell survival compared to WT (Figure S2B). This is in stark contrast to the effects seen in HeLa cells, where staurosporine treatment and starvation increase apoptotic rates and induces PARP1 cleavage ( and [Citation13]).
Figure 2. Defective lysosome function impairs autophagy-mediated clearance in VTRNA1-1 KO cells. (A) IC50 values of Huh-7 WT and KO cells after 24 h of staurosporine treatment. (B) Representative immunoblots for PARP1 (FL, full-length) and ACTB/β-actin in Huh-7 and HeLa cells after Staurosporine IC30 treatment (24 h, 5 μM). HeLa cells were used as control for apoptosis induction. (C) Representative immunoblots for LAMP1, SQSTM1/p62, LC3B-I, LC3B-II (o. e., overexposed) and ACTB/β-actin in Huh-7 cells under complete culture medium (10% FBS – Ctrl) and treatment with chloroquine (CQ – 20 μM, 4 h) or bafilomycin A1 (BafA, 100 nM, 4 h). (D) Top panel, mean ± SD of lysosomal pH values measured by flow cytometry in Huh-7 pre-incubated in culture medium supplemented with FITC-dextran (0.1 mg/mL, 72 h) followed starvation (0.1% FBS) for 24 h and high starvation (HS, with low glucose, without amino acids and FBS) for 6 h. n = 3. Bottom panel, illustrative histograms from flow cytometry showing the shift of FITC-dextran emission wavelength. (E) Representative immunoblots for GST-BHMT of total lysate obtained from Huh-7 cells transiently transfected with the GST-BHMT construct and either maintained in complete culture media or starving media supplemented with leupeptin and E64d and without essential amino acids and FBS for 6 h. Red Ponceau staining served as loading control. P values < 0.05 were considered statistically significant and are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
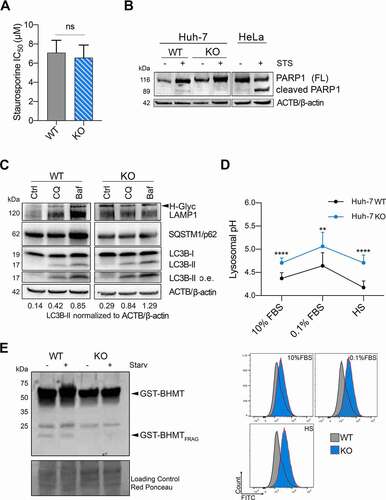
A recent study suggested that VTRNA1-1 acts as a regulator of autophagy by interacting with SQSTM1/p62 (sequestosome 1), which is involved in targeting substrates of the autophagy machinery, preventing its self-oligomerization in Huh-7 cells [Citation9]. As already shown by Horos et al., VTRNA1-1-dependent impairment of the autophagic flux can be detected by treating Huh-7 cells with the two late-stage autophagy inhibitors, chloroquine (CQ) and bafilomycin A1 (BafA). Western blot analysis revealed an increased amount of MAP1LC3B/LC3 (microtubule- associated protein 1 light chain 3 beta) expression in KO cell lines compared to WT (, S2C). Conversion of LC3B-I to LC3B-II is the hallmark for autophagosome formation. Interestingly, we detected a pronounced LC3B-II protein accumulation in cells lacking VTRNA1-1 already at basal level, under normal culture conditions, suggesting that loss of this ncRNA induces autophagy impairment (). Of note, no significant changes in SQSTM1/p62 levels were apparent under control conditions in the absence of VTRNA1-1 ().
Autophagosomes are intermediate structures in a very dynamic process, where their intracellular localization results from a balance between their generation and their conversion rate into autolysosomes [Citation36]. Thus, autophagosome accumulation may derive from either autophagy induction or impaired lysosomal clearance [Citation28]
To identify the phenomenon causing autophagosome accumulation in VTRNA1-1 KO cells, we first aimed to examine lysosomal compartment structural proteins focusing on the expression levels of LAMP1 (lysosomal associated membrane protein 1). Interestingly, LAMP1 levels were increased, together with an enhanced expression of its hyperglycosylated (H-Glyc) form only in VTRNA1-1 KO cells (). Further, in variance to WT cells we found that CQ- and BafA-induced upregulation of LAMP1, a clear sign of a larger accumulation of undigested autolysosomes, was not detectable in the absence of VTRNA1-1 (). An increased expression of H-Glyc LAMP1 has been reported to be linked with intrinsic lysosomal compartment instability [Citation37]. Moreover, in addition to lysosomal compartment stability, lysosome-mediated cellular clearance processes require the concerted action of hydrolases and the intralumenal acidification machinery [Citation28]. Interestingly, in VTRNA1-1 KO cells we detected a significant increase in lysosomal pH toward more alkaline values (). Taking advantage of the GST-BHMT assay, previously developed for measuring autophagic degradation capabilities [Citation38], we next assessed the lysosomal proteolytic activity in the cells. In line with our previous observations, the ability of lysosomes to degrade GST-BHMT was markedly reduced in absence of VTRNA1-1, indicating an impaired lysosomal proteolytic activity (). Conversely, in HeLa cells, where VTRNA1-1 was shown to confer apoptosis resistance [Citation8,Citation13], neither changes in lysosomal pH nor in lysosomal proteolytic activity were apparent when VTRNA1-1 was depleted (Figure S2D and E).
Collectively, our data demonstrate that VTRNA1-1 has a crucial role in Huh-7 in modulating lysosomal function, especially for regulating their intralumenal pH and proteolytic activity.
