ABSTRACT
The role of meiotic proteasome-mediated degradation has been extensively studied. At the same time, macroautophagy/autophagy only emerged recently as an essential regulator for meiosis progression. Our recent publication showed that autophagy in meiotic cells exhibits a temporal pattern distinct from that in quiescent cells or mitotic cells under prolonged starvation. Importantly, autophagic activity oscillates during meiotic cell divisions, i.e., meiosis I and meiosis II, which can accelerate meiotic progression and increase sporulation efficiency. Our in vitro and in vivo assays revealed that the conserved phosphatase Cdc14 stimulates autophagy initiation during meiotic divisions, specifically in anaphase I and II, when a subpopulation of active Cdc14 relocates to the cytosol and interacts with phagophore assembly sites (PAS) triggering the dephosphorylation of Atg13 to stimulate Atg1 kinase activity and autophagy. Together, our findings reveal a mechanism for the coordination of autophagy activity in the context of meiosis progression.
Autophagy, a conserved lysosomal degradation pathway, is a prerequisite for pre-meiotic DNA duplication and meiosis entry. In 2020, in collaboration with Dr. Soni Lacefield at Indiana University and Dr. Vlad Denic at Harvard University, we discovered that acute inhibition of autophagy during yeast meiotic divisions leads to delayed onset of meiosis II, an extra round of spindle pole body (SPB, equivalent to the mammalian centrosome) replication, and abnormal chromosome segregation that collectively prevent meiosis exit and gametogenesis. Thus, we proposed that meiosis should maintain a certain level of autophagy during meiotic divisions.
In our recent publication [Citation1], using time-lapse fluorescence microscopy (FM), we found that upon meiosis entry cells rapidly upregulate and maintain the level of the autophagy rate-limiting factor Atg8, and its vacuolar delivery (indication of autophagy flux). This pattern differs from that of quiescence or mitosis under starvation conditions, in which the Atg8 level and autophagy flux initially increase, but are eventually downregulated. Interestingly, autophagy levels indicated by vacuolar mNeonGreen-Atg8 delivery present high single-cell variation during meiotic divisions. Cells with higher levels of autophagic flux during meiotic divisions complete meiosis faster. Consistently, increasing autophagy by expressing a gain-of-function Atg13 allele (Atg13[8SA]) remarkably accelerates meiosis progression and increases sporulation efficiency. These findings demonstrate that autophagy modulation is a driving force for meiosis progression.
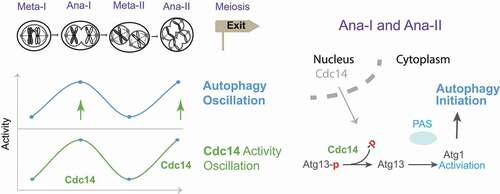
Although the Atg8 level is upregulated upon meiosis entry and maintained during meiotic divisions, we determined that autophagy initiation oscillates during meiotic divisions. Autophagy initiation peaks occur at anaphase I and II, as measured by the analysis of Atg1 activation, formation of the PAS (marked by Atg1 and Atg13 puncta), and autophagosome biogenesis (marked by cytosolic Atg8 puncta). Among the many processes that are triggered at anaphase I and II, such as nuclear division or spindle disassembly, careful analysis of single-cell time series showed that the nucleolar release of the phosphatase Cdc14 presents a clear temporal correlation with the peaks of autophagy initiation. This result led us to hypothesize that Cdc14's periodic activation and relocation from the nucleolus to the cytosol at anaphase I and II could drive the oscillations in autophagy initiation during meiotic divisions.
We found that cytosolic Cdc14 is recruited to the Atg1-Atg13-containing autophagy initiation sites (PAS). Furthermore, Cdc14 dephosphorylates Atg13 in vivo and in vitro, which stimulates autophagy by increasing the Atg1-Atg13 interaction and Atg1 activation. Bona fide Cdc14 docking sites were found by mutagenesis analysis of the putative docking sites for Cdc14 on Atg13. The Atg13(PxL3m) mutant, in which the Cdc14 docking site (-P476-X477-L478-) is mutated, reduces the Cdc14-Atg13 interaction in vitro, the formation of Atg13 puncta (PAS), and the colocalization of Cdc14 with Atg13 in vivo during meiotic divisions. The Atg13(PxL3m) allele presents reduced autophagy, decreased meiotic kinetics, and sporulation efficiency, which can be almost fully rescued by additionally mutating six predicted Cdc14-targeted phosphorylation sites (S-P sites) to alanine (Atg13[6SA]) mimicking the result of Cdc14-mediated Atg13 dephosphorylation. In particular, PxL3 site on Atg13 is conserved and located in an intrinsically disordered region (IDR), which likely offers the best access to the six S-P sites due to their physical proximity and the structural flexibility. It is noteworthy that our biochemical data were orthogonally confirmed by using a deep-learning convolutional neural network to analyze images of meiotic cells expressing Cdc14-mNeonGreen and mScarlet-Atg13. This automated approach allowed us to analyze the cytoplasmic spatial distribution of Cdc14 and Atg13 at 19,060 PAS sites. Despite the dynamic behavior of Cdc14 during meiotic divisions and the ephemeral duration of anaphase I and II, the algorithmic analysis found that 9.7% of the PAS are enriched for Cdc14 during meiotic nuclear divisions.
Collectively, our data shows that Cdc14 dephosphorylates Atg13 at the PAS site to upregulate autophagy during anaphase I and II, which benefits meiotic progression (). At this point, it is unclear how meiosis-tailored autophagy mechanistically facilitates meiosis. Conceptually, autophagy can recycle cellular contents to drive synthesis of cellular components during the starvation conditions cells face during meiosis. In addition, autophagy can also be required to eliminate protein aggregates or complex structures during the dramatic transitions of the meiotic program from nuclear divisions to packaging organelles into spores. However, why does meiosis upregulate autophagy in the cytoplasm specifically at anaphase I and II? Previously, we found that autophagy degrades Rim4, a meiosis specific RNA-binding protein that sequesters a subset of mid-late meiotic mRNAs (e.g., the B-type cyclin CLB3 for meiosis II) to prevent their premature translation before meiosis II. Interestingly, rapid Rim4 degradation enables the timely translation of Rim4-sequestered mRNAs at anaphase I, which is temporally correlated with Cdc14-mediated autophagy upregulation. We have also reported over-duplication of SPB post-anaphase II in cells that failed to exit meiosis due to meiotic autophagy inhibition. Thus, Cdc14-enhanced autophagy might provide a link between meiotic progression and the autophagy functions required to precisely time Rim4 degradation-mediated meiotic translational control and SPB dynamics at meiotic anaphase I and anaphase II. Whether these events are mechanistically coordinated remains a key question. Last, this study provides clear evidence that a phosphatase (Cdc14) from the nucleus can transiently activate autophagy in the cytoplasm, while the potential of Cdc14 to regulate autophagy selectivity is yet to be explored.
Figure 1. Model of Cdc14-mediated meiotic autophagy oscillation. Left, cytosolic Cdc14 oscillation stimulates autophagy oscillation at anaphase I (Ana-I) and anaphase II (Ana-II); Right, nuclear Cdc14 relocates to the cytosol to stimulate autophagy by dephosphorylating Atg13 and activating Atg1 at the PAS.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Feng W, Argüello-Miranda O, Qian S, et al. Cdc14 spatiotemporally dephosphorylates Atg13 to activate autophagy during meiotic divisions. The Journal of Cell Biology. 2022;221(5). DOI:https://doi.org/10.1083/jcb.202107151
