ABSTRACT
Massive infiltrated and enriched decidual macrophages (dMφ) have been widely regarded as important regulators of maternal-fetal immune tolerance and trophoblast invasion, contributing to normal pregnancy. However, the characteristics of metabolic profile and the underlying mechanism of dMφ residence remain largely unknown. Here, we observe that dMφ display an active glycerophospholipid metabolism. The activation of ENPP2-lysophosphatidic acid (LPA) facilitates the adhesion and retention, and M2 differentiation of dMφ during normal pregnancy. Mechanistically, this process is mediated through activation of the LPA receptors (LPAR1 and PPARG/PPARγ)-DDIT4-macroautophagy/autophagy axis, and further upregulation of multiple adhesion factors (e.g., cadherins and selectins) in a CLDN7 (claudin 7)-dependent manner. Additionally, poor trophoblast invasion and placenta development, and a high ratio of embryo loss are observed in Enpp2±, lpar1−/− or PPARG-blocked pregnant mice. Patients with unexplained spontaneous abortion display insufficient autophagy and cell residence of dMφ. In therapeutic studies, supplementation with LPA or the autophagy inducer rapamycin significantly promotes dMφ autophagy and cell residence, and improves embryo resorption in Enpp2± and spontaneous abortion mouse models, which should be dependent on the activation of DDIT4-autophagy-CLDN7-adhesion molecules axis. This observation reveals that inactivation of ENPP2-LPA metabolism and insufficient autophagy of dMφ result in resident obstacle of dMφ and further increase the risk of spontaneous abortion, and provides potential therapeutic strategies to prevent spontaneous abortion.
Abbreviations: ACTB: actin beta; ADGRE1/F4/80: adhesion G protein-coupled receptor E1; Atg5: autophagy related 5; ATG13: autophagy related 13; BECN1: beclin 1; CDH1/E-cadherin: cadherin 1; CDH5/VE-cadherin: cadherin 5; CFSE: carboxyfluorescein succinimidyl ester; CLDN7: claudin 7; CSF1/M-CSF: colony stimulating factor 1; CSF2/GM-CSF: colony stimulating factor 2; Ctrl: control; CXCL10/IP-10: chemokine (C-X-C) ligand 10; DDIT4: DNA damage inducible transcript 4; dMφ: decidual macrophage; DSC: decidual stromal cells; ENPP2/ATX: ectonucleotide pyrophosphatase/phosphodiesterase 2; Enpp2±: Enpp2 heterozygous knockout mouse; ENPP2i/PF-8380: ENPP2 inhibitor; EPCAM: epithelial cell adhesion molecule; ESC: endometrial stromal cells; FGF2/b-FGF: fibroblast growth factor 2; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GPCPD1: glycerophosphocholine phosphodiesterase 1; HE: heterozygote; HIF1A: hypoxia inducible factor 1 subunit alpha; HNF4A: hepatocyte nuclear factor 4 alpha; HO: homozygote; ICAM2: intercellular adhesion molecule 2; IL: interleukin; ITGAV/CD51: integrin subunit alpha V; ITGAM/CD11b: integrin subunit alpha M; ITGAX/CD11b: integrin subunit alpha X; ITGB3/CD61: integrin subunit beta 3; KLRB1/NK1.1: killer cell lectin like receptor B1; KRT7/cytokeratin 7: keratin 7; LPA: lysophosphatidic acid; LPAR: lysophosphatidic acid receptor; lpar1−/−: lpar1 homozygous knockout mouse; LPAR1i/AM966: LPAR1 inhibitor; LY6C: lymphocyte antigen 6 complex, locus C1; LYPLA1: lysophospholipase 1; LYPLA2: lysophospholipase 2; Lyz2: lysozyme 2; MAP1LC3B: microtubule associated protein 1 light chain 3 beta; MARVELD2: MARVEL domain containing 2; 3-MA: 3-methyladenine; MBOAT2: membrane bound O-acyltransferase domain containing 2; MGLL: monoglyceride lipase; MRC1/CD206: mannose receptor C-type 1; MTOR: mechanistic target of rapamycin kinase; NP: normal pregnancy; PDGF: platelet derived growth factor; PLA1A: phospholipase A1 member A; PLA2G4A: phospholipase A2 group IVA; PLPP1: phospholipid phosphatase 1; pMo: peripheral blood monocytes; p-MTOR: phosphorylated MTOR; PPAR: peroxisome proliferator activated receptor; PPARG/PPARγ: peroxisome proliferator activated receptor gamma; PPARGi/GW9662: PPARG inhibitor; PTPRC/CD45: protein tyrosine phosphatase receptor type, C; Rapa: rapamycin; RHEB: Ras homolog, mTORC1 binding; SA: spontaneous abortion; SELE: selectin E; SELL: selectin L; siCLDN7: CLDN7-silenced; STAT: signal transducer and activator of transcription; SQSTM1: sequestosome 1; TJP1: tight junction protein 1; VCAM1: vascular cell adhesion molecule 1; WT: wild type.
Introduction
Early pregnancy loss occurs in 10% of all clinically recognized pregnancies. And 80% of pregnancy losses occur in the first trimester [Citation1]. Approximately 1–3% of women have recurrent spontaneous abortion [Citation2]. A variety of investigations have been carried out to identify causes of early pregnancy loss, including genetic causes, uterine anomaly, infectious etiologies, endocrine disturbance, thrombophilia and immune factors [Citation3]. However, the cases of a large proportion of spontaneous abortion remain unexplained.
Immune dysfunction at the maternal-fetal interface is closely related to the onset of unexplained spontaneous abortion. Macrophages account for about 20% of total leukocytes within the human decidua [Citation4]. Within gestational age, the number of decidual macrophages (dMφ) varies and is highest in the first and second trimester [Citation5]. Human decidua-resident macrophage homeostasis plays a decisive role in the establishment and maintenance of normal pregnancy, contributing mainly to the trophoblast invasion, spiral artery remodeling and embryo-friendly immunological environment during early pregnancy [Citation6,Citation7]. Macrophage depletion induces fetal resorption in pregnant mouse model [Citation8]. It has been proposed that the dominant factor conferring tissue-resident macrophage identity and self-maintenance capacity would be the niche competition rather than origin [Citation9]. The niche of decidua-resident macrophage would directly affect the stability of decidual microenvironment, whereas the biological events and mechanisms involved need to be further studied.
Immune cells are migratory and leukocyte recruitment through modified blood vessels and into affected tissues is generally achieved by rapid migration of inflammatory cells out of circulation [Citation10]. Macrophages can migrate from the peritoneal cavity directly into the adjacent liver-bypassing circulation [Citation11]. Tissue-resident macrophages present programs for cell migration and adhesion and mediate vascular repair through physical adhesion and mechanical traction [Citation12]. Tissue cells, as well as cancer cells, express specific adhesion molecules to attract macrophages, restore function, and thrive in the leukocyte-rich microenvironments [Citation13–15]. Decidua, the maternal bed embracing the embryo and the major locations of dMφ, is an important site of fetal-maternal immune cross-talk and maintains the pregnancy and maternal and offspring’s health [Citation7,Citation16].
Macrophages are highly polarizable and plastic. Metabolic reprogramming of macrophages plays a predominant role in regulating their phenotype but also their plasticity [Citation17]. Succinate, itaconate, lactic acid and lysophosphatidic acid (LPA) are all representative examples of reprogramming metabolites in macrophage differentiation as energy metabolism regulators and/or signal transduction molecules [Citation18–22]. Phosphatidylcholine (PC) is hydrolyzed by PLA (phospholipase) to produce lysophosphatidylcholine (LysoPC). Under the catalysis of ENPP2/autotaxin/ATX (ectonucleotide pyrophosphatase/phosphodiesterase 2; a metabolic enzyme with lysophospholipase D activity), LPA, a potent bioactive lysophospholipid which is metabolized by PLPP1 (phospholipid phosphatase 1) and MGLL (monoglyceride lipase), is generated from LPC. LPA binds to specific cell-surface G protein-coupled receptors, including six identified receptors, LPAR1 to LPAR6 [Citation23], and the intracellular receptor PPARG/PPARγ peroxisome proliferator activated receptor gamma) [Citation24]. LPAR1 and LPAR3 are crucial for decidualization, implantation, embryo spacing and vascularization during pregnancy [Citation25–27]. Additionally, autophagy has also been reported to be involved in menstruation, decidualization, implantation, trophoblast and placenta development [Citation28–31]. Tissue-specific metabolic state of macrophages is a feature of niche adaptation [Citation32]. However, studies on the metabolic characteristics and autophagy level of dMφ and its possible roles in spontaneous abortion are still scarce.
Therefore, it is urgent to study the metabolic characteristics of dMφ and its roles in dMφ residence and homeostasis during early pregnancy. Specifically, we focused on the aberrant LPA metabolism-autophagy axis-mediated unstable dMφ niche in the pathogenesis of spontaneous abortion, and further explored the possible intervention strategies in this study.
Results
dMφ exhibit an active glycerophospholipid metabolic profile and are rich in LPA
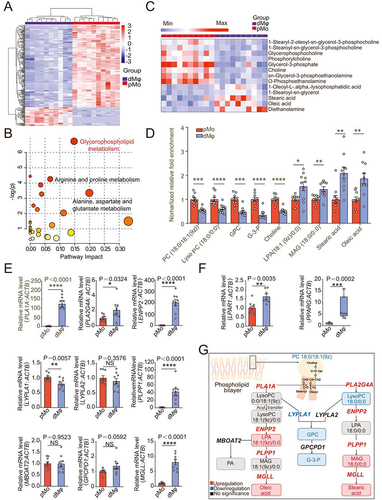
To access the metabolite profiles of dMφ, the purified peripheral blood monocytes (pMo) and dMφ from women with normal early pregnancy were collected and analyzed by metabolomics. As shown, dMφ presented completely different metabolic profiles compared with pMo (), especially the greatest enrichment of glycerophospholipid metabolism pathway (). LPA and its downstream metabolites were enriched, while levels of its upstream related metabolites were decreased in dMφ, suggesting that LPA metabolism is active, and LPA is enriched in dMφ (, D). Further analysis showed that the levels of most of the enzymes associated with LPA metabolism in dMφ were significantly higher than those of pMo, including PLA1A, PLA2G4A, ENPP2, PLPP1, and MGLL (). In particular, the level of ENPP2, the gene encoding the key metabolic enzyme of LPA production in dMφ, was about 200 times higher than that in pMo (). The inhibition of ENPP2 led to the decrease of LPA in macrophage in vitro (Figure S1A). Therefore, LPA is an important intermediate metabolite in these differential metabolites related to the glycerophospholipid metabolism pathway. LPA receptors (LPAR1 and PPARG) were significantly high-expressed in dMφ (, Figure S1B, C). However, there was no significant change in the expression of ENPP2 in stromal cells during decidualization (Figure S1D), and it was much lower than those of dMφ (Figure S1E). These data suggest that dMφ presents an active ENPP2-mediated LPA metabolism () with the enrichment of LPA and high levels of LPA receptors at the maternal-fetal interface.