VTRNA1-1 levels influence TFEB nuclear translocation and affect TFEB-driven CLEAR network gene expression
Lysosomal function is closely monitored to respond and adapt to environmental stimuli by its master TFEB regulator. Under basal conditions, TFEB is located in the cytoplasm and under specific stimuli, such as starvation, it is rapidly translocated into the nucleus. The cellular localization and activity of TFEB are mainly controlled by phosphorylation, which depends predominantely on MTORC1 and MAPK1/ERK2 [Citation39,Citation40]. To gain further mechanistic insight into the molecular events regulating TFEB activation, we tested whether its upstream regulators might be influenced by VTRNA1-1 expression. While we did not observe any difference in MTOR phosphorylation (), VTRNA1-1 depletion resulted in a clear basal increase of MAPK pathway activation ( and S3A). We observed a higher phosphorylation level of MAPK1/3, its upstream kinases MAP2K1/MEK1-MAP2K/MEK2 as well as RPS6KA1/90RSK (ribosomal protein S6 kinase A1), its downstream target ( and S3A). Importantly, the elevated markers of phosphorylation returned to WT-levels in the complementation cells ( and S3A) thus suggesting that re-introducing VTRNA1-1 restores signaling. Surprisingly, the MAPK pathway was also regulated in HeLa VTRNA1-1 KO cells, but in an opposite manner (Figure S3B). Our data suggests that the loss of VTRNA1-1 leads to a hyperactivation of MAPK1/3, which might in turn prevent TFEB nuclear translocation. We indeed found the amount of nuclear TFEB accumulation to be significantly lower in KO cells compared to the WT ( and S3C). Other transcription factors of the same family known to regulate starvation-induced lysosomal biogenesis and autophagy [Citation41,Citation42] were either lowly expressed in Huh-7 and thus below the western blot detection limit (MITF), or did not show changes in protein expression or altered localization upon VTRNA1-1 removal (TFE3) (). These findings are in accordance with a previous study demonstrating no nuclear translocation of TFE3 or MITF in Huh-7 cells under the applied starvation conditions [Citation43]. Therefore, our results reveal VTRNA1-1 levels to specifically influence TFEB as the major transcription factor for lysosome biogenesis.
Figure 3. VTRNA1-1/vtRNA1-1 plays a key role in TFEB nuclear translocation and TFEB-driven CLEAR network genes expression. (A) Representative immunoblots for p-MTOR, total MTOR and GAPDH in Huh-7 WT and VTRNA1-1 KO cells under normal culture conditions (10% FBS). (B) Left, representative immunoblots for p-MAPK1/ERK2-MAPK3/ERK1, total MAPK1/3 and ACTB/β-actin in Huh-7 WT, VTRNA1-1 KO and complementation cells under normal culture conditions (10% FBS). Right, quantification of p-MAPK1/3 obtained by normalizing the p-MAPK1/3 and total MAPK1/3 levels by ACTB/β-actin followed by the ratio of p-MAPK1/3 over total MAPK1/3. Values are expressed as the mean values of three (for WT and KO) or two (for complementation) independent experiments ± SD normalized to WT level. (C) Left, representative nuclear/cytoplasm fractionation immunoblots for TFEB, TUBA/α-tubulin, histone H3 in Huh-7 WT and VTRNA1-1 KO cells under high starvation for 3 h. Right, quantification of TFEB nuclear levels normalized to histone H3. Values are expressed as the mean values of three independent experiments ± SD. (D) Representative nuclear-cytoplasm fractionation immunoblots for TFE3, GAPDH, histone H3 in Huh-7 WT and VTRNA1-1 KO cells under high starvation for 3 h. (E) Real-time qPCR data (mean ± SEM) of TFEB, LAMP1, CTSD, CLCN7 and ATP6V0D2 mRNA levels in Huh-7 VTRNA1-1 KO cells grown under complete medium (10% FBS – Ctrl) and starving culture conditions (0.1% FBS) for 24 h, normalized to Huh-7 WT (n = 3) and VTRNA1-1 KO-derived tumors normalized to WT-transplanted ones (WT n = 4, KO n = 9). (F) Real-time qPCR mean ± SEM data of SQSTM1/p62 in Huh-7 KO cells and derived tumors normalized to Huh-7 WT and WT-transplanted tumors respectively. P values < 0.05 were considered statistically significant and are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
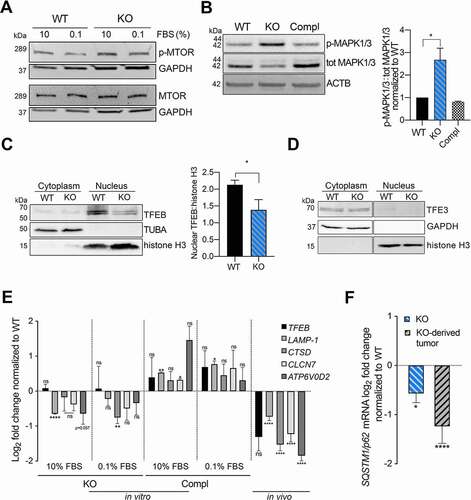
TFEB orchestrates lysosomal expression and regulation (CLEAR) network genes, involved in lysosomal biogenesis, lysosome-to-nucleus signaling and lipid catabolism [Citation28]. Therefore, we analyzed the expression of known TFEB target genes in KO cells compared to WT at basal level and under serum-deprived culture conditions, where especially tumor cells more heavily rely on their intracellular catabolic machinery. In fact, the recycling of intracellular constituents provides alternative energy sources during periods of metabolic stress to maintain homeostasis and viability [Citation44]. While the mRNA levels of TFEB remained unchanged, its target genes, such as LAMP1, CLCN7 (chloride voltage-gated channel 7), CTSD (cathepsin D) and ATP6V0D2 (ATPase H+ transporting V0 subunit d2) were downregulated in the absence of VTRNA1-1 (). Upon complementation, Huh-7 cells restored VTRNA1-1 levels (), concomitantly with an increased expression of CLEAR network genes (). Overexpressing a TFEB gain-of-function mutant with pronounced nuclear localization characteristics ([Citation45] and Figure S3D) in the VTRNA1-1 KO cells rescued expression of CLEAR network genes (Figure S3E). These observations substantiate the interdependence of VTRNA1-1 expression, TFEB nuclear localization and lysosome activity. Consistently, we also confirmed that the CLEAR network genes were downregulated in tumors in VTRNA1-1 KO-injected mice compared to the WT control (), indicating that the previously observed altered lysosome function in the VTRNA1-1 KO cells () can be attributed to the downregulation of CLEAR gene expression. Of note, SQSTM1/p62 expression is also under the transcriptional control of TFEB [Citation46] and in fact, in line with our previous findings, its mRNA levels were significantly downregulated in VTRNA1-1 KO cells compared to WT in cell culture as well as in the in vivo mouse model ().
