ABSTRACT
Macroautophagy/autophagy-related protein Atg8/LC3 is important for autophagosome biogenesis and required for selective degradation of various substrates. In our recent study, we performed a yeast two-hybrid screening to identify proteins that interact with Atg8a, the Drosophila homolog of Atg8/LC3. The screening identified several Atg8a-interacting proteins. These proteins include: i) proteins which have already been experimentally verified to bind Atg8a, such as Atg1, DOR, ref(2)P and key (Kenny); ii) proteins for which their mammalian homologs interact with Atg8-family members, like Ank2, Atg4, and Nedd4; and iii) several novel Atg8a-interacting proteins, such as trc/STK38 and Tak1. We showed that Tak1, as well as its co-activator, Tab2, both interact with Atg8a and are substrates for selective autophagic clearance. We also determined that SH3PX1 interacts with Tab2 and is necessary for the effective regulation of the immune-deficiency (IMD) pathway. Our findings suggest a mechanism for the regulatory interactions between Tak1-Tab2-SH3PX1 and Atg8a, which contribute to the fine-tuning of the IMD pathway.
Macroautophagy is an evolutionarily conserved process, whereby cells generate nutrients by degrading own intracellular constituents. A group of specific interactions between cargo and autophagy-related proteins is key in promoting the targeted uptake of certain autophagy substrates over others. The most well-characterized among such interactions is when substrates that contain specific sequences known as LC3-interacting regions (LIRs) bind to the LIR-docking site (LDS) of the autophagy-related (Atg) protein Atg8 (LC3/GABARAP subfamilies in mammals). Considerable interest and effort are invested into characterizing the Atg8 interactome, and, by extension, studying the regulatory effect of autophagy on cellular processes that such Atg8-interacting proteins participate in.
We conducted a high-throughput yeast 2-hybrid (Y2H) screen using Drosophila 3rd instar larvae library to identify the Atg8a-interactome [Citation1]. We identified 34 Atg8a-interacting proteins in total. These include proteins that have been experimentally verified to bind Atg8-family members (8 proteins), as well as novel interactors for which a direct association with Atg8a has not been previously reported (26 proteins). Prominent examples of known Drosophila Atg8a-interactors include the autophagy proteins Atg1, Atg4 and the adaptor protein ref(2)P (refractory to sigma P). In addition, the Y2H results showed that the fruit fly homologs of ANK2, TP53INP2/DOR and NEDD4 (Ank2, DOR and Nedd4, respectively) among others, are also candidate Atg8a-binding proteins. ANK2, TP53INP2/DOR and NEDD4 have been characterized in mammals to associate with LC3/GABARAP-family members via LIR-LDS-dependent interactions. With regard to the group of novel Atg8a-interacting members, we first selected trc (tricornered), as its mammalian homolog STK38 (serine/threonine kinase 38) associates with the autophagy-related protein BECN1 (Atg6 in Drosophila) and both STK38 and trc depletion impair autophagosome formation. By employing GST (glutathione S-transferase) affinity-isolation assays as well confocal imaging of Drosophila tissue we observed that trc associates with Atg8a, both in vitro and in vivo, but in a LIR-independent manner.
We next focused our attention on Tak1 (TGF-β activated kinase 1). Tak1 is the apical kinase in the immune-deficiency (IMD) pathway of the innate immune response and forms a complex with its co-activator, Tab2 (TAK1-associated binding protein 2), to convey its downstream signaling effects. Using a proteomics-based approach we also determined that Tab2 associates with sorting nexin SH3PX1 (SH3 and PX domain containing 1) and corroborated their interaction further in GST affinity-isolation assays. SH3PX1 promotes autophagy by participating in autophagosome formation and has already been shown to interact with Atg8a.
The interactions between Tak1, Tab2, SH3PX1 and Atg8a prompted us to investigate their relationship further. We showed that both Tak1 and Tab2 interact with Atg8a in vitro using GST affinity-isolation assays. The interaction between Tab2 and Atg8a does not seem to be LIR-LDS dependent, whereas for Tak1, the binding to Atg8a is conveyed by the LIR motif bearing the sequence EGWVVI between amino-acid positions 667–672. We found in addition that both Tab2 and Tak1 are also substrates for autophagic clearance.
