ABSTRACT
HTT (huntingtin) is a 350-kDa protein of unknown function. While HTT moves bidirectionally within axons and HTT loss/reduction causes axonal transport defects, the identity of cargo-containing vesicles that HTT helps move remain elusive. Previously, we found an axonal retrogradely moving HTT-Rab7 vesicle complex; however, its biological relevance is unclear. Using Drosophila genetics, in vivo microscopy, membrane isolation and pharmacological inhibition, here we identified that adaptors Hip1 and Rilpl aid the retrograde motility of LAMP1-containing HTT-Rab7 late endosomes, not autophagosomes. Reduction of Syx17 and chloroquine- or bafilomycin A1-mediated pharmacological inhibition, but not reduction of Atg5, disrupted the in vivo motility of these vesicles. Further, because HTT-Rab7 vesicles colocalized with long-distance signaling components (BMP signaling: tkv-wit, injury: wnd) and move in a retrograde direction after Drosophila nerve crush, we propose that these vesicles likely traffic damage signals following axonal injury. Together, our findings support a previously unknown role for HTT in the retrograde movement of a Rab7-LAMP1-containing signaling late endosome.
Abbreviations
Atg5: Autophagy-related 5; Atg8a: Autophagy-related 8a; AL: autolysosome; AP: autophagosome; BAF1: bafilomycin A1; BDNF: brain derived neurotrophic factor; BMP: bone morphogenetic protein; Cyt-c-p: Cytochrome c proximal; CQ: chloroquine; DCTN1: dynactin 1; Dhc: dynein heavy chain; EE: early endosome; DYNC1I1: dynein cytoplasmic 1 intermediate chain 1; HD: Huntington disease; HIP1/Hip1: huntingtin interacting protein 1; HTT/htt: huntingtin; iNeuron: iPSC-derived human neurons; IP: immunoprecipitation; Khc: kinesin heavy chain; KIF5C: kinesin family member 5C; LAMP1/Lamp1: lysosomal associated membrane protein 1; LE: late endosome; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MAP3K12/DLK: mitogen-activated protein kinase kinase kinase 12; MAPK8/JNK/bsk: mitogen-activated protein kinase 8/basket; MAPK8IP3/JIP3: mitogen-activated protein kinase 8 interacting protein 3; NGF: nerve growth factor; NMJ: neuromuscular junction; NTRK1/TRKA: neurotrophic receptor tyrosine kinase 1; NRTK2/TRKB: neurotrophic receptor tyrosine kinase 2; nuf: nuclear fallout; PG: phagophore; PtdIns3P: phosphatidylinositol-3-phosphate; puc: puckered; ref(2)P: refractory to sigma P; Rilpl: Rab interacting lysosomal protein like; Rip11: Rab11 interacting protein; RTN1: reticulon 1; syd: sunday driver; SYP: synaptophysin; SYT1/Syt1: synaptotagmin 1; STX17/Syx17: syntaxin 17; tkv: thickveins; VF: vesicle fraction; wit: wishful thinking; wnd: wallenda
Introduction
The Huntington disease (HD) protein, HTT (huntingtin), is ubiquitously expressed; however, its neuronal function remains unknown despite being essential for development [Citation1]. Although more than 350 binding partners for HTT have been identified with functions across a wide-range of cellular processes including gene expression, metabolism, protein turnover, endocytosis, and trafficking [Citation2–6], the molecular mechanisms by which HTT and its binding partners function remain elusive. Previously, we showed that HTT moves bi-directionally within axons by interacting with kinesin 1 and dynein [Citation7], and htt can mediate the motility of several Rab-GTPase-containing vesicles, suggesting that htt can serve as a scaffolding protein for vesicular trafficking. Reduction of htt disrupts the bi-directional movement of Rab3, Rab4, Rab11, Rab19, the retrograde motility of Rab7 and enhanced the anterograde motility of Rab2 [Citation8–10]. Therefore, while it is likely that htt can differentially facilitate the axonal motility of Rab-GTPase-containing vesicles, the functional significance of these distinct HTT-Rab containing axonal complexes remain ambiguous.