In summary these data provided evidence that VTRNA1-1 plays a role in establishing lysosome function, where its absence lead to a marked loss of autolysosome proteolytic activity, decreased TFEB nuclear translocation and subsequent reduced expression of CLEAR network genes.
Removal of VTRNA1-1 potentiates the cytotoxicity of sorafenib in vitro and in vivo
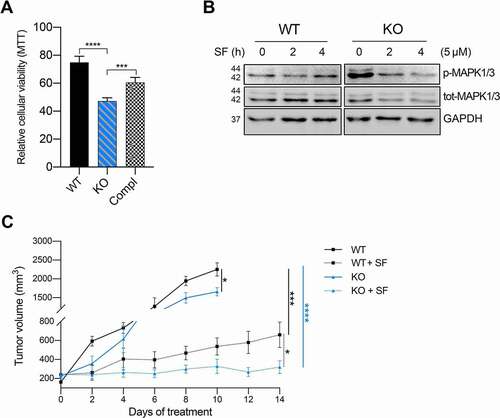
Our data provide evidence that loss of VTRNA1-1/vtRNA1-1 interferes with lysosome function, thus leading to a deficiency in the autophagic machinery. Weak base anticancer compounds can be sequestered in the acidic lumen of lysosomes by cation trapping and consequently are no longer able to reach their target, thereby reducing their effectiveness [Citation47]. It has been reported that the cytotoxic potential of sorafenib (SF), the first-line treatment option for advanced hepatocellular carcinoma patients and a known tyrosine kinase inhibitor of the MAPK pathway [Citation48], is significantly decreased due to its passive lysosomal compartment trapping [Citation37]. Therefore, we next investigated if loss of VTRNA1-1 leads to lysosomal malfunction and thus consequently to increased drug sensitivity. When treated with the IC30 dose of SF, Huh-7 KO cells showed a significantly decreased viability compared to WT cells after treatment (). This increased SF cytotoxicity indeed depends on VTRNA1-1 levels since the rescued cell line behaved similarly to the WT cells (). Interestingly, we found that SF markedly downregulates MAPK1/3 phosphorylation in a time-dependent manner in Huh-7 KO cells compared to the WT (), suggesting a higher amount of drug being able to reach its intracellular molecular target due to a decreased lysosomotropism. On the other hand, the lysosomotropic compound LLOMe (L-leucyl-L-leucine methyl ester), which is an acidic pH-driven lysosome-specific membrane damage inducer, showed a markedly reduced cytotoxicity in VTRNA1-1 KO cells (Figure S4A). Huh-7 WT cells viability was more strongly decreased compared to VTRNA1-1 KO cells after LLOMe administration (Figure S4A). We observed a significantly lower LLOMe-induced lysosomal pH alkalinization in Huh-7 VTRNA1-1 KO cells compared to the WT (Figure S4B) further confirming a weaker LLOMe lysosomal compartment tropism and trapping of LLOMe, mainly due to the previously shown elevated alkaline intralumenal pH observed for the KO cells (). Corroborating our in vitro data, SF treatment significantly impaired tumor growth and progression in the Huh-7 KO-transplanted mice as compared to animals transplanted with the WT control cells ().
Figure 4. Removal of VTRNA1-1 potentiates the cytotoxicity of sorafenib in vitro and in vivo. (A) Mean ± SD relative viability of Huh-7 WT, KO and complementation cells, after sorafenib IC30 treatment (24 h, 14 μM) measured with the MTT assay. Values were normalized to untreated cells, n = 3. (B) Representative immunoblots for p-MAPK1/3, total MAPK1/3 and GAPDH in Huh-7 WT and VTRNA1-1 KO cells in presence or absence of sorafenib (5 μM) as indicated. (C) Tumor volume of Huh-7-transplanted xenografts mice treated with vehicle and sorafenib (SF – 100 mg/kg – daily oral gavage) for 14 days (SF n = 6/group; vehicle WT n = 4, KO n = 5). P values < 0.05 were considered statistically significant and are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
These data exhibit a role of VTRNA1-1 in lysosomal compartment functionality, which seems to involve in the positive modulation of the pro-tumorigenic MAPK cascade activation (). Cumulatively we found that VTRNA1-1 plays a significant role in determining the intracellular fate and potency of lysosomotropic compounds and thus affects their molecular target site distribution and antitumor effectiveness.