Figure 1. Decidual macrophages exhibit an active glycerophospholipid metabolic profile and are rich in LPA. (a) Heatmap of differential metabolites in metabolomics for pMo (n = 10) and dMφ (n = 10) from normal early pregnant women. (b) Enriched metabolic pathways of differential metabolites in (a) was obtained by topology analysis. (c and d) Differential metabolites in glycerophospholipid metabolism pathway between pMo and dMφ. (e) Relative mRNA expression of metabolic enzymes in glycerophospholipid metabolism pathway between pMo and dMφ by RT-PCR. (f) Transcriptional levels of LPAR1 and PPARG in pMo and dMφ by RT-PCR. (g) Summary of glycerophospholipid metabolism characteristics in dMφ from normal early pregnant women (inside the box: metabolites, outside the box: metabolic enzymes). Data were presented as mean ± SEM or median and quartile and analyzed by t test or Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS: no significance.
Macrophage with aberrant LPA metabolism induces spontaneous abortion by impairing trophoblast invasion and placenta development
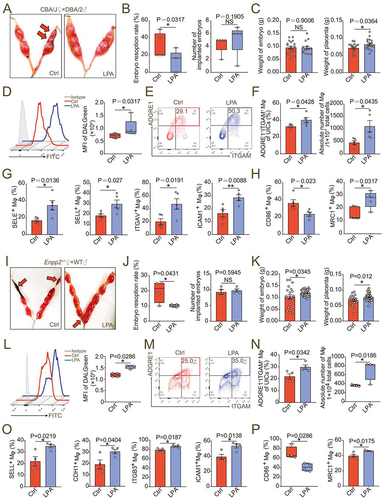
To clarify the possible role of the ENPP2-LPA metabolism in normal pregnancy, enpp2 knockout mouse models were established. Because homozygous knockouts are lethal, the pregnancy outcomes of heterozygous (Enpp2±) mice and wild-type (WT) mice were observed. Compared with WT pregnant mice, as shown, Enpp2± pregnant mice (were mated with Enpp2± mice ♂) at 13.5th day of pregnancy showed adverse pregnancy outcomes (), the overall embryo resorption rate was significantly increased (), and the weight and size of embryos and placenta were significantly reduced (). To eliminate the interference of heterozygous mating embryo genotypes on the resorption rate, we identified the embryo genotypes and found that almost all of embryos with enpp2−/− genotypes were resorbed (, F left). Of note, the resorption rate of WT embryos in Enpp2± pregnant mice was still significantly higher than that of WT pregnant mice (were mated with WT mice ♂) ( right), and the size of the embryo and placenta was also smaller than that of WT pregnant mice (). However, there was no difference of placenta and embryo sizes between WT embryos and Enpp2± embryos from same Enpp2± pregnant mice, suggesting that the heterozygous loss of Enpp2 did not influence the development of placenta and embryo (). Interestingly, the infiltration of placenta trophoblast cells decreased significantly in decidua of Enpp2± pregnant mice (), indicating that heterozygous loss of Enpp2 in uterus should contribute to the impaired invasiveness of trophoblasts. Excluding the effects of embryo genotypes, similarly, there were also increased embryo loss, decreased embryo and placenta weight, and/or impaired infiltration of placenta trophoblast cells in lpar1−/− pregnant mice (Figure S1F-K) and PPARG inhibitor (PPARGi)-treated pregnant mice (Figure S1L-Q). Additionally, high rates of embryo loss, decreased embryo and placenta weight of pregnant mice with macrophage depletion were observed in the transfer group of macrophages from Enpp2± mice compared with the transfer of macrophage from WT mice (Figure S2A-E). These data suggest that pregnant mice with the inactive Enpp2-LPA metabolism or LPA receptor signaling of uterus were prone to spontaneous abortion.
Figure 2. Macrophage with aberrant LPA metabolism induces spontaneous abortion by impairing trophoblast invasion and placenta development. (a) Photograph of uterus of wild-type (WT) and Enpp2± mice (red arrow: resorption point). (b) Photograph of embryos and placentas of WT (WT♀×WT♂) and Enpp2± (Enpp2±♀×Enpp2±♂) pregnant mice and the genotypes of embryos for Enpp2± pregnant mice. (c) Numbers of resorptive and non- resorptive embryos of WT and Enpp2± mice (left, red number on the top of the columns: the total resorption rate) and comparison of resorption rates of total embryos (right, n = 8). (d and e) The weight or size of embryos and placentas of WT and Enpp2± mice. (f) Numbers of resorptive and non-resorptive embryos different genotypes of Enpp2± mice (left, red number on the top of the columns: the total resorption rate) and comparison of the resorption rates of WT embryos from WT (n = 9) and Enpp2± (n = 7) pregnant mice (right). (g) Comparison of the sizes of WT embryos from WT pregnant mice and WT or HE embryos form Enpp2± pregnant mice. (h) Diffusion and invasion of KRT7+ trophoblasts into uterus of WT (n = 8) or Enpp2±(n = 8) pregnant mice (white solid triangles: uterine glandular epithelium cells). (i and j) Relative depth of trophoblast invasion into uterus: the ration of KRT7+ trophoblast depth (Line 1/L1 in red) to the total depth (Line 2/L2 in Orange) (white solid triangles: uterine glandular epithelium cells). (k) Schematic diagram of the operation of the invasion assays of JEG3 cells in the co-culture system with M0 macrophages or M2 macrophages. (l) Matrigel invasion assays for evaluation of the invasiveness of JEG3 cells in the co-culture system (48 h) with control or LPA-treated M0 macrophages (24 h). (m) Statistical graph of numbers of invasive JEG3 cells from (l). (n) Matrigel invasion assays for evaluation of the invasion of JEG3 cells in the co-culture system (48 h) with control, ENPP2 inhibitor, LPAR1 inhibitor or PPARG inhibitor-treated M2 macrophages macrophage (24 h). (o) Statistical graph of numbers of invasive JEG3 cells from (n). Data were presented as mean ± SEM or median and quartile and analyzed by t test, Mann-Whitney U test or one-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS: no significance.
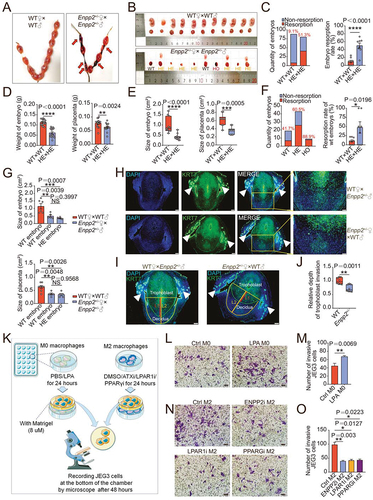
Further analysis showed that M2 macrophages treated with CSF1/M-CSF and IL10 had high levels of ENPP2 (Figure S3A). To further clarify the possible mechanism of Enpp2 deficiency in spontaneous abortion, macrophage and JEG3 cells was cocultured in vitro to imitate the crosstalk between decidual macrophage and trophoblasts at the maternal-fetal interface (, Figure S3B). As shown, LPA-treated M0 macrophages stimulated the invasion and proliferation of JEG3 cells (, M, Figure S3C-D), while inhibition of ENPP2, LPAR1 or PPARG in M2 macrophages inhibited the invasion and proliferation of JEG3 cells (, O, Figure S3E, F). Additionally, LPA promoted the secretion of several pro-angiogenic factors by macrophage, especially IL8 and CXCL10/IP-10 (chemokine (C-X-C) ligand 10) and growth factors, FGF2/b-FGF (fibroblast growth factor 2) and PDGF (platelet derived growth factor)-BB (Figure S3G). Therefore, the activated ENPP2-LPA metabolism of dMφ contributes to growth and invasion homeostasis of trophoblast cells and placenta developments, and these cytokines (IL8, IL10, FGF2, and PDGF-BB) should be involved in these processes.
LPA promotes the adhesion and enrichment of M2-polarized dMφ in decidua
To further explore the potential role of ENPP2-LPA metabolism in dMφ, firstly, RNA-sequence was performed to evaluate the difference of ENPP2-LPAlow pMo and ENPP2-LPAhigh dMφ from women during normal early pregnancy (). Compared with pMo, dMφ exhibited different phenotypes (). Enrichment analysis of GO term and KEGG pathway showed that significantly upregulated genes of dMφ were mainly enriched in the cell adhesion, biological adhesion, focal adhesion, and so on (). Among these, there were 137 adhesion-related different genes (Figure S4), suggesting that these molecules are important for macrophage localization and enrichment in the decidua. Interestedly, LPA significantly promoted macrophage adhesion to DSCs in a dose-dependent manner (, D). While PF-8380 (an ENPP2 inhibitor) or LPA receptor inhibitors were pretreated, their adhesion and residence in the stromal cell layer was significantly inhibited ().
Figure 3. LPA promotes the adhesion and enrichment of M2-polarized macrophages in decidua. (a) Heatmap of differential genes in RNA-seq for pMo (n = 3) and dMφ (n = 3) from women with normal pregnancy. (b) Enriched KEGG and GOTERM pathways of differential gene clustering from (a). (c and d) Adhesion assays for control or LPA-treated (24 h) PKH-26-labeled red-fluorescent M0 macrophages to PKH-67-labeled green-fluorescent DSCs (n = 5). (e and f) Adhesion assays for PKH-26-labeled red-fluorescent M2 macrophages treated with vehicle (n = 10), ENPP2 inhibitor (n = 7), LPAR1 inhibitor (n = 6) or PPARG inhibitor (n = 6) for 24 h to PKH-67-labeled green-fluorescent DSCs. (g and h) The residence and number of ADGRE1+ macrophages in the uterus of WT (n = 5) and Enpp2± (n = 4) mice by immunofluorescence assays. (i and j) Flow cytometry assays for detection of the proportion in PTPRC+ immune cells and the absolute numbers of ADGRE1+ITGAM+ uterine macrophages per 10,000 cells in uterus of WT (n = 8) or Enpp2± (n = 8) pregnant mice. (k and l) Adhesion molecules and differentiation markers of PTPRC+ADGRE1+ITGAM+ uterine macrophages of WT (n = 8) or Enpp2± (n = 8) pregnant mice in flow cytometry assays. Data were presented as mean ± SEM or median and quartile and analyzed by t test, Mann-Whitney U test or one-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001.
