ABSTRACT
During an animal’s life, many cells undergo apoptosis, a form of genetically programmed cell death. These cells are swiftly engulfed by other cells through phagocytosis and subsequently degraded inside phagosomes. Phagocytosis and macroautophagy/autophagy are two different cellular events: whereas phagocytosis is a cell-eat-cell event, autophagy, or “self-eating”, occurs within one cell, resulting in the enveloping of protein aggregates or damaged organelles within double-membrane autophagosomes. Despite this critical difference, these two events share common features: (1) both are means of safe garbage disposal; (2) both phagosomes and autophagosomes fuse to lysosomes, which drive the degradation of their contents; and (3) both events facilitate the recycling of biological materials. Previously, whether autophagosomes per se directly participate in the degradation of apoptotic cells was unknown, although autophagy proteins were implicated in apoptotic cell clearance. We recently discovered that autophagosomes fuse with phagosomes and contribute to the degradation of apoptotic cells.
Canonical autophagosomes fuse with phagosomes that carry apoptotic cells
We study the engulfment and degradation of apoptotic cells in the nematode Caenorhabditis elegans, a small roundworm whose body is transparent and whose entire cell lineage has been mapped. We use fluorescence time-lapse microscopy to monitor membrane-trafficking events occurring in engulfing cells that carry phagosomes in living embryos [Citation1]. C. elegans has two orthologs of mammalian LC3, named LGG-1 and LGG-2. Four lines of evidence indicate that LC3-labeled vesicles fuse with phagosomes, and that these vesicles are double-membrane autophagosomes, not the single-membrane LC3-associated phagocytosis (LAP) vesicles reported to facilitate phagosome maturation in mammalian cells: (1) The membrane-bound mCherry-LGG-1 and -LGG-2 reporters accumulate inside the phagosomal lumen after being seen enriched on phagosomal surfaces, a phenomenon that is only possible when the inner vesicle of a double-membrane organelle is deposited into the phagosomal lumen after the outer membrane is fused to the phagosomal membrane (); (2) multiple genes that are required for autophagosome biogenesis, including those dispensable for the biogenesis of LAP vesicles (atg-13 and epg-8/atg-14), are essential for generating the LC3-labeled vesicles that fuse with phagosomes; (3) these LC3+ vesicles that fuse with phagosomes are also labeled with ATG-9, an autophagosomal membrane protein not known to appear on LAP vesicles; and (4) double-membrane vesicles are observed on phagosomal surfaces by transmission electron microscopy. Our observations indicate that besides early endosomes and lysosomes, autophagosomes are a novel kind of intracellular organelle that are recruited to the phagosomal surfaces and subsequently fuse with phagosomes ().
Figure 1. A diagram illustrating how autophagosomes and lysosomes are recruited to the surfaces of phagosomes and subsequently fuse with phagosomes. C. elegans CED-6 and DYN-1 are members of the CED-1 pathway. CED-6, a phosphotyrosine-binding (PTB) domain-containing protein and a homolog of mammalian GULP1, is an adaptor for the phagocytic receptor CED-1. DYN-1 encodes the large GTPase dynamin. The LGG/LC3 markers label both the outer and inner membranes of autophagosomes. Once the outer membranes fuse with the phagosomal membrane, the inner vesicles are deposited into the phagosomal lumen, resulting in the accumulation of the mCherry-LGG-1/LGG-2 reporters inside the phagosomal lumen. On the contrary, the fusion of the single-membrane lysosomes with phagosomes results in the retention of any lysosomal membrane markers on the phagosomal membrane. The question mark depicts that it is unclear, after autophagosome-phagosome fusion, how long the LGG/LC3 markers would remain on the phagosomal membrane.
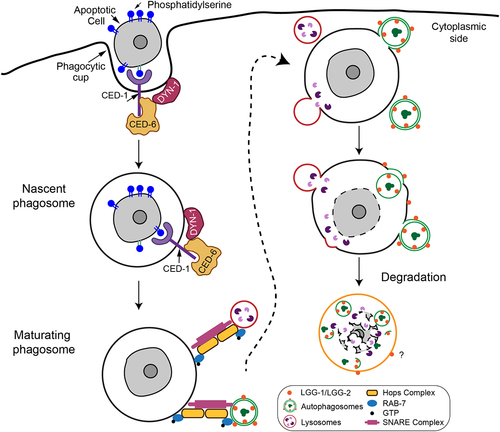
The phagocytic receptor CED-1 drives the incorporation of autophagosomes into phagosomes
The small GTPase RAB7 and its effector, the HOPS complex, act as a membrane tethering factor for the autophagosome-lysosome and phagosome-lysosome fusion events. Loss-of-function mutations of rab-7 and vps-18, the latter of which encodes a HOPS subunit, block the fusion between autophagosomes and phagosomes, resulting in the drastic accumulation of LC3+ puncta on the phagosomal surfaces. RAB-7 and the HOPS complex thus specifically drive the fusion but not recruitment of autophagosomes with phagosomes ().