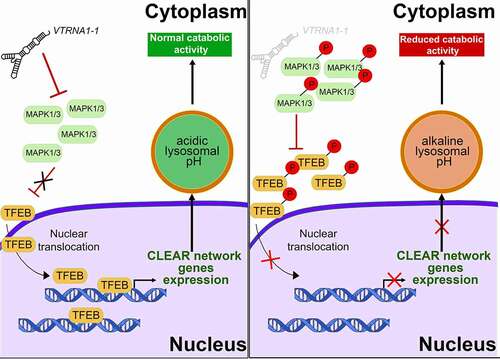
Figure 5. Model for lysosome and autophagy mediated clearance regulation. In VTRNA1-1/vtRNA1-1 WT cells low MAPK1/3 phosphorylation level allows TFEB nuclear translocation leading to CLEAR network genes expression and ensuring intracellular catabolic compartment stability and function (left). Reduced VTRNA1-1 levels (right) lead to the hyperphosphorylation of MAPK1/3 and TFEB, and consequently to its reduced nuclear translocation, leading to a downregulation of the CLEAR network genes and lysosomal compartment dysfunction.
Discussion
The human genome encodes on chromosome 5 four vault RNA paralogs, VTRNA1-1, VTRNA1-2, VTRNA1-3 and VTRNA2-1 [Citation5]. These ncRNAs are around 100 residues long and they received their name based on an early observation that they are integral to the vault particle, a gigantic hollow RNP with a molecular mass of 13 MDa [Citation1]. Despite decades of dedicated research, the biological function of the vault particle or the vault RNAs remained enigmatic [Citation2,Citation49]. More recently, it was demonstrated that only a minor fraction of vault RNA is actually associated with the vault complex [Citation4,Citation50] thus suggesting cellular functions of these ncRNAs beyond the vault complex. Accumulating evidence reveals a key role of the small non-coding VTRNA1-1/vtRNA1-1 in conferring cellular resistance against cell death via chemoresistance [Citation7,Citation12], viral defense and apoptosis [Citation4,Citation8,Citation13,Citation51]. This led us to further investigate the involvement of VTRNA1-1 in tumorigenesis and cancer progression. In agreement with its previously described function, we observed a modulation of VTRNA1-1 expression in response to apoptosis induced by physical and mechanical stress in primary human hepatocytes (Figure S1 A, C). We further describe that VTRNA1-1 is involved in tumor cell proliferation, tumorigenesis and chemoresistance, via its newly described role in supporting the function of the intracellular catabolic machinery (, and 4A-C). However, we did not observe that the sensitivity of Huh-7 cells to apoptosis was altered by the lack of VTRNA1-1 (, B and Figure S2A, B). This is in clear contrast to several other immortalized cell lines, such as the Burkitt Lymphoma cell lines (BL2, BL41), the breast cancer model HS578T, the human embryonic kidney cells (HEK293), the lung carcinoma cells A549 and HeLa cells [Citation8,Citation13]. It is safe to conclude that high VTRNA1-1 levels are correlated with pro-survival characteristics in numerous human cell lines of different tissue origins (Figure S5). Thus, it is possible that the well-documented pro-survival biological function of VTRNA1-1 manifests itself differently and is cell line or tissue dependent and a generalized mechanism of VTRNA1-1 should not be based on data obtained in one cellular system. A recent study in Huh-7 cells reported that VTRNA1-1 was able to contribute in the regulation of the autophagic flux, by directly interacting with the autophagy receptor protein SQSTM1/p62 [Citation9]. In our study, we were able to confirm that the loss of VTRNA1-1 leads to elevated LC3B-II levels suggesting autophagosome accumulation ( and Figure S2C). Previously these observations were interpreted as evidence for an increase of the autophagic flux in the absence of VTRNA1-1 [Citation9]. Our in vitro and in vivo experiments provide further insight into VTRNA1-1 biology and elaborate the suggested role of SQSTM1/p62 as post-transcriptional regulator of autophagy [Citation9]. Although the decreased steady-state level of SQSTM1/p62 was initially attributed to the lost interaction with VTRNA1-1 [Citation9], we found that depletion of VTRNA1-1 resulted in transcriptional downregulation of several catabolism-related genes, including also SQSTM1/p62 ( and F). Our data demonstrated that reduced expression of SQSTM1/p62 is transcriptionally controlled and it is not necessarily dependent on its interaction with VTRNA1-1.