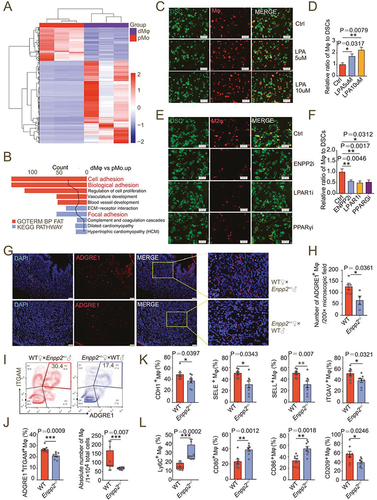
In the uterus of Enpp2± mice on 7.5th day of pregnancy, the proportion and the number of decidua-resident macrophages in the uterus were significantly lower than those of WT mice (). In contrast, the percentage of NKT cells was increased markedly (Figure S5). The expression of adhesion molecules on the surface of PTPRC+ADGRE1+ ITGAM+ macrophages was decreased, including CDH1, SELE, SELL and ITGAV/integrin αV (). Moreover, the levels of the decidual macrophage migratory molecules Ly6C and M1 polarizing molecules CD80 and CD86 were significantly increased, while the level of M2 polarizing indicator CD209 decreased (). Similar results were observed in the uterus of lpar1−/− pregnant mice (Figure S6A-F) or PPARGi-treated pregnant mice on 7.5th day of gestation (Figure S6G-L). More importantly, transferring macrophage from WT mice led to more M2 like macrophage residence and higher expression of adhesion molecules in pregnant mice with macrophage depletion, compared with the transfer group of macrophages from Enpp2± mice (Figure S6M-P). These results indicate that LPA metabolism can trigger dMφ adhesion and enrichment, and M2 differentiation in decidua.
Autophagy is critical to LPA-induced adhesion of dMφ in decidua
To investigate the possible mechanism of LPA metabolism in dMφ adhesion and residence, ENPP2 inhibitor-treated, LPAR1 inhibitor-treated and control macrophages were collected for RNA-Seq and statistics of pathway enrichment of differential genes showed that the LPA metabolism and receptor signaling were strongly correlated with cell autophagy (Figure S7A). Autophagy is a highly evolutionarily conserved process for the recycling and degradation of cytoplasmic constituents. Autophagy plays an important role in embryogenesis, implantation, placentation and maintenance of pregnancy [Citation30,Citation33]. Compared with pMo from normal early pregnant women, dMφ presented high levels of the number of autophagosomes and autolysosomes (). More notedly, LPA obviously accelerated the formation of autolysosomes in macrophages as well as an autophagy inducer rapamycin (). Flow cytometry assays confirmed that LPA could induce macrophage autophagy in a dose-dependent manner (). In contrast, treatment with ENPP2i or LPAR1i decreased the number of autophagosomes in macrophages (Figure S7B), and the expression of autophagy-related genes MAP1LC3B and BECN1 (Figure S7C).
Figure 4. Autophagy is critical to LPA-induced adhesion of dMφ in decidua. (a) Autophagic structures under transmission electron microscopy in pMo (n = 6) and dMφ (n = 6). (b) Numbers of autophagic structures in (a). (c) DALGreen for autophagy detection of M0 macrophages dyed with PKH26 red fluorescence and treated with vehicle, rapamycin, LPA (5 μM or 10 μM) for 48 h. (d and e) Flow cytometry assays for autophagy detection of M0 macrophages treated with vehicle or LPA (5 μM or 10 μM) for 48 h. (f) Transcription level of DDIT4 in pMo (n = 8) and dMφ (n = 8) by RT-PCR. (g) Western-blot for DDIT4, RHEB, ATG13, BECN1, LC3I/II or SQSTM1 of M0 macrophages treated with LPA (10 μM) for 0, 6, 12 or 24 h. (h-k) Immunofluorescence detection for MTOR and MAP1LC3B expression of M0 macrophages treated with LPA (10 μM) for 0, 6, 12 or 24 h. (l and m) Adhesion assays for control, LPA-treated (10 μM for 24 h), 3-MA-treated (10 mM for 24 h) or both 3-MA (10 mM for 24 h-pretreatment) and LPA-treated (10 μM for 24 h) PKH-26-labeled red-fluorescent dMφ to green-fluorescent DSCs (n = 5). (n) Flow cytometry assays for adhesion molecule expression of control, LPA-treated (10 μM for 24 h) or 3-MA (10 mM for 24 h-pretreatment) and LPA-treated (10 μM for 24 h) dMφ. Data were presented as mean ± SEM or median and quartile and analyzed by t test, Mann-Whitney U test or one-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS: no significance.
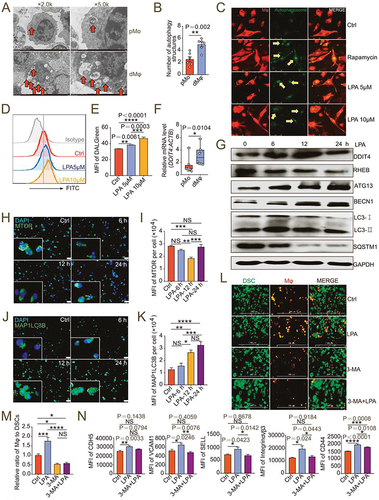
DDIT (DNA damage-inducible transcript) 4 is a negative regulator of MTOR (mechanistic target of rapamycin kinase) signaling pathway and involved in autophagy regulation [Citation34]. As an important differential gene in the RNA-Seq results, we also found that DDIT4 was highly expressed in dMφ (). LPA upregulated the expression of DDIT4 (), whereas ENPP2i or LPAR1i downregulated it, which was consistent with the results of transcriptome sequencing (Figure S8A). Additionally, LPA inhibited the expression of a downstream molecule RHEB (), thereby inhibiting the expression and phosphorylation of MTOR (, I, Figure S8B, C) and promoting the expression of downstream ATG (autophagy-related proteins autophagy-related protein)13, BECN1 and LC3B (). And expression of autophagy substrate protein SQSTM1 decreased (). When LPAR1 and PPARG signals were blocked, the MTOR signal was activated, autophagy was inhibited, and levels of autophagy-related proteins such as LC3B, ATG13 and BECN1 also decreased accordingly (Figure S8A, D-P). These results suggest that LPA induces autophagy of dMφ possibly in a DDIT4-dependent manner.
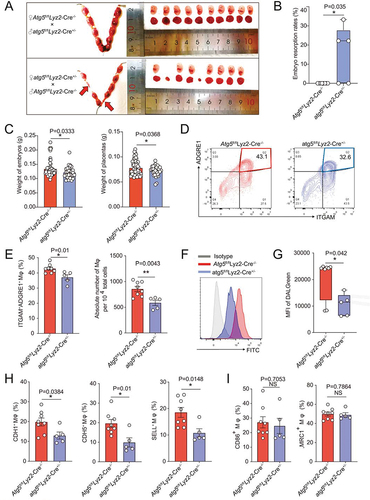
Then we explored whether autophagy was involved in the regulation of decidual macrophage adhesion and residence by LPA. Adhesion ratio of DSCs by macrophages pre-treated with 3-MA (3-methyladenine, an autophagy inhibitor) in vitro was significantly inhibited (). The stimulatory effects of LPA on macrophage adhesion and the expression of adhesion molecules were also restricted due to the inhibition of autophagy by 3-MA (). After intraperitoneal injection of 3-MA, the number and proportion in pregnant mice of uterine macrophages were decreased (Figure S9A-D), the diffusion and invasion of placental extravillous trophoblasts was suppressed and the relative depth of implantation was reduced (Figure S9E-G). More importantly, in atg5fl/flLyz2-Cre± pregnant mice, we observed higher embryo resorption rates, lower embryo and placenta weight, and less residence of M2 like macrophage with low autophagy and adhesion molecules levels, which were evaluated by flow cytometry assays (). These results confirm that LPA promotes dMφ adhesion and enrichment in an autophagy-dependent manner.
Figure 5. Atg5fl/fl Lyz2-Cre± pregnant mice show adverse pregnancy outcomes and reduced uterine macrophage residence. (a and b) Photograph (red arrow: absorption point) and analysis of embryo absorption rates and numbers of implanted embryos of uterus from atg5fl/fl Lyz2-Cre± (mated with male Atg5fl/fl Lyz2-Cre−/− mice, n = 8) and Atg5fl/fl Lyz2-Cre−/− (mated with male atg5fl/fl Lyz2-Cre± mice, n = 5) pregnant mice. (c) Comparison of weight of embryos and placentas of atg5fl/fl Lyz2-Cre± (n = 8) and Atg5fl/fl Lyz2-Cre−/− (n = 5) pregnant mice. (d and e) The residence and number of ADGRE1+ macrophages in the uterus of atg5fl/fl Lyz2-Cre± (n = 8) and Atg5fl/fl Lyz2-Cre−/− (n = 5) pregnant mice by immunofluorescence assays. (f and g) Flow cytometry assays for autophagy detection of ADGRE1+ ITGAM+ uterine macrophages of atg5fl/fl Lyz2-Cre± (n = 8) and Atg5fl/fl Lyz2-Cre−/− (n = 5) pregnant mice. (h and i) Adhesion molecules and differentiation markers of PTPRC+ ADGRE1+ ITGAM+ uterine macrophages of atg5fl/fl Lyz2-Cre± (n = 8) and Atg5fl/fl Lyz2-Cre−/− (n = 5) pregnant mice in flow cytometry assays. Data were presented as mean ± SEM or median analyzed by t test. *P < 0.05, **P < 0.01, NS: no significance.
LPA-autophagy-mediated adhesion and residence of dMφ are dependent on CLDN7
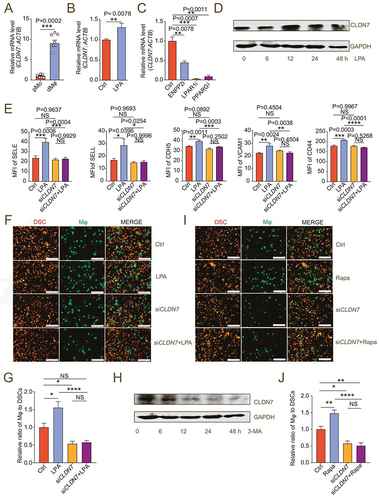
Tissue-resident macrophages physically adhere and stick via various adhesion molecules to reside locally and perform their functions [Citation12]. Claudins are a family of cell adhesion molecules mainly localized at apical tight junctions [Citation35]. The result of RNA-sequence analysis showed that CLDN7 was a differentially expressed gene of macrophage between NC and ENPP2i groups. Claudin-7 has been reported to play a key role in intestinal epithelial tissue integrity and immune homeostasis [Citation36,Citation37]. We observed that the level of differentially expressed gene CLDN7 in dMφ was significantly higher than that in pMo (). The expression of CLDN7 in macrophages was increased by the administration of LPA and was significantly decreased by ENPP2i or LPA receptor inhibition (). LPA promoted the expression of CLDN7 in macrophages (). Knocking down CLDN7 in macrophages significantly inhibited the high levels of adhesion molecules on macrophages and high adhesion ratio to DSCs induced by exogenous LPA (, Figure S9H-I), indicating that LPA-mediated adhesion of dMφ is dependent on the CLDN7. The suppression of autophagy by 3-MA led to the downregulation of CLDN7 level in macrophages (). Conversely, the autophagy inducer rapamycin had a strong promotion effect on adhesion of macrophages (). However, CLDN7-silenced macrophages did not respond to rapamycin (). These data above indicate that CLDN7 induced by autophagy is a key link in the promotion of dMφ adhesion by the LPA.