To study the physiological effect that the inactivation of the Tak1 LIR motif may have on immune response, we used tak1 LIR mutant flies created by CRISPR to examine the mRNA levels of signature IMD-regulated anti-microbial peptide (AMP) genes, whose transcription follows the activation status of the IMD pathway. In qPCR assays we observed that young as well as older adult Tak1 LIR mutant flies present with persistently elevated levels of the AMP genes studied, compared to controls. This finding underscored that the LIR motif of Tak1 is necessary for the efficient regulation of the IMD pathway.
We concluded our work by integrating our findings into the proposal of a working model regarding how the interactions between Tak1, Tab2, SH3PX1 and Atg8a could help shape IMD signaling (). Based on our observations, we posit that selective autophagy is upregulated following IMD activation and removes Tak1-Tab2 signaling complexes through the interaction of both Tak1 and Tab2 with Atg8a. In this context, SH3PX1 is an essential mediator that associates with Tab2 and Atg8a and further tethers the complex on phagophores. Our findings suggest that both the LIR motif of Tak1, as well as SH3PX1 are indispensable for the efficient removal of the Tak1 complex from the IMD cascade, as loss of either results in IMD overactivation. In our graphic representation of the model, we depict each member of the tripartite Tak1-Tab2-SH3PX1 complex as interacting with a different Atg8a molecule. However, of the three, only Tak1 is so far verified to associate with Atg8a in a LIR-LDS-dependent manner and likely occupies the cognate site on Atg8a. We observed that Tab2 does not rely on the LDS site of Atg8a for binding and neither is a LIR-dependent interaction for SH3PX1-Atg8a known to date. As such, it is possible that some or all members can bind to different sites on the same Atg8a moiety. Furthermore, SH3PX1 is reported to transiently associate with Atg8a during autophagosome formation and is not itself an obligatory substrate for autophagy. This, together with its association with Tab2 as we showed, and our observations that Tab2 does not accumulate as strongly compared to Tak1 upon autophagy inhibition, invite an interesting possibility that perhaps dissociation of the Tak1-Tab2 complex could also be taking place. Our findings that all three proteins are capable in vitro of binding directly to Atg8a, may perhaps allude to a failsafe mechanism, that increases the likelihood of Tak1 being targeted for degradation and removed from active complexes.
Figure 1. A working model for regulation of the IMD pathway by selective autophagy via the Tak1-Tab2-SH3PX1-Atg8a interactions. Based on findings from the current study, we posit with this 2D schematic representation, that both components of the Tak1-Tab2 complex interact with Atg8a. Tak1 binds Atg8a via its functional LIR motif, whereas Tab2 does not necessarily rely on a LIR motif to interact with Atg8a. SH3PX1 binds Tab2 and Atg8a and likely further stabilizes the Tak1-Tab2 complex on the phagophore membrane. Both Tak1ʹs LIR motif and SH3PX1, seem to be equally required for the efficient sequestration of the complex by phagophores, as in either’s absence the IMD pathway is overactivated based on our current observations.
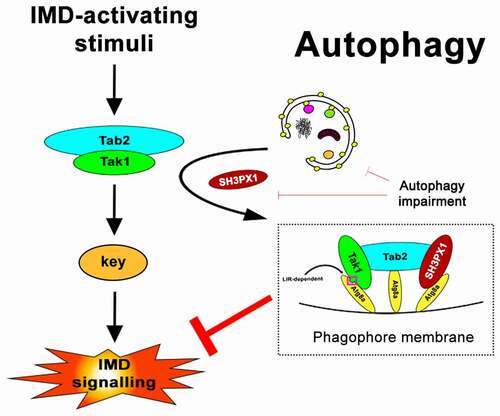
In summary, the combined interactions among Tak1, Tab2, SH3PX1 with Atg8a can promote the removal of the Tak1-Tab2 complex by autophagy and aid in termination of IMD signaling. Our study highlights the physiological importance of selective autophagy in the innate immune response of metazoans and demonstrates the plasticity of its participating regulators.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Tsapras P, Petridi S, Chan S, et al. Selective autophagy controls innate immune response through a TAK1/TAB2/SH3PX1axis. Cell Rep. 2022;38:110286.