Lysosomes and autophagy are two tightly linked and evolutionarily conserved processes that provide a constant supply of cellular energy mainly through recycling of damaged organelles and proteins [Citation27]. As previously described by Horos et al., VTRNA1-1 depletion leads to mature autophagosomes accumulation, suggesting an increased autophagic flux [Citation9]. However, autophagosomes accumulation, in the absence of any specific chemical modulator of the flux progression, may come from either autophagy induction or impaired lysosomal autophagy-mediated clearance. Lysosomes are intracellular vesicles responsible for such catabolic and clearance processes in cell homeostasis, and their function requires the concerted action of hydrolases, acidification pumps for maintaining acidic the intralumenal pH and lysosomal membrane proteins to ensure the integrity of this specific vesicular compartment [Citation28]. Investigating the lysosome-mediated clearance process, we demonstrated that deletion of VTRNA1-1 caused an alkalinization of the intralumenal lysosomal pH, leading to a marked impairment of their proteolytic and degradative activity (). LAMPs are structural proteins important for maintaining lysosomal membrane integrity and stability by segregating the acidic environment of the lumen from the rest of the cell [Citation52]. Under normal conditions, LAMP1 glycosylation protects the inner lysosomal membrane from cleavage and degradation from the low pH of the intralumenal environment [Citation53]. Knockout of VTRNA1-1 leads to an impairment of CQ- and BafA-induced LAMP1 accumulation (). Additionally, an already stressed and intrinsically instable status of the catabolic compartment in VTRNA1-1 KO cells is indicated by the increased glycosylation of LAMP1 in untreated growth conditions (). Concurrently, VTRNA1-1 depletion was indeed able to induce a clear switch of the lysosome intralumenal pH toward a more alkaline environment, affecting the catabolic and proteolytic activity (). These findings seem to attribute the autophagosomes accumulation evident in VTRNA1-1 KO cells to the lysosomal compartment dysfunction.
The expression and activity of the lysosomal components are tightly coordinated to allow it to function under physiological and pathological conditions by the CLEAR genes network, as well as by TFEB nuclear translocation and transcriptional control [Citation54]. Investigating the potential cause for the compromised lysosomal function, we uncovered that TFEB nuclear translocation was decreased in VTRNA1-1 KO cells, with a significant downregulation of the CLEAR network genes in vitro and in vivo (). Genetic complementation of VTRNA1-1 in KO cells restored not only cellular proliferation () but also CLEAR gene expression (), further suggesting their interdependence with VTRNA1-1 levels. Given the well-known tight link between the MAPK pathway and TFEB activation [Citation39], we found that VTRNA1-1 depletion markedly induced an increased phosphorylation of the downstream effector of the MAPK kinase cascade MAPK1/3 (), which finally suggested that VTRNA1-1 KO triggers a compensatory overactivation of the MAPK pathway (). Similarly, in HeLa cells we recently demonstrated that deletion of VTRNA1-1, but not of the paralog VTRNA1-3, modulated several signaling pathways, including the MAPK cascade [Citation13]. The link between vault RNA expression and MAPK signaling was recently substantiated in cultured mouse cortical neurons [Citation55]. Further, we demonstrated that all these phenomena reported here can potentiate the antitumor effect of conventional chemotherapeutic drugs. Specifically, the vast majority of chemotherapeutic agents, due to their hydrophobic and weak base chemical characteristics, often get passively sequestered within the acidic lumen of lysosomes, becoming unable to reach their intracellular molecular target [Citation56]. Interestingly, the lack of VTRNA1-1 and the consequently elevated alkaline lysosomal pH lead to a diminished cytotoxicity of the lysosomal disruptor LLOMe, mainly due to its decreased lysosomotropism (Figure S4). On the other hand, the similarly reduced lysosomotropism of sorafenib observed in the KO cells in vitro and in vivo revealed an increased availability of sorafenib at its target site and therefore a stronger antitumor effect ().
Taken together, our study uncovered the involvement of VTRNA1-1/vtRNA1-1 in tumor cell proliferation and tumorigenesis, demonstrating its crucial role to maintain the catabolic compartment and chemoresistance (). This novel pro-survival function may identify vtRNA1-1 as a new therapeutic target to overcome chemo-insensitivity caused by passive lysosomal sequestration of antitumor drugs and augment tumor-patients outcomes.
Materials and methods
Cell culture
Huh-7 (kindly provided by Matthias Hentze; EMBL, Heidelberg, Germany) and HeLa cells (ATCC®-CCL-2™) were cultured in low glucose (5 mM) DMEM (Thermo Fisher, 21,331,046) supplemented with 10% heat inactivated FBS (10,082,147, Thermo Fisher) and 100 U/ml PenStrep glutamine (Thermo Fisher, 10,378,016). The VTRNA1-1 KO and WT Huh-7 cell lines were kindly provided by M. Hentze (EMBL, Heidelberg, Germany). We derived a Huh-7 VTRNA1-1 complementation cell line using lentiviral transduction (below) in the Huh-7 KO cells. After successful lentiviral transduction, positively infected cells were selected by treating them with 1 μg/ml puromycin for at least 1 week. Single cell clones were obtained by serial dilution. Normal liver tissue was obtained from surgical resections from consented patients at the University Hospital of Bern. Hepatocytes were then isolated by enzymatic perfusion as previously described [Citation57] and kept in culture at 37°C in a humidified incubator with 5% of CO2.
Patient-derived tissues
Primary human liver tissue for hepatocyte isolation [Citation57] and HCC tumors from liver were obtained from patients of the University Hospital Bern (Inselspital). Informed consent was obtained prior to surgery in compliance with the local ethics regulations and under approval of local ethics commission (Project-ID Nr. 2019–00157).
Lentiviral transduction
Lentiviral particles were generated in human HEK 293 T cells (kindly provided by M. Ruepp, King’s College London), which were transiently transfected with lentiviral plasmids containing cDNAs coding for VTRNA1-1, together with the packaging plasmids pSPAX and the envelope plasmid pVSV-G (kindly provided by S. Geley, Medical University Innsbruck). After 48 h and 72 h lentiviral supernatant was collected, sterile filtered (Sigma-Aldrich, Whatman Puradisc FP30, 0.2 mM) and supplemented with polybrene (Sigma, 107,689) to a final concentration of 4 μg/ml and added to the target cells overnight.