Figure 6. LPA/autophagy-mediated adhesion and residence of dMφ is dependent on CLDN7. (a) Transcription level of CLDN7 in pMo (n = 7) and dMφ (n = 6) by RT-PCR. (b) Transcription level of CLDN7 in control or LPA-treated (10 mM for 12 h) M0 macrophages by RT-PCR. (c) Transcription level of CLDN7 in control, ENPP2 inhibitor (100 nM), LPAR1 inhibitor (100 nM) or PPARG inhibitor (3.3 nM)-treated M2 macrophages by RT-PCR (12 h). (d) Western-blot for CLDN7 expression of M0 macrophages after treatment with LPA (10 μM) for 0, 6, 12, 24 or 48 h. (e) Flow cytometry assays for adhesion molecule expression of control, LPA-treated (10 μM for 24 h), CLDN7-silenced (siCLDN7) or LPA-treated (10 μM for 24 h) siCLDN7 macrophages. (f and g) Adhesion assays of control, LPA-treated (10 μM for 24 h), siCLDN7 or LPA-treated (10 μM for 24 h) siCLDN7 green-fluorescent dMφ to PKH-26-labeled red-fluorescent DSCs (n = 5). (h) Western-blot for levels of CLDN7 of M2 macrophage after treatment with 3-MA (10 mM) for 0, 6, 12, 24 or 48 h. (i and j) Adhesion assays of control, rapamycin-treated (2 μM for 24 h), siCLDN7 or rapamycin-treated (2 μM for 24 h) siCLDN7 green-fluorescent dMφ to red-fluorescent DSCs (n = 5). Data were presented as mean ± SEM and analyzed by t test or one-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS: no significance.
Patients with unexplained spontaneous abortion display insufficient LPA metabolism and dMφ enrichment
Compared with the decidua of women in normal early pregnancy (NP), the levels of ENPP2, and LPAR1 and PPARG in dMφ in patients with unexplained spontaneous abortion (SA) were significantly decreased (). Additionally, the autophagy level and the levels of autophagy-related genes MAP1LC3B and BECN1 of dMφ in SA group were lower than in NP group (). Conversely, the expression and phosphorylation level of MTOR was high (). There was a lower density of CD68+ macrophages in the decidua of SA patients (), and the number and proportion of PTPRC+ CD14+ dMφ in DICs were also reduced (). Accordingly, the mRNA and protein levels of adhesion molecules (CDH5, SELE, SELL, ICAM2 and VCAM1) and CLDN7 in dMφ were declined in SA patients ().
Figure 7. dMφ in spontaneous abortion exhibit weakened LPA-autophagy axis. (a) Transcription level of ENPP2, LPAR1 and PPARG in dMφ of women in normal early pregnancy (NP, n = 10 or 9) or with unexplained spontaneous abortion (SA, n = 10 or 9) by RT-PCR. (b and c) Flow cytometry for autophagy detection of dMφ of women in NP (n = 6) or with SA (n = 5). (d) Autophagic structures under transmission electron microscopy in dMφ from NP women (n = 5) or SA patients (n = 3). (e) Numbers of autophagic structures in (d). (f) Transcriptional level of autophagy-related genes (MAP1LC3B, BECN1 and MTOR) of dMφ of women in NP (n = 9) or with SA (n = 9) by RT-PCR. (g and h) Immunofluorescence detection for MAP1LC3B and pMTOR expression of dMφ from NP women (n = 6) or SA patients (n = 6). Data were presented as mean ± SEM or median and quartile and analyzed by t test or Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001.
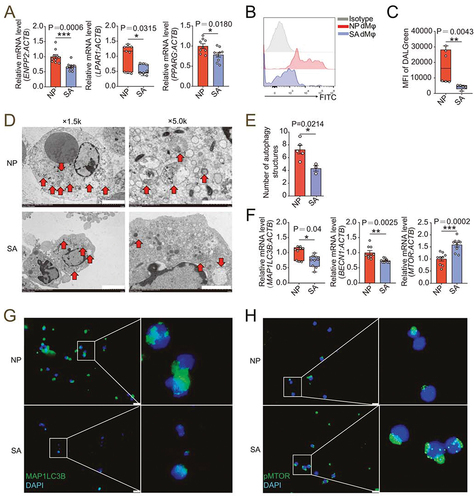
Figure 8. dMφ in spontaneous abortion shows abnormal residence and reduced adhesion. (a) Immunohistochemistry assays for numbers of resident CD68+ dMφ (red arrows) from women in NP (n = 4) or with SA (n = 4). (b) Average numbers of dMφ per 400× microscope field in the images from (a). (c) Numbers of dMφ per 10,000 total decidua single cells from NP (n = 6) or SA (n = 5) women flow cytometry assays. (d) Proportion of CD14+ macrophages in PTPRC+ decidual immune cells in NP (n = 6) or SA (n = 5) group in flow cytometry assays. (e) Relative mRNA level of adhesion-related genes in NP (n = 9) or SA (n = 9) dMφ, including CDH5, SELE, SELL, VCAM1, ICAM2 and CLDN7 by RT-PCR. (f and g) Expression of adhesion molecules on NP (n = 6) or SA (n = 5) dMφ in flow cytometry assays. (h and i) Photograph (red arrow: resorption point) and embryo resorption rates of uterus from normal pregnant (NP) or spontaneous abortion-prone mouse (SA) models. (j-l) Proportion of ADGRE1+ITGAM+ uterine macrophages in PTPRC+ uterus immune cells and numbers per 10,000 uterine cells in NP (n = 8) or SA mouse models (n = 8) in flow cytometry assays. (m and n) Adhesion molecules and differentiation markers of PTPRC+ ADGRE1+ ITGAM+ uterine macrophages of NP (n = 8) or SA (n = 8) mice in flow cytometry assays. Data were presented as mean ± SEM or median and quartile and analyzed by t test or Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
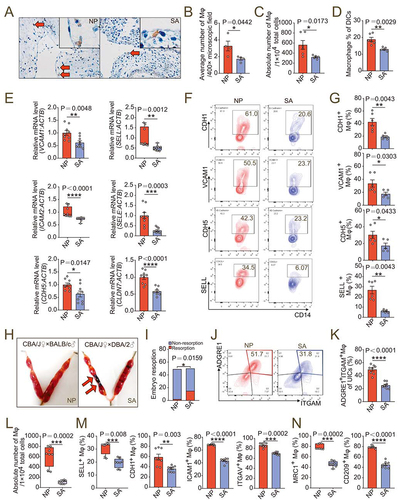
We then established normal pregnancy mouse models (CBA/J♀×BALB/c♂) and abortion mouse models (CBA/J♀×DBA/2♂) (). The proportion and number of PTPRC+ ITGAM+ ADGRE1+ macrophages in uterine of aborted mice decreased (), and the adhesion molecules were downregulated (). In particular, the uterine macrophages of normal pregnant mice had a M2 polarized phenotype, and the proportion of MRC1+ and CD209+ uterine macrophages of aborted pregnant mice decreased (), which echoed our previous research results [Citation8]. Therefore, the enrichment and function maintenance of dMφ, especially M2 macrophages, play a key role in establishing a normal pregnancy. However, the insufficient LPA metabolism and autophagy of dMφ contribute to the occurrence of spontaneous abortion by triggering the insufficient infiltration of macrophage in decidua.
LPA prevents pregnancy loss by promoting autophagy-mediated macrophage residence in decidua
Owing to the positive regulatory effects of LPA on the autophagy-mediated cell residence and M2 differentiation of dMφ, the potential values of rapamycin and LPA in preventing spontaneous abortion were evaluated in vivo. As expected, LPA and low-dose rapamycin effectively improved embryo loss, increased implanted embryo numbers and weight, and/or placenta weight of abortion mouse models (, Figure S10A-C). Additionally, both LPA and rapamycin markedly promoted macrophage autophagy and enrichment, and upregulated the expression of adhesion molecules in uterus of abortion mouse models (, Figure S10D-G). Different from LPA, rapamycin did not influence on macrophage differentiation (, Figure S10H). Similarly, LPA and rapamycin reversed embryo loss, increased embryo weight and/or placenta weight, and promoted macrophage autophagy, adhesion and enrichment, and/or M2 differentiation in uterus of Enpp2± pregnant mice (, Figure S10I-P). Taken together, these findings indicate that LPA and autophagy inducer rapamycin promote autophagy-mediated dMφ residence and enrichment in decidua, which contributes to trophoblast invasion and placenta development, and further decreases the risk of spontaneous abortion.
Figure 9. LPA prevents pregnancy loss by promoting autophagy-mediated macrophage residence in decidua. (a and b) Photograph (red arrow: resorption point) and analysis of embryo resorption rates and numbers of implanted embryos of uterus from SA mice intraperitoneally injected with 1% PBS (Ctrl, n = 5) or LPA (n = 5). (c) Comparison of weight of embryos and placentas of SA mice intraperitoneally injected with 1% PBS (Ctrl, n = 5) or LPA (n = 5). (d) Flow cytometry assays for autophagy detection of ADGRE1+ ITGAM+uterine macrophages of SA mice intraperitoneally injected with 1% PBS (Ctrl, n = 5) or LPA (n = 5). (e and f) Flow cytometry assays for detection of the proportion of ADGRE1+ITGAM+uterine macrophages in PTPRC+ immune cells and the absolute numbers per 10,000 cells in uterus of 1% PBS (Ctrl, n = 5)/LPA (n = 5)-treated SA mice. (g and h) Adhesion molecules and differentiation markers of PTPRC+ ADGRE1+ ITGAM+ uterine macrophages of 1% PBS (Ctrl, n = 5)/LPA (n = 5)-treated SA mice in flow cytometry assays. (i and j) Photograph (red arrow: resorption point) and embryo resorption rates and numbers of implanted embryos of uterus from Enpp2± pregnant mice intraperitoneally injected with 1% PBS (Ctrl, n = 4) or LPA (n = 4). (k) Comparison of weight of embryos and placentas of Enpp2± pregnant mice intraperitoneally injected with 1% PBS (Ctrl, n = 4) or LPA (n = 4). (l) Flow cytometry assays for autophagy detection of ADGRE1+ ITGAM+ uterine macrophages of Enpp2± pregnant mice intraperitoneally injected with 1% PBS (Ctrl, n = 4) or LPA (n = 4). (m and n) Flow cytometry assays for detection of the proportion of ADGRE1+ ITGAM+ uterine macrophages in PTPRC+ immune cells and the absolute numbers per 10,000 cells in uterus of 1% PBS (Ctrl, n = 4)/LPA (n = 4)-treated Enpp2± pregnant mice. (o and p) Adhesion molecules and differentiation markers of PTPRC+ ADGRE1+ ITGAM+ uterine macrophages of 1% PBS (Ctrl, n = 4)/LPA (n = 4)-treated Enpp2± pregnant mice in flow cytometry assays. Data were presented as mean ± SEM or median and quartile and analyzed by t test or Mann-Whitney U test. *P < 0.05, **P < 0.01, NS: no significance.
Discussion
Decidual macrophages show a phenotype typical for homeostatic M2 macrophages induced by CSF1/M-CSF and IL10 [Citation38]. They play a role diversely in the recognition and clearance of infections, the clearance of apoptotic debris and tissue remodeling [Citation39]. Here, we observed the number and proportion of local macrophages in decidua of SA patients and aborted mice were significantly less than those of NP women and normal pregnant mice. Macrophages adoptively transferred by caudal vein can reach and stay in the uterus of pregnant mice [Citation8], suggeting that monocyte of peripheral blood should be an important source of dMφ during pregnancy. Compared with pMo, dMφ highly express more than 100 adhesion molecules, helping them to reside locally in decidua. Inadequate dMφ adhesion and residence of women with SA, implying that the instability of dMφ is related to the pathogenesis of the disease.