C. elegans CED-1 is a phagocytic receptor that resides on the surfaces of engulfing cells and recognizes phosphatidylserine, an “eat me” signal exposed on the apoptotic cell surfaces. MEGF10, the mammalian CED-1 homolog, acts in the brain to phagocytose apoptotic neurons and unnecessary synapses. Remarkably, CED-1 plays two separate roles in apoptotic cell clearance. CED-1 initiates the engulfment of apoptotic cells once detecting the “eat me” signal; CED-1 also promotes phagosome maturation after engulfment is completed. CED-1 leads a signaling pathway that recruits RAB-5 and RAB-7, two RAB GTPases, to phagosomal surfaces. It also drives the transient production and turnover of PtdIns3P, a second messenger, in the phagosomal membranes. Together, the RAB GTPases and PtdIns3P drive the incorporation of early endosomes and lysosomes, which provide the digestive enzymes and acidic pH to degrade phagosomal cargos, into the phagosomal lumen. By analyzing the mutant embryos of ced-1 and other members in the pathway (), we found that the ced-1 pathway is pivotal for the recruitment of autophagosomes to phagosomes. In addition, the essential role of CED-1 in recruiting RAB-7 to the phagosomal surfaces indicates that this pathway is also pivotal for the fusion of autophagosomes with phagosomes.
What do autophagosomes bring to phagosomes?
Blocking autophagosome biogenesis slows down the degradation of apoptotic cells significantly. As a result, phagosomes persist inside engulfing cells in atg mutants, including unc-51, atg-13 and epg-8, the C. elegans homologs of mammalian ULK1, ATG13, and ATG14, respectively, which are dispensable for LAP vesicle biogenesis. This phenomenon demonstrates that autophagosomes, not LAP vesicles, facilitate the degradation of apoptotic cells. We observed, in atg mutants, modest defects in the acidification of the phagosomal lumen and the degradation of the apoptotic cell chromosomal DNA. In the same mutants, the lysosome-phagosome fusion occurs at a normal pace. Our further observations indicate that autophagosomes and lysosomes contribute to the degradation of phagosomes in a parallel and additive manner.
Presumably, autophagosomes provide specific material(s) to the phagosomal lumen and/or membrane through the fusion of their outer membranes with the phagosomal membrane and the deposition of the inner vesicles into the phagosomal lumen (). Currently, the identities of such material(s) remain unknown. Future investigation will reveal novel molecules or novel functions of known molecules that are delivered from autophagosomes to phagosomes and that facilitate phagosomal degradation.
Summary and Future Directions
Previously, we thought that autophagosomes only degrade materials within their own cell. Now we demonstrate that they also help clear other cells. This discovery raises many questions. First, it is unclear why, in C. elegans, no single-membrane LAP vesicles are detected to interact with phagosomes, unlike what has been reported for mammalian cells. Based on the evolutionary conservation of molecular mechanisms that control phagocytosis and autophagy in metazoans, we propose that in mammalian cells, besides LAP vesicles, canonical autophagosomes also contribute to apoptotic cell clearance. Second, do autophagosomes also facilitate the degradation of phagosomal cargos that are not apoptotic cells, for example, phagocytosed pathogens? Third, what autophagosomal material(s) aid the degradation of phagosomal cargos? Fourth, what controls the breakdown of the inner vesicle inside the phagosomal lumen? Further investigating the crosstalk between autophagosomes and phagosomes in multiple experimental systems will answer these questions and reveal novel and evolutionarily conserved mechanisms that ensure the clearance of unwanted cells and substances.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Pena-Ramos O, Chiao L, Liu X, et al. Autophagosomes fuse to phagosomes and facilitate the degradation of apoptotic cells in Caenorhabditis elegans. eLife. 2022;11:e72466. PMC8769646.