Transfection and treatments
Transfections were done using Lipofectamine 3000 (Thermo Fisher, L3000008) for plasmid DNA. Doxorubicin (Sigma-Aldrich, D1515) was diluted in water to 5 mM and used at 1 μM for 48 h. Bafilomycin A1 (BafA; Sigma-Aldrich, B1793) was diluted in DMSO to 200 μM and used at 100 nM for 4 h. Chloroquine (CQ; Sigma-Aldrich, C6628) was diluted in DMSO to 50 mM and used at 20 μM for 4 h. Rapamycin (Sigma-Aldrich, R0395) was diluted in ethanol to 2 mM and used at 200 nM for 4 h. Leu-Leu methyl ester hydrobromide (LLOMe; Sigma-Aldrich, L7393) was diluted in DMSO to 1 M and used from 0.001 to 1.4 mM for 6 h. Sorafenib tosylate (MedChemExpress, HY-10201A) was used from 5 to 14 μM for 24 h in vitro and 100 mg/kg in vivo daily. FITC-dextran (Sigma-Aldrich, FD40S) was directly diluted in the medium to 0.1 mg/mL. MTT (Thermo Fisher, M6494) was diluted in PBS (137 mM NaCl, 2.7 mM KCl, 12.5 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) to 5 mg/mL. For high starvation, cells were washed twice with PBS and starved in low glucose DMEM lacking amino acids (USBiological, D9800-13) and serum. To overexpress the TFEB variant with pronounced nuclear localization characteristics [Citation45] the plasmid pCIP-caTfeb (Addgene, 79,013; deposited by Reuben Shaw) was transfected into Huh-7 VTRNA1-1 KO cells.
Cell proliferation and viability assay
Cell proliferation was detected using 3-(4,5-dimethylthazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Thermo Fisher, M6494) or via automated cell counting. For the MTT assay approximately 5 x 103 and 1.5 × 104 cells/well, for proliferation or viability respectively, were seeded into 96-well plates and allowed to attach overnight. After 24, 48, 72 and 96 h, cell viability was assessed using MTT assay. The MTT solution (5 mg/mL) was added to each well for 3 h. The resulting formazan crystals were dissolved in DMSO and the optical density was measured at 570 nm using a Tecan infinite M1000Pro plate reader. Three independent experiments were conducted in triplicates. To assess proliferation rates via automated cell counting, 1 × 105 cells/well were seeded on 6 well plates. After 24 h, 48 h and 72 h cells were washed twice with PBS, detached using trypsin and counted using a TC10™ Automated Cell Counter (Bio-Rad).
Colony formation assay
Approximately 5 × 102 cells were placed in a 6-well plate in DMEM containing 10% FBS for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet in 20% methanol for 30 min. Each assay was conducted in triplicate.
RNA extraction and quantitative real-time RT-qPCR (qPCR)
RNA was isolated from cells as well as tissue samples by Tri-reagent (Zymo Research, R2050-1-200) following the manufacture’s protocol. Reverse transcription (RT) of total RNA was carried out using SuperScript™ IV One-Step RT-PCR System (Invitrogen, 18,090,010), according to the manufacturer’s protocol, using random primer hexamers for cDNA amplification. Subsequent quantitative PCR (qPCR) on the cDNA was carried out using GoTaq® qPCR Master Mix (Promega, A6002), according to the manufacturer’s protocol (Table S1 lists the used primers). The QIAgility robot was used to pipette all the reagents into the qPCR reactions and qPCR amplification was carried out using Rotor Gene 6000 (QIAGEN), according to the manufacturer’s instructions. qPCR analyses were done in Roboticx software (QIAGEN), and differential mRNA transcript abundances were calculated using the ΔΔCt method as described [Citation58].
Northern blot
2–10 µg total RNA was separated on an 8% denaturing polyacrylamide gel (7 M Urea, in 1x TBE buffer). The gel was subsequently electroblotted onto a nylon membrane (Amersham Hybond N+; GE Healthcare, RPN203B) as described [Citation59]. The membranes were hybridized using 32P-labeled probes listed in Table S1 and exposed to phosphorimaging screens. An autoradiogram was developed using a Typhoon FLA1000 phosphorimager.
Western blot
Total protein cell lysates were obtained using RIPA buffer (10 mM Tris/Cl, pH 8, 1 mM EDTA, 1% Triton X-100 [Sigma-Aldrich, T8787], 0.1% deoxycholate [Sigma-Aldrich, D6750], 0.1% SDS, 140 mM NaCl) containing protease inhibitor cocktail (Sigma-Aldrich, 11,697,498,001), phosphatase inhibitors (Thermo Scientific, A32957) and 0.5% NP-40 (Sigma-Aldrich, 56,741). For nuclear-cytoplasmic fractionation, cells were treated as described previously [Citation60] . Western blots were performed according to standard protocols using the antibodies listed in .