Subsequently, we analyzed the characteristics of dMφ from the perspective of cellular metabolism, and observed the enrichment of ENPP2-LPA in dMφ, which are consistent with other tissue-resident macrophages (i.e., monocyte‒derived macrophages in broncho alveolar lavage, intestinal macrophage, and microglial cells) [Citation40–42]. Physiological and molecular processes initiated during normal pregnancy are complex but highly organized. These events are primarily coordinated by estrogen and progesterone and perfectly modulated by the molecular dialogue that originates locally from the mother, embryo or both governing the orderly chronological transitions [Citation43]. ENPP2 was reported as an estrogen-responsive gene in rat hippocampus [Citation44], suggesting that the local microenvironment at the maternal-fetal interface in early pregnancy, especially the endocrine system should play important roles in the regulation of ENPP2-LPA. However, the exact mechanism remains to be further studied.
As an intermediate metabolite in the glycerophospholipid metabolism, LPA is the smallest and simplest phospholipid molecule. It has been reported to be involved in the development and homeostasis of macrophages. LPA enhances macrophage-derived foam cell formation via LPAR1-LPAR3-AKT activation [Citation45]. LPA converts murine or human monocytes into macrophages, in which process, AKT-MTOR signaling is involved and PPARG is the key transcriptional regulator [Citation22]. A previous report by microarray data analysis indicated genes (i.e., PPARG) expressed by ITGAX/CD11cHi dMφ might be associated with lipid metabolism [Citation46]. Here, we found LPA transmembrane receptor LPAR1 and intracellular receptor PPARG are highly expressed in dMφ from NP women, compared with pMo. Notably, enriched LPA caused by active glycerophospholipid metabolism in dMφ promotes their adhesion and M2 differentiation through LPAR1 and PPARG. The inhibition of LPA enrichment or dysfunction resulted in severe pregnancy loss, and significant impairments of embryo growth, placental development, and trophoblast invasion. Additionally, there are marked differences in resorption of WT and HE embryos in the same Enpp2± pregnant mice, suggesting that ENPP2-LPA may be involved in the crosstalk between trophoblast and macrophage, which needs to be further research.
Autophagy is activated by a range of environmental conditions, such as hypoxia and starvation, but also hormones, cytokines, growth factors and receptor engagement [Citation47]. Signaling for autophagy induced by CSF1/M-CSF or CSF2/GM-CSF can promote the survival of differentiated macrophages from monocytes [Citation48]. Interestingly, our data indicate that decidua-resident macrophages display active autophagy during normal early pregnancy, and autophagy regulates the adhesion of macrophages to decidua, which enlarges the role of autophagy in the homeostasis of immune cells.
A key regulator of autophagy in mammalian cells is a kinase complex MTORC1 that receives multiple upstream signals and monitors nutrient and growth factor availability to control diverse biological processes including protein translation and bioenergetic organization [Citation49]. Activated MTORC1 leads to suppression of autophagy through phosphorylation of proximal components of the autophagy machinery including ULK1 (unk-51 like kinase 1) and ATG13 [Citation50]. DDIT4 is one of the essential regulators of MTORC1 activity in response to hypoxia and energy stress [Citation51]. LPA stimulates a pseudohypoxic response via RAC-mediated activation of NADPH oxidase and generation of reactive oxygen species in ovary cancer cells, resulting in activation of HIF1A [Citation52]. At the maternal-fetal interface, LPA promoted the expression of DDIT4, inhibit the downstream RHEB, and thereby suppressed the level of MTOR, ultimately leading to an increase in autophagy levels of dMφ. Therefore, our data reveal a new and essential mechanism by which LPA signals are transduced to active autophagy possibly through the metabolic regulator DDIT4. Hypoxic tumor-associated macrophages strongly upregulate the expression of DDIT4, and DDIT4-mediated MTOR inhibition hinders glycolysis in tumor-associated macrophages and curtails their excessive angiogenic response, with consequent formation of abnormal blood vessels [Citation53]. Here, we observed LPA promoted the expression of growth factors and pro-angiogenic factors, and LPA-treated macrophages had positive effects on the growth and invasion of trophoblasts. These processes should be due to the regulation of LPA on the DDIT4-MTOR signaling and autophagy. Hypoxia results in overactivation of LPAR1 through selective inhibition of G protein-coupled receptor kinase 2 expression in fetal neural progenitor cells [Citation54]. Therefore, there should be a two-way positive feedback loop between LPA signaling and hypoxia. It should be emphasized that the LPA signaling-DDIT4-autophagy axis contributes to the enrichment and homeostasis of dMφ, and unique hypoxic environment at maternal-fetal interface may be a vital link throughout this regulatory process.
Cell adhesion molecules play a fundamental role in in human implantation stage [Citation55]. CLDN7 is one of the most dominant claudins expressed in the intestine. Deletion of CLDN7 led to the disruption of intestinal architecture with mucosal ulcerations, epithelial sloughing, and inflammation [Citation36]. Especially, expression of cell-cell adhesion molecules of the ileum and colon, including CLDN1, CLDN2, CLDN3, CLDN8, CLDN15 and CLDN25, MARVELD2, TJP1, EPCAM and CDH1 in the cKO colon are significantly decreased compared with the control group, suggesting a core role of CLDN7 in the normal barrier function of intestinal mucosa [Citation37]. In our study, CLDN7 is also found to be the core molecule of LPA-autophagy-mediated cell adhesion of dMφ. Studies have shown that HNF4A upregulates CLDN7 expression in colonic epithelial cells [Citation56], and HNF4A can induce autophagosome formation and MAP1LC3/LC3-II protein levels by activation of UKL1 [Citation57]. Moreover, ULK1 is critical for induction of macroautophagy/autophagy and has a more specific role in mitophagy in response to hypoxia [Citation58]. The evidence above suggests that the regulation of CLDN7 by LPA-autophagy should be dependent on the HNF4A-ULK1, which needs further study.
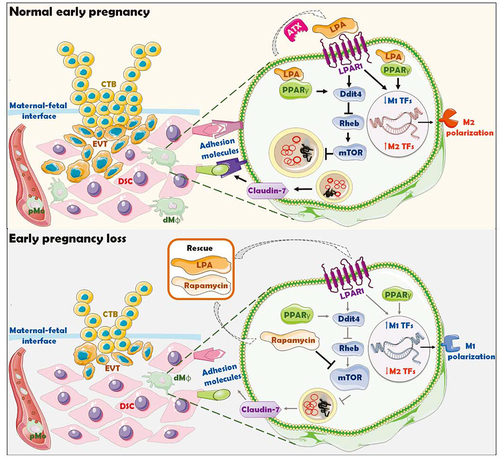
Therefore, as shown in and , this study indicates that the enrichment and activation of ENPP2-LPA metabolism at the maternal-fetal interface, promotes the expression of adhesion molecules and M2 differentiation on dMφ via the LPA receptors-DDIT4-autophagy-CLDN7 axis, thereby facilitating the adhesion and retention of macrophage in decidua, trophoblast invasion, fetal and placental development of early pregnancy. Inactivation of ENPP2-LPA metabolism and insufficient autophagy of dMφ results in the decrease of dMφ residence, increasing the risk of spontaneous abortion. These results help to elucidate the immune metabolism mechanism of dMφ residence and development during early pregnancy and the mechanism of spontaneous abortion caused by dMφ dysfunction. Excitingly, supplement with LPA or an autophagy inducer rapamycin significantly prevents the risk of spontaneous abortion. Thus, the potential therapeutic values of LPA and rapamycin in spontaneous abortion should be emphasized due to the activation of DDIT4-autophagy-CLDN7-adhension molecules-mediated dMφ residence and trophoblast invasion during early pregnancy. Considering that autophagy and LPA have been reported to have various negative impacts like cancer promotion [Citation52,Citation59–61], the potential risks of LPA in cancer promotion are still not be ignored.
Figure 10. Schematic roles of inactive lysophosphatidic acid metabolism-autophagy axis in increasing spontaneous abortion risk by restricting dMφ residence. The growth and invasion of trophoblasts is the key to the establishment and maintenance of normal pregnancy, which is accompanied by the residence and development of dMφ. The enrichment and activation of ENPP2-LPA metabolism promotes the expression of adhesion molecules on dMφ via the LPA receptors-DDIT4-RHEB-MTOR-autophagy-CLDN7 axis, thereby facilitating the adhesion and retention of macrophage in decidua. Decidual-resident macrophages further promote trophoblast invasion, fetal and placental development of early pregnancy. Inactivation of ENPP2-LPA metabolism and insufficient autophagy of dMφ results in the decrease of dMφ residence, increasing the risk of spontaneous abortion. LPA and rapamycin have potential therapeutic values in spontaneous abortion due to the activation of DDIT4-autophagy-CLDN7-adhension molecules-mediated dMφ residence and trophoblast invasion during early pregnancy.
Materials and methods
Mice
Female CBA/J mice (5- to 7-weeks-old), male DBA/2 mice and male BALB/c mice were purchased from Beijing Huafukang Biotechnology Co., Ltd. (China). After 3 weeks of adaptive feeding, 8- to 10-weeks-old female CBA/J mice were mated with male BALB/c mice or male DBA/2 mice (2: 1) to construct a normal pregnancy mouse model or abortion mouse model. All knockout mice were constructed from Shanghai Model Organisms Center, Inc. (China). Due to homozygous lethality, female Enpp2 heterozygous (Enpp2±) mice of childbearing age were mated with male Enpp2± or wild-type (WT) mice. In order to exclude the effect of embryo genotype, female homozygous lpar1 knockout (lpar1−/−) mice were mated with male WT mice to obtain pregnant mice in the experimental group, while female WT pregnant mice were mated with male lpar1−/− mice to obtain the control mice. C57 mice of childbearing age were purchased from Shanghai Jiesijie Experimental Animal Co., Ltd. (China). From day 0 of pregnancy, C57 pregnant mice were injected intraperitoneally with a PPARG inhibitor (GW9662, 1 mg/kg; MCE, HY-16578) daily, an autophagy inhibitor 3-methyladenine (3-MA, 10 mg/kg; Sigma-Aldrich, 5142–23-4) every 3 days (day 0, 4, 7 or 0, 4, 7, 10, 13) or vehicle (DMSO, 1%; Sigma-Aldrich, 67–68-5). CBA/J pregnant mice (mated with male DBA/2 mice) or Enpp2± pregnant mice (mated with male WT mice) were injected intraperitoneally with oleoyl-L-α-lysophosphatidic acid (10 nmol, every 3 days; Sigma-Aldrich, L7260) or low-dose rapamycin (0.04 mg/kg, every 3 days; MCE, HY-10219) from day 0 of pregnancy. Female WT mice (mated with male WT mice) were depleted of macrophages by liposome intraperitoneal injection on the 1.5th (200 μL liposomes; FormuMax, F70101C-AC) and 4.5th day (100 µL liposomes; FormuMax, F70101C-AC) of gestation and adoptively transferred on the 5.5th day of gestation with ADGRE1-positive or ADGRE1-positive PKH-26 (Sigma-Aldrich, MIDI26)-labeled macrophages sorted by anti-ADGRE1 MicroBeads UltraPure (Miltenyi, 130–110-443) from the spleen of WT mice or Enpp2± mice. Female atg5fl/flLyz2-Cre± mice were mated with male Atg5fl/flLyz2-Cre−/− mice and female Atg5fl/flLyz2-Cre−/− were mated with male atg5fl/flLyz2-Cre± mice. On the 7.5th day of pregnancy, the numbers of embryos implanted were recorded. The uterus and embryo tissues were fixed in 4% paraformaldehyde for subsequent sectioning. The uterine tissues were in stripped and collected and digested with 50% type IV collagenase (Sigma-Aldrich, 9001–12-1) for 1 h and filtered into a single cell suspension for flow cytometry assays. On the 13.5th day of pregnancy, the uterus of the pregnant mice was taken, the embryo resorption was observed, the uterine tissues were separated, and the embryo and placenta were weighed, sized, or genotyped. In particular, the embryo resorption rate was calculated by dividing the number of embryos resorbed by total number of embryos (×100%) of each pregnant mouse. The Animal Care and Use Committee of Fudan University approved all animal protocols.