Table 1. Antibodies used in this study
GST-assay
Huh-7 and HeLa cells were transiently transfected with the GST-BHMT plasmid as previously shown by Mercer et al. [Citation61]. Briefly, BHMT transfected cells were cultured in starvation DMEM medium lacking the essential amino acids (Arg, Cys, His, Ile, Leu, Lys, Met, Phe, Thr, Trp, Tyr, and Val) (USBiological, D9800-27) and FBS for 6 h in the presence of E64d (6 mM; Sigma-Aldrich, E8640-250UG) and leupeptin (11 mM; Sigma-Aldrich, L-0649). Afterward, proteins from the cells were extracted with RIPA buffer. Total protein of cell extracts was determined by Bradford method (ThermoScientific, 23,227) and GST‑BHMT was precipitated with glutathione agarose (Sigma, G4510). Precipitates were then mixed in SDS‑PAGE sample buffer, resolved by SDS‑PAGE and visualized by western blotting and chemiluminescence.
Lysosomal pH measurements by flow cytometry
Lysosomal pH measurements were based on an adjusted protocol recently published by Gavini et al [Citation37]. In brief, HCC cell lines were seeded and cultured in cell culture medium containing FITC-Dextran (0.1 mg/mL; Sigma-Aldrich, FD40S) for three days. After this period FITC-Dextran was removed by aspirating the medium and fresh cell culture medium was added with the respective treatments as indicated. Lysosomal pH from samples belonging to the standard curve scale (pH ranging from 4 to 6) and experimental-treated ones were analyzed by FACS. Triplicate samples were taken for each condition analyzed.
Subcutaneous HCC cell line xenograft mouse models
Eight- to 12-week-old male rag2−/- il2rg/γc−/- mice (in house breeding) were used for the subcutaneous xenograft model. 2 × 106 HCC cells were injected in a 1:1 ratio with Matrigel® (BD Biosciences, 356,234). HCC patient-derived specimens were transplanted subcutaneously. Tumor volume was measured using a digital caliper thanks to a modified ellipsoid formula: volume = (4/3) × π × (length/2) × (width/2) × (height/2). Upon reaching a volume of 250 mm3, animals were randomly divided into different groups as described: administration of the vehicle (50% Cremophor EL and 50% ethyl alcohol mixture at 12–24 mg/mL – control group and sorafenib (100 mg/kg) daily by oral gavage. The tumor volume and the body weight were recorded every second day for fourteen days. At the end of experiment, the entire tumor was carefully removed, and some snap-frozen samples were stored in liquid nitrogen and kept at −80°C until further use. Other parts of the tissue were fixed in 4% formaldehyde for histological analysis. All animal experiments were conducted in accordance to Swiss Guidelines of Care and Use of Laboratory Animals.
Immunohistochemistry
Tissue samples were fixed in 4% formaldehyde, processed, embedded in paraffin and sectioned with Leica Microtome. Sections were stained using MKI67/Ki67 antibody (Abcam, ab16667). For each sample five randomly chosen fields were imaged by Leica camera and analyzed by ImageJ software. Coverslips were mounted with VECTASHIELD Antifade Mounting Medium and images were taken using an automated inverted microscope (Leica DMSI4000 B).
Statistical analysis
Images were quantified with ImageJ. Data are displayed as mean ± SD or SEM; Student’s t test was used; n values are indicated in the respective Figure legends. p-values are indicated in the Figure, p < 0.05 was considered statistically. GraphPad Prism v8 was used to create plots.
Supplemental Material
Download Zip (2.2 MB)Acknowledgments
We would like to thank Matthias Hentze for providing the Huh-7 VTRNA1-1 knockout cell line and for valuable discussions. We further thank Adrian Keogh for providing human primary hepatocytes.
Disclosure statement
The authors declare no competing interests.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol. 1986;103(3):699–709.
- Berger W, Steiner E, Grusch M, et al. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66(1):43–61.
- Kickhoefer VA, Poderycki MJ, Chan EKL, et al. The La RNA-binding protein interacts with the vault rna and is a vault-associated protein. J Biol Chem. 2002;277(43):41282–41286.
- Nandy C, Mrázek J, Stoiber H, et al. Expression of a novel human vault RNA. J Mol Biol. 2009;388(4):776–784.
- Stadler PF, Chen JJL, Hackermüller J, et al. Evolution of Vault RNAs. Mol Biol Evol. 2009;26(9):1975–1991.
- Lee K-S, Park J-L, Lee K, et al. Nc886, a non-coding RNA of anti-proliferative role, is suppressed by CpG DNA methylation in human gastric cancer. Oncotarget. 2014;5(11):3944–3955.
- Chen J, OuYang H, An X, et al. Vault RNAs partially induces drug resistance of human tumor cells MCF-7 by binding to the RNA/DNA-binding protein PSF and inducing oncogene GAGE6. PLoS One. 2018;13(1):e0191325.
- Amort M, Nachbauer B, Tuzlak S, et al. Expression of the vault RNA protects cells from undergoing apoptosis. Nat Commun. 2015;6:7030.
- Horos R, Büscher M, Kleinendorst R, et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019;176(5):1054–1067.e12.
- Persson H, Kvist A, Vallon-Christersson J, et al. RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11(10):1268–1271.
- Gopinath SCB, Matsugami A, Katahira M, et al. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucleic Acids Res. 2005;33(15):4874–4881.
- Gopinath SCB, Wadhwa R, Kumar PKR. Expression of noncoding vault RNA in human malignant cells and its importance in mitoxantrone resistance. Mol Cancer Res. 2010;8:1536–1546.
- Bracher L, Ferro I, Pulido-Quetglas C, et al. Human VtRNA1-1 levels modulate signaling pathways and regulate apoptosis in human cancer cells. Biomolecules. 2020;10(4):614.