Tissues
Normal decidual tissues (n = 68) and peripheral blood samples (n = 28) were from women in normal first trimester pregnancy and abortion decidual tissues (n = 28) were from women with unexplained recurrent spontaneous abortion excluding genetic or endocrine factors for selective termination (age, 23‐35 years old; gestational age, 7‐9 weeks). Endometrial tissues (n = 10) were collected from women with normal pregnancy history of reproductive ages (28–40 years old), who were diagnostic curettage or undergoing hysterectomy for benign reasons unrelated to endometrial dysfunction. All samples were evaluated by a histopathologist to identify the cyclic phase as secretory phase and exclude endometrial pathology. None of them had received hormonal medication in the 6 months before the surgical procedure. Tissue collection was all approved by the Ethical Committee and conducted in Obstetrics and Gynecology Hospital of Fudan University. All procedures were obtained with the consent of the patients.
After surgery, the decidual tissues or endometrial tissues were collected under sterile conditions and transported to the laboratory on ice within 30 min in Dulbecco’s modified Eagle’s medium (DMEM)/F‐12 (HyClone, SH30023.01) with 10% fetal bovine serum (FBS; Gibco, 10,091,148) for further isolating DSCs, DICs or endometrial stromal cells (ESCs). Peripheral blood samples were taken sterilely in heparinized Hanks’ buffer solution (Gibco, 24,020,117) before anesthesia and were transported to the laboratory on ice for sorting of peripheral blood mononuclear cells (PBMCs) and pMo.
Cells
Primary DSCs and DICs were isolated and purified form collected decidual tissues. Tissues were washed in Ca2+Mg2+-free PBS (HyClone, SH30256.01), minced and then digested with 5% DNA enzymes (3000IU; Sigma-Aldrich, 9003–98-9) and 20% collagenase type IV (0.1%; Sigma-Aldrich, 9001–12-1) at 37°C with constant agitation. After 30 min, the enzymatic reaction was stopped by DMEM high-glucose medium (HyClone, SH30243.01) containing 20% FBS. The suspension was then filtered through sterile gauze pads (pore diameter sizes: 100, 200, and 400 mesh). The filtered suspension was collected and centrifuged at 400 g for 10 min to acquire cell pellet, which was then suspended in DMEM/F12 (HyClone, SH30023.01) and centrifuged on a discontinuous gradient of 20%, 40%, and 60% Percoll bulk standard for 30 min at 800 g. Percoll bulk standard consists of 90% Percoll (Amersham, GE Healthcare Life Sciences, 17–0891-01) and 10% 10× PBS (HyClone, SH30258.01). DSCs or DICs were acquired from the 20%/40% interface or the 40%/60% interface respectively.
The endometrial tissues were minced into 2 mm pieces and then digested with 10% collagenase type IV (0.1%; Sigma-Aldrich, 9001–12-1) and constant agitation for 40 min at 37°C. The tissue pieces were filtered through sterile gauze pads (pore diameter sizes: 100, 200 and 400 mesh). After centrifugation, the supernatant was discarded, and the cells were resuspended in DMEM/F-12 (HyClone, SH30023.01) containing 10% FBS, plated on culture flasks and incubated in a humidified incubator with 5% CO2 at 37°C. The ESCs were allowed to adhere for 20 min. The culture medium was replaced every 2–3 days.
PBMCs were isolated from the blood samples of first trimester pregnant women by Ficoll‐Hypaque (Sigma-Aldrich, 26,873–85-8) density gradient centrifugation at 800 g for 30 min. According to the manufacturer’s instructions (Miltenyi Biotec, 130–050-201), pMo from PBMCs and dMφ from DICs were obtained through positive selection by an CD14+ cell isolation kit.
The human monocyte cell line U937 was purchased from American Type Culture Collection (ATCC, CRL-3253) and cultured with RPMI-1640 medium (HyClone, SH30027.01) containing 10% FBS. U937 cells seeded in 6-well plate (5 × 105/well) were treated with recombinant human (rHs) macrophage colony stimulating factor (CSF1, 50 ng/mL; Pepro Tech, AF-300-25-100) to induce M0 macrophages and both rHs-CSF1 (50 ng/mL) and rHs-IL10 (50 ng/mL; Pepro Tech, AF-200-10-100) for 6 days to induce M2 macrophages according to previous report [Citation38]. For microscope or confocal scanning, macrophages were induced to be adherent with phorbol 12-myristate 13-acetate (PMA, 100 mg/mL; Sigma-Aldrich, 16,561–29-8) for 24 h.
Transcriptome sequencing
Human pMo and dMφ enriched and purified during the first trimester of normal pregnancy were subjected to transcriptome sequencing (RNA-Seq). M2 macrophages in the control group and M2 macrophages treated with an ENPP2 inhibitor (PF8380, 100 nM; MCE, HY-13344) or a LPAR1 inhibitor (AM966, 100 nM; MCE, HY-15277) for 12 h were collected and subjected to RNA-Seq. After total RNA was extracted according to standard instructions, quality assessment was performed by Agilent 2100. The libraries were established before sequencing. After the library quality inspection was qualified, the Illumina Hiseq4000 was used for sequencing, and the sequencing read length is double-ended 2 × 150bp. RNA-seq reads were cleaned using fastq-mcf and mapped to the genome using STAR. Read counts per gene were obtained from the aligned reads using htseq-count and DESeq2 was used for normalization and differential gene expression analysis. To rank the genes, log2-transformed fold-change values were used. Differential genes or differential metabolites were enriched by the GO ontology or KEGG pathway in gene set enrichment analysis.
Metabolomics testing (LC-MS analysis)
Human pMo and dMφ enriched and purified during the first trimester of normal pregnancy were subjected to metabolomics testing. The cells were homogenized and lysed by sonication (30,000 Hz) in pre-chilled PBS buffer. The proteins in the supernatant after centrifugation (x 1,000 g) with cold 85% acetone repeatedly were harvested as prepared free metabolites-free proteome. Chromatography was carried out in LC-MS detection (Thermo, Ultimate 3000LC, Orbitrap Elite). The separation was done on a Hypergod C18 column (100 mm × 4.6 mm, 3 um particle size), with a column flow rate 0.3 mL/min and separated on the analytical column (Acclaim PepMap C18, 75 mm × 15 cm) with a linear gradient. The analytical column was heated to 40°C using an AgileSLEEVE column heater (Analytical Sales and Services). And the analytical column was equilibrated to 98% Mobile Phase A (0.1% formic acid/3% acetonitrile) and 2% Mobile Phase B (0.1% formic acid/90% acetonitrile) and maintained at a constant column flow of 0.3 mL/min. Sample analysis was performed applying SIEVE software (Thermo).
Based on the human metabolic pathway information provided by KEGG (www.kegg.jp/kegg/pathway.html), the MetaboAnalyst 3.0 tool (http://www.metaboanalyst.ca/) was used to obtain significantly enriched metabolic pathways. p < 0.05 was used as the threshold of significant enrichment, and combined with the impact factor of pathway topology analysis (Impact), the key pathway with the highest correlation with metabolite difference was found.
Cell transfection
Recombinant lentivirus of siRNA targeting CLDN7 (siCLDN7, Genechem, 1366 NM_001307) or control lentivirus with GFP was from Shanghai Genechem Co., LTD. (China). U937 cells were treated with phorbol 12-myristate 13-acetate (PMA, 100 mg/mL; Sigma-Aldrich, 16,561–29-8) for 24 h to induce adherent macrophages. Macrophages were seeded in 6-well culture plates (5 × 105 cells/well) and subsequently infected with siCLDN7 or control lentivirus. Twelve h after infection, fresh complete culture medium was used to continue the culture. The infection efficiency was evaluated by GFP fluorescence intensity and RT-PCR 96 h after infection.
Immunohistochemistry and immunofluorescence
All paraffin sections were technically supported by Wuhan Servicebio Technology Co., Ltd. (China). Paraffin sections of decidua tissues or pregnant mouse uterus were dehydrated in graded ethanol. Then the sections were incubated with 3% hydrogen peroxide (H2O2) and 1% bovine serum albumin/TBS to block endogenous peroxidase activity. Human decidua samples were subsequently incubated with rabbit anti-human CD68 (1:200; Abcam, ab125212), or Rabbit IgG, polyclonal – isotype control (1:200; Abcam, ab171870) overnight at 4°C in a humid chamber. After washing three times with TBS, the sections were incubated and the signal was visualized with GTvision TMIII Immunohistochemical Detection Kit (Gene Tech, GK500710). The sections were counterstained with hematoxylin (Biotech Well Biotechnology, WH2013).
Mouse uterus samples were incubated with rat anti-mouse ADGRE1 antibody (1:200; Abcam, ab6640) or mouse anti-mouse keratin (KRT)-7 antibody (1:1000; Abcam, ab181598) overnight at 4°C in a humid chamber. After washing three times with TBS, the sections were incubated goat anti-rat IgG H&L (Alexa Fluor® 594) preadsorbed antibody (1:400; Abcam, ab150160) or goat anti-mouse IgG H&L (Alexa Fluor® 488) preadsorbed antibody (1:400; Abcam, ab150113). And the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime, C1002).
LPA-treated (10 μM; 0, 6, 12 or 24 h) M0 macrophages and PF8380-treated (100 nM), AM966-treated (100 nM) or PPARG inhibitor (GW9662)-treated (3.3 nM) (0, 6, 12 or 24 h) M2 macrophages were seeded in a flat-bottom 24-well plate with cover glasses at the bottom (2 × 105/well). The collection of decidual macrophages from NP or SA women was as described above. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 (Sigma‐Aldrich, 9036–19-5) and blocked with 10% BSA in PBS. The cells were incubated with rabbit anti-human MAP1LC3B antibody (1:200; Novus, NB100-2220), rabbit anti-human MTOR antibody (1:400; Cell Signaling Technology, 2983) or rabbit anti-human MTOR (phospho S2448)/pMTOR (1:100; Abcam, ab131538) at 4°C overnight. After washing three times with TBS, the cells were incubated with goat anti-rabbit IgG H&L (Alexa Fluor® 488) preadsorbed antibody (1:400; Abcam, ab150077). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Beyotime, C1002).
All the images were taken by microscope (Olympus) and the expression levels of factors were quantified using integrated optical density values generated by the Image-Pro Plus 6.0 software (Media Cybernetics). The experiments were performed in triplicate.
Real time (RT)-RCR
Human pMo, dMφ, PF8380/AM966/GW9662-treated (12 h) M2 macrophages, oleoyl-L-α-lysophosphatidic acid sodium salt (LPA, 10 μM, 12 h Aladdin, 22,556–62-3) treated M0 macrophages, and macrophages that were treated by LPA for 12 h with pretreatment with PF8380 for 4 h were collected. U937 cells were treated with PBS (1‰), recombinant human CFS1 (50 ng/mL; Pepro Tech, AF-300-25-100) or recombinant human CSF1 (50 ng/mL; Pepro Tech, AF-300-25-100) and IL10 (50 ng/mL; Pepro Tech, AF-200-10-100) for 6 days.
Total RNA of macrophages or mouse uterus tissues weas extracted using the TRIzol® reagent (Invitrogen, 15596018). RNA was reverse-transcribed into cDNA using the Prime Script RT reagent Kit (Takara, RR036A). RT-PCR was performed using SYBR Premix Ex Taq (Takara, RR820A) and analyzed using an ABI Prism 7900 Fast Sequence Detection system (Thermo Fisher Scientific). The primer sequences of these genes are provided in Supplementary table S1. Each sample was analyzed in three replicate wells and the fold change in the transcriptional expression of the above genes was calculated using the 2−ΔΔCt method. Relative mRNA expression levels were normalized to ACTB. The experiments were performed in triplicate.
Flow cytometry assays
A total of 1 × 106 cells was resuspended in PBS buffer and then the cells were incubated at room temperature for 30 min in 50 μL of staining buffer with 1 μg/mL of relevant antibodies or isotype IgG antibodies. After staining cells with antibodies with cell membrane molecules, cells were fixed, permeabilized (BD Biosciences), and stained with antibodies with intracellular molecules. The details of all antibodies are shown in . The labeled cells were washed twice with cold PBS. The samples were analyzed by using a Beckman CytoFLEX S flow cytometer (Beckman Coulter) and Cellquest software (Beckman). The experiments were performed in triplicate.
Table 1. Antibodies for flow cytometry assays.
Western blotting (WB)
Macrophages treated with LPA (10 μM), AM966 (100 nM), GW9662 (3.3 nM) or 3-MA (10 mM) with different times were washed in PBS and centrifuged for 20 min at 10,000 × g at 4°C. The pellet was resuspended in high efficiency cell tissue rapid lysis buffer (RIPA, Beyotime, P0013B) containing 1% phenylmethanesulfonylfluoride (PMSF, Beyotime, ST506) proteinase. Protein concentrations were quantified using the BCA protein assay kit (Beyotime, P0012). Cell lysates were boiled for 10 min at 95°C and then were stored at −80°C. Total proteins (25 μg) were electrophoresed in SDS-PAGE gels (Epizyme Biotech, PG113) using a Miniprotein III system (Bio-Rad, 1,658,033) and were transferred to PVDF membranes (Millipore, ISEQ00010) for 90 min, followed by incubation with 5% skimmed milk in PBST (Beyotime, P0222) for 1 h and overnight incubation with rabbit anti-human DDIT4/REDD1 antibody (1:1000; Abcam, ab191871), rabbit anti-human RHEB antibody (1:1000; Abcam, ab25873), rabbit anti-human ATG13 antibody (1:1000; Cell Signaling Technology, 13,468), rabbit anti-human BECN1 antibody (1:1000; Cell Signaling Technology, 3495), rabbit anti-human MAP1LC3B antibody (1:1000; Abcam, ab131538), mouse anti-human SQSTM1/p62 antibody (1:1000; Cell Signaling Technology, 88,588), rabbit anti-human claudin-7/CLDN7 antibody (1:1000; Abcam, ab27487) or rabbit anti-human GAPDH antibody (1:3000; Affinity, AFA0911) at 4°C. Then PVDF membranes were washed three times with PBST and were incubated at room temperature for 1 h in peroxidase-conjugated goat anti-rabbit or anti-mouse IgG secondary antibodies (1:5000; Bioworld Technology, BS10003). Thereafter the membranes were washed three times and processed for chemiluminescence using the Immobilon Western Chemiluminescent HRP Substrate Kit (Millipore, WBKLS0100). The experiments were performed in triplicate.
Transmission electron microscopy detection
Human pMo, dMφ, and M2 macrophages were collected and fixed in 2.5% glutaraldehyde and post-fixed in 1% osmium tetroxide. The samples were dehydrated in an ascending series of alcohols, and then embedded in epoxy resin (Sigma-Aldrich, 45,345). The sections were cut, stained with uranyl acetate and lead citrate, and then examined under a Philips CM120 transmission electron microscope (Philips, Amsterdam, Netherlands). Autophagosomes are found in the middle stage of autophagy which are a double-layered vacuole structure containing cytoplasmic components, such as mitochondria, endoplasmic reticulum and ribosomes. Autolysosomes are seen in the late stage of autophagy with monolayer membrane and cytoplasmic components have been degraded.
DALGreen autophagy detection
M0 macrophages were seeded on μ-slide 8 well (Ibidi, 80827) and cultured overnight. The cells were washed with culture medium and then incubated at 37°C for 30 min with 250 μL of 1 μmol/L DALGreen working solution (DALGreen-Autophagy Detection; Dojindo, D675). After washed with the culture medium twice, the cells were treated with DMSO (1%; Sigma-Aldrich, 67–68-5), rapamycin (2 μM; Sigma-Aldrich, 53123–88-9), LPA (5 μM or 10 μM; Aladdin, 22556–62-3) for 48 h. The cells were washed with Hanks’ buffer solution twice and then DALGreen was observed by confocal fluorescence microscopy. Or the cells were washed with PBS, treated with trypsin and centrifuged. The pellets were suspended in Hanks’ buffer solution, and detected by flow cytometry. Fluorescence images were obtained using confocal microscopy (Leica) at an excitation wavelength of 488 nm and a 500–563 nm emission filter. These data were obtained using flow cytometer (Beckman) at an excitation wavelength of 405 nm and a 485–535 nm emission filter. The experiments were performed in triplicate.
Cell adhesion assay
The dMφ, M0 macrophages or M2 macrophages pre-treated with LPA, rapamycin, 3-MA, PF8380, AM966, GW9662 for 24 h or CLDN7 knockdown lentivirus (siCLDN7) were collected to study their adhesive capacity to DSCs. The day before the collection of macrophages, DSCs were labeled with green fluorescence using PKH67 green fluorescent cell linker kit (Sigma-Aldrich, MIDI67) or red fluorescence using PKH26 red fluorescent cell linker kit (Sigma-Aldrich, MIDI26) for general cell membrane labeling according to the manufacturer’s protocol then seeded into a 48-well plate at a density of 5 × 104 cells/mL. Macrophages labeled with red fluorescence using PKH26 red fluorescent cell linker kit or with GFP-green fluorescence were co-cultured with PKH67‐labeled or PKH26-labeled DSCs at a density of 1 × 105 cells/mL. After 6 h, the culture medium was removed and the rest of the cells were washed with PBS once to leave the adherent macrophages. Next, the cells were photographed immediately using an Olympus BX51 + DP70 fluorescence microscope (Olympus). The images were taken in 5 randomly selected fields in every sample at a magnification of 100× and the cells were counted in every photograph. The numbers of adherent macrophages and DSCs were quantified using the Image-Pro Plus 6.0 software (Media Cybernetics). After calculating the ratio of macrophages to DSCs, the relative ratio was obtained for statistics based on the control group. The experiments were performed in triplicate.
Liquid bead suspension chip array
The culture supernatant was harvested, centrifuged to remove cellular debris, and stored at −80°C. Liquid bead suspension chips (Bio-Plex Pro Human Cytokine GrpIPanel 27-plex, LXSAHM-09) were customized by Shanghai Huaying Biomedical Technology Co., Ltd. (China).
Enzyme linked immunosorbent assay (ELISA)
The culture supernatant of M2 macrophages after treated with DMSO (1%) or ENPP2 inhibitor (100 nM for 12, 24 or 48 h) was harvested. The levels of LPA were detected by Human LPA Elisa Kit (CUSABIO, CSB-EQ028005HU) according to the standard directions. The experiments were performed in triplicate.
Cell proliferation and matrigel invasion assays
The HLA-G positive choriocarcinoma cell line JEG3, which was applied to investigate the evolution of placentation and regulating on classical and non-classical MHC class I molecules [Citation62,Citation63], was used in this research for trophoblast cell proliferation and Matrigel invasion assays in vitro. JEG3 was purchased from American Type Culture Collection (ATCC, HTB-36) and cultured with DMEM/F12 medium (HyClone, SH30023.01) containing 10% FBS. In addition, M0 macrophages were treated with LPA (10 Μm; Aladdin, 22,556–62-3) or PBS for 24 h, and M2 macrophages were treated with ENPP2 inhibitor (PF8380, 100 nM; MCE, HY-13344), LPAR1 inhibitor (AM966, 100 nM; MCE, HY-15277) or PPARG inhibitor (3.3 nM; MCE, HY-16578) for 24 h.
Pre-treated macrophages were collected and then indirectly co-cultured with JEG3 cells dyed by CFSE according to the standard protocols in the CFSE Cell Division Tracker Kit (Biolegend, 423,801) for 48 h by using Transwell chamber (0.4 μm; Corning, 3413). Through flow cytometry analysis, the proliferation of JEG3 cells in 48-h co-culture system with LPA-treated M0 macrophages, ENPP2 inhibitor or LPA receptor inhibitors-treated M2 macrophages were analyzed based on the median fluorescence intensity of CFSE. The experiments were performed in triplicate.
JEG3 cells in serum- and tertiary antibody-free culture medium were resuspended into the Transwell chamber (8 μm; Corning, 3422), and pre-treated macrophages were resuspended into the well plate below the chamber. After 48-hour, the invasive ability of JEG3 cells was analyzed by a Matrigel (Corning, 356,234) Transwell invasion assays in the indirect co-culture system. The experiments were performed in triplicate.
Statistical analysis
All analyses were conducted by SPSS 25 (IBM). Normally distributed data with uniform variance were analyzed by a t test between two groups or one-way ANOVA test among multiple groups. Other data were analyzed by a Mann‐Whitney U test or a Kruskal-Wallis test for two or multiple samples. Each experiment was repeated 3 times, and data were presented as mean ± standard error (SEM) for normally distributed data or median and quartile for others. P < 0.05 was considered to indicate a statistically significant difference.