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–221.
- Mei J, Hao L, Wang H, et al. Systematic characterization of non-coding RNAs in triple-negative breast cancer. Cell Prolif. 2020;53:e12801.
- Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055.
- Oberbauer V, Schaefer MR. TRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes (Basel). 2018;9(12):607.
- Zhu L, Ge J, Li T, et al. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37.
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–1806.
- Cristodero M, Polacek N. The Multifaceted Regulatory Potential of TRNA-Derived Fragments. Non-coding RNA Investig. 2017;1:7.
- Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552(7683):57–62.
- Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian MRNA-binding proteins. Cell. 2012;149(6):1393–1406.
- Shao Y, Sun Q, Liu X, et al. TRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90(5):730–738.
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112(29):E3816–E3825.
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293.
- Inguscio V, Panzarini E, Dini L. Autophagy contributes to the death/survival balance in cancer photodynamic therapy. Cells. 2012;1(3):464–491.
- Settembre C, Fraldi A, Medina DL, et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296.
- Zhitomirsky B, Assaraf YG. Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat. 2016;24:23–33.
- Olsavsky Goyak KM, Laurenzana EM, Omiecinski CJ, et al. Methods Mol. Biol 2010;640:115–138.
- Elaut G, Henkens T, Papeleu P, et al. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab. 2006;7(6):629–660.
- Bailly-Maitre B, De Sousa G, Zucchini N, et al. Spontaneous apoptosis in primary cultures of human and rat hepatocytes: molecular mechanisms and regulation by dexamethasone. Cell Death Differ. 2002;9(9):945–955.
- Vinken M, Maes M, Oliveira AG, et al. Their cultures in liver apoptosis research. Arch Toxicol. 2014;88:199–212.
- Dutta C, Day T, Kopp N, et al. BCL2 suppresses PARP1 function and nonapoptotic cell death. Cancer Res. 2012;72(16):4193–4203.
- Sjogren AKM, Liljevald M, Glinghammar B, et al. Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch Toxicol. 2014;88:1427–1437.
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326.
- Gavini J, Dommann N, Jakob MO, et al. Verteporfin-induced lysosomal compartment dysregulation potentiates the effect of sorafenib in hepatocellular carcinoma. Cell Death Dis. 2019;10(10):749.
- Dennis PB, Mercer CA. Chapter 7 the GST-BHMT assay and related assays for autophagy. Methods Enzymol. 2009;452:97–118.
- Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433.
- Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via MTOR and TFEB. Embo J. 2012;31(5):1095–1108.
- Pastore N, Vainshtein A, Klisch TJ, et al. TFE 3 regulates whole‐body energy metabolism in cooperation with TFEB. EMBO Mol Med. 2017;9(5):605–621.
- Möller K, Sigurbjornsdottir S, Arnthorsson AO, et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep. 2019;9(1):1055.
- Bartel K, Pein H, Popper B, et al. Connecting lysosomes and mitochondria - a novel role for lipid metabolism in cancer cell death. Cell Commun Signal. 2019;17(1):87.
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967.
- Young NP, Kamireddy A, Van Nostrand JL, et al. Specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 2016;30(5):535–552.
- Palmieri M, Impey S, Kang H, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20(19):3852–3866.
- Zhitomirsky B, Assaraf YG. Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget. 2017;8(28):45117–45132.
- Zhang Z, Zhou X, Shen H, et al. Phosphorylated ERK is a potential predictor of sensitivity to sorafenib when treating hepatocellular carcinoma: evidence from an in vitro study. BMC Med. 2009;7:41.
- Büscher M, Horos R, Hentze MW. ‘High vault-age’: non-coding RNA control of autophagy. Open Biol. 2020;10(2):190307.
- Kickhoefer VA, Rajavel KS, Scheffer GL, et al. Vaults are up-regulated in multidrug-resistant cancer cell lines. J Biol Chem. 1998;273(15):8971–8974.
- Li F, Chen Y, Zhang Z, et al. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 2015;43(21):10321–10337.
- Li Y, Chen B, Zou W, et al. The lysosomal membrane protein SCAV-3 maintains lysosome integrity and adult longevity. J Cell Biol. 2016;215(2):167–185.
- Mareninova OA, Sendler M, Malla SR, et al. Lysosome-associated membrane proteins (LAMP) maintain pancreatic acinar cell homeostasis: LAMP-2-deficient mice develop pancreatitis. CMGH. 2015;1(6):678–694.
- Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483.
- Wakatsuki S, Takahashi Y, Shibata M, et al. Small noncoding vault RNA modulates synapse formation by amplifying MAPK Signaling. J Cell Biol. 2021;220(2):e201911078.
- Zhitomirsky B, Assaraf YG. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosomedependent cancer multidrug resistance. Oncotarget. 2015;6(2):1143–1156.
- Portmann S, Fahrner R, Lechleiter A, et al. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12(4):499–508.
- Rao X, Huang X, Zhou Z, et al. An improvement of the 2ˆ(-Delta Delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3(3):71–85.
- Gebetsberger J, Zywicki M, Künzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in haloferax volcanii. Archaea. 2012;2012:260909.
- Suzuki K, Bose P, Leong-Quong RY, et al. REAP: a two minute cell fractionation method. BMC Res Notes. 2010;3:294.
- Mercer CA, Kaliappan A, Dennis PB. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy. 2008;4(2):185–194.