Supplemental Material
Download Zip (7.7 MB)Acknowledgments
We are grateful to Prof. Shi-Min Zhao from Institute of Metabolism and Integrative Biology (IMIB), School of Life Sciences, Fudan University, for guidance and help.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Musallam R, Salem N, Al Halol R, et al. Management of pregnancy loss in the first trimester: a retrospective audit. Lancet. 2018;391(Suppl 2):S34. DOI:10.1016/S0140-6736(18)30400-8.
- Garrido-Gimenez C, Alijotas-Reig J. Recurrent miscarriage: causes, evaluation and management. Postgrad Med J. 2015;91(1073):151–162.
- Alijotas-Reig J, Garrido-Gimenez C. Current concepts and new trends in the diagnosis and management of recurrent miscarriage. Obstet Gynecol Surv. 2013;68(6):445–466.
- Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31(1):387–411.
- Williams PJ, Searle RF, Robson SC, et al. Decidual leucocyte populations in early to late gestation normal human pregnancy. J Reprod Immunol. 2009;82(1):24–31. DOI:10.1016/j.jri.2009.08.001.
- Faas MM, Spaans F, De Vos P. Monocytes and macrophages in pregnancy and pre-eclampsia. Front Immunol. 2014;5:298.
- Mori M, Bogdan A, Balassa T, et al. The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin Immunopathol. 2016;38(6):635–649. DOI:10.1007/s00281-016-0574-0.
- Meng YH, Zhou WJ, and Jin LP, et al. RANKL-mediated harmonious dialogue between fetus and mother guarantees smooth gestation by inducing decidual M2 macrophage polarization. Cell Death Dis. 2017;8(10):e3105.
- Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol. 2017;17(7):451–460.
- Okabe Y, Medzhitov R. Wormhole travel for macrophages. Cell. 2016;165(3):518–519.
- Wang J, Kubes P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell. 2016;165(3):668–678.
- Liu C, Wu CA, Yang QF, et al. Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity. 2016;44(5):1162–1176. DOI:10.1016/j.immuni.2016.03.008.
- De Schepper S, Verheijden S, Aguilera-Lizarraga J, et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell. 2018;175(2):400–415 e413. DOI:10.1016/j.cell.2018.07.048.
- Liou GY, Doppler H, Necela B, et al. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5(1):52–63. DOI:10.1158/2159-8290.CD-14-0474.
- Shechter R, Miller O, Yovel G, et al. Recruitment of beneficial m2 macrophages to injured spinal cord is orchestrated by remote brain choroid plexus. Immunity. 2013;38(3):555–569. DOI:10.1016/j.immuni.2013.02.012.
- Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19(5):548–556.
- Van den Bossche J, O’Neill LA, Menon D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 2017;38(6):395–406.
- Colegio OR, Chu NQ, Szabo AL, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. DOI:10.1038/nature13490.
- O’Neill LAJ, Artyomov MN. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat Rev Immunol. 2019;19(5):273–281.
- Tannahill GM, Curtis AM, Adamik J, et al. Succinate is an inflammatory signal that induces IL-1 beta through HIF-1 alpha. Nature. 2013;496(7444):238–242. DOI:10.1038/nature11986.
- Kaya B, Doñas C, Wuggenig P, et al. Lysophosphatidic acid-mediated gpr35 signaling in CX3CR1(+) macrophages regulates intestinal homeostasis. Cell Rep. 2020;32(5):107979. DOI:10.1016/j.celrep.2020.107979.
- Ray R, Rai V. Lysophosphatidic acid converts monocytes into macrophages in both mice and humans. Blood. 2017;129(9):1177–1183.
- Liu S, Murph M, Panupinthu N, et al. ATX-LPA receptor axis in inflammation and cancer. Cell Cycle. 2009;8(22):3695–3701. DOI:10.4161/cc.8.22.9937.
- McIntyre TM, Pontsler AV, Silva AR, et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc Natl Acad Sci U S A. 2003;100(1):131–136. DOI:10.1073/pnas.0135855100.
- Ye X, Hama K, Contos JJ, et al. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435(7038):104–108. DOI:10.1038/nature03505.
- Aikawa S, Kano K, Inoue A, et al. Autotaxin–lysophosphatidic acid– LPA 3 signaling at the embryo-epithelial boundary controls decidualization pathways. EMBO J. 2017;36(14):2146–2160. DOI:10.15252/embj.201696290.
- Chen SU, Lee H, Chang DY, et al. Lysophosphatidic acid mediates interleukin-8 expression in human endometrial stromal cells through its receptor and nuclear factor-kappaB-dependent pathway: a possible role in angiogenesis of endometrium and placenta. Endocrinology. 2008;149(11):5888–5896. DOI:10.1210/en.2008-0314.
- Tsukamoto S, Kuma A, Murakami M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321(5885):117–120. DOI:10.1126/science.1154822.
- Oestreich AK, Chadchan SB, Medvedeva A, et al. The autophagy protein, FIP200 (RB1CC1) mediates progesterone responses governing uterine receptivity and decidualization. Biol Reprod. 2020;102(4):843–851. DOI:10.1093/biolre/ioz234.
- Nakashima A, Cheng SB, Ikawa M, et al. Evidence for lysosomal biogenesis proteome defect and impaired autophagy in preeclampsia. Autophagy. 2020;16(10):1771–1785.
- Shen HH, Zhang T, Yang HL, et al. Ovarian hormones-autophagy-immunity axis in menstruation and endometriosis. Theranostics. 2021;11(7):3512–3526. DOI:10.7150/thno.55241.
- Caputa G, Castoldi A, Pearce EJ. Metabolic adaptations of tissue-resident immune cells. Nat Immunol. 2019;20(7):793–801.
- Lu H, Yang HL, Zhou WJ, et al. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy. 2021;17(9):2511–2527. DOI:10.1080/15548627.2020.1833515.
- Ellisen LW. Growth control under stress - MTOR regulation through the REDD1-TSC pathway. Cell Cycle. 2005;4(11):1500–1502.
- Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16(4):181–188.
- Ding L, Lu Z, Foreman O, et al. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology. 2012;142(2):305–315. DOI:10.1053/j.gastro.2011.10.025.
- Tanaka H, Takechi M, Kiyonari H, et al. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut. 2015;64(10):1529–1538. DOI:10.1136/gutjnl-2014-308419.
- Svensson J, Jenmalm MC, Matussek A, et al. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J Immunol. 2011;187(7):3671–3682. DOI:10.4049/jimmunol.1100130.
- Svensson-Arvelund J, Ernerudh J. The role of macrophages in promoting and maintaining homeostasis at the fetal-maternal interface. Am J Reprod Immunol. 2015;74(2):100–109.
- Amaral RF, Geraldo LHM, Einicker-Lamas M, et al. Microglial lysophosphatidic acid promotes glioblastoma proliferation and migration via LPA 1 receptor. J Neurochem. 2021;156(4):499–512. DOI:10.1111/jnc.15097.
- Kim SJ, Howe C, Mitchell J, et al. Autotaxin loss accelerates intestinal inflammation by suppressing TLR4-mediated immune responses. EMBO Rep. 2020;21(10):e49332. DOI:10.15252/embr.201949332.
- Sinclair KA, Yerkovich ST, Hopkins PM, et al. The autotaxin-lysophosphatidic acid pathway mediates mesenchymal cell recruitment and fibrotic contraction in lung transplant fibrosis. J Heart Lung Transplant. 2021;40(1):12–23. DOI:10.1016/j.healun.2020.10.005.
- Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18(12):1754–1767.
- Takeo C, Ikeda K, Horie-Inoue K, et al. Identification of Igf2, Igfbp2 and Enpp2 as estrogen-responsive genes in rat hippocampus. Endocr J. 2009;56(1):113–120. DOI:10.1507/endocrj.K08E-220.
- Chen L, Zhang J, Deng X, et al. Lysophosphatidic acid directly induces macrophage-derived foam cell formation by blocking the expression of SRBI. Biochem Biophys Res Commun. 2017;491(3):587–594. DOI:10.1016/j.bbrc.2017.07.159.
- Houser BL, Tilburgs T, Hill J, et al. Two unique human decidual macrophage populations. J Immunol. 2011;186(4):2633–2642. DOI:10.4049/jimmunol.1003153.
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–364.
- Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19(3):170–183.
- Zoncu R, Efeyan A, Sabatini DM. MTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35.
- Kim J, Kundu M, Viollet B, et al. AMPK and MTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–U171. DOI:10.1038/ncb2152.
- Sofer A, Lei K, Johannessen CM, et al. Regulation of MTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25(14):5834–5845. DOI:10.1128/MCB.25.14.5834-5845.2005.
- Ha JH, Radhakrishnan R, Jayaraman M, et al. LPA induces metabolic reprogramming in ovarian cancer via a pseudohypoxic response. Cancer Res. 2018;78(8):1923–1934. DOI:10.1158/0008-5472.CAN-17-1624.
- Wenes M, Shang M, Di Matteo M, et al. Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 2016;24(5):701–715. DOI:10.1016/j.cmet.2016.09.008.
- Herr KJ, Herr DR, Lee CW, et al. Stereotyped fetal brain disorganization is induced by hypoxia and requires lysophosphatidic acid receptor 1 (LPA1) signaling. Proc Natl Acad Sci U S A. 2011;108(37):15444–15449. DOI:10.1073/pnas.1106129108.
- Feng Y, Ma X, Deng LW, et al. Role of selectins and their ligands in human implantation stage. Glycobiology. 2017;27(5):385–391. DOI:10.1093/glycob/cwx009.
- Farkas AE, Hilgarth RS, Capaldo CT, et al. HNF4alpha regulates claudin-7 protein expression during intestinal epithelial differentiation. Am J Pathol. 2015;185(8):2206–2218. DOI:10.1016/j.ajpath.2015.04.023.
- Lee DH, Park SH, and Ahn J, et al. Mir214-3p and Hnf4a/Hnf4alpha reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy. 2020;7(9):2415–2431.
- Deng R, Zhang HL, and Huang JH, et al. MAPK1/3 kinase-dependent ULK1 degradation attenuates mitophagy and promotes breast cancer bone metastasis. Autophagy. 2020;17(10):3011–3029.
- Hope JM, Wang FQ, and Whyte JS, et al. LPA receptor 2 mediates LPA-induced endometrial cancer invasion. Gynecol Oncol. 2009;112(1):215–223
- Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25(5):1037–1043.
- Xia H, Green DR, Zou W. Autophagy in tumour immunity and therapy. Nat Rev Cancer. 2021;21(5):281–297.
- Zhang X, Pavlicev M, Jones HN, et al. Eutherian-specific gene TRIML2 attenuates inflammation in the evolution of placentation. Mol Biol Evol. 2020;37(2):507–523. DOI:10.1093/molbev/msz238.
- Friedrich M, Vaxevanis CK, Biehl K, et al. Targeting the coding sequence: opposing roles in regulating classical and non-classical MHC class I molecules by miR-16 and miR-744. J Immunother Cancer. 2020;8(1):e000396. DOI:10.1136/jitc-2019-000396.