Upon endocytic internalization into an early endosome (EE), proteins and lipids are sorted for recycling or degradation. Cargo destined to be degraded remain in an EE, which then mature to a late endosome (LE), marked by RAB7/Rab7 [Citation11,Citation12]. In neurons, LEs are thought to be retrogradely moved from distal regions of the axon to the cell body for either lysosome-mediated degradation [Citation13,Citation14] and/or long-distance signaling [Citation15,Citation16]. Neurons are suggested to maintain a spatial gradient of LE/lysosome sub-populations, marked by Rab7 and/or LAMP1/Lamp1, with highly degradative lysosomes being enriched near the cell body while pre-degradative LEs move from distal regions of the axons and dendrite to the cell body for degradation [Citation14,Citation17,Citation18]. In parallel, ATG5/Atg5 (autophagy related 5)-dependent phagophore (PG) expansion [Citation19,Citation20] forms a nascent autophagosome (AP) in distal axons, marked by Atg8/MAP1LC3/LC3 (Autophagy-related 8/microtubule associated protein 1 light chain 3) [Citation21,Citation22]. An AP can form an amphisome through STX17/Syx17 (syntaxin 17)-dependent fusion [Citation23,Citation24] with an LE, which then are proposed to retrogradely move to the cell body for degradation upon fusing with a lysosome to form an autolysosome (AL [Citation14,Citation25]). HTT has been proposed to be present in macroautophagy/autophagy during cargo loading of an elongating PG [Citation26] and during the transport of an AP [Citation27]. However, since Atg8/LC3 is present in all autophagic compartments from the PG to the AL [Citation28], this ambiguity creates a challenge in isolating HTT/htt during autophagy.
Following neuronal injury, retrograde movement is critical for damage signaling [Citation29] and axonal regeneration [Citation30]. In fact, an anterograde-to-retrograde conversion has been observed following rat sciatic nerve crush [Citation31]. In response to axonal injury, retrogradely moving damage signals such as MAP3K12/DLK/wnd (mitogen-activated protein kinase kinase kinase 12/wallenda) and/or MAPK8/JNK (mitogen-activated protein kinase 8)-containing vesicles promote upregulation of pro-regenerative genes essential for axon regeneration [Citation29,Citation32–34]. Interestingly, Rab7/RAB7 associates with syd/MAPK8IP3/JIP3 (sunday driver/mitogen-activated protein kinase 8 interacting protein 3)-containing endosomes that traffic MAPK/JNK [Citation29,Citation35] and with retrogradely moving MAP3K12/DLK-containing vesicles [Citation36]. Although HTT was recently shown to be upregulated during mouse corticospinal motor neuron regeneration following spinal cord injury [Citation37], and has been proposed to be an injury-response signaling hub in axotomized retinal ganglion cells [Citation38], how HTT functions during axonal injury remains unknown.
Because Rab7 is present with LEs and during autophagy, here we test the hypothesis that the putative retrogradely moving HTT-Rab7 vesicle complex is a LE and/or is part of autophagy. We found that the retrograde motility of the HTT-Rab7 complex is aided by HTT or Rab7-associated proteins Hip1 (Huntingtin-interacting protein 1) and Rilpl (Rab interacting lysosomal protein like), and that this complex is distinct from the bidirectionally-moving HTT-RAB4/Rab4 synaptic vesicle complex we previously isolated [Citation10]. HTT-Rab7 vesicles are likely LAMP1/Lamp1-containing LEs, and not APs, since the motility of HTT-Rab7 vesicles were impaired by loss/reduction of LE-AP fusion mediated by mutant Syx17 as well as pharmacological inhibition (chloroquine or bafilomycin A1) of lysosome acidification and lysosome-AP fusion. Further, the retrogradely moving HTT-Rab7 vesicles colocalize with long-distance signaling components (the BMP [bone morphogenetic protein]- receptors tkv [thickveins] and wit [wishful thinking]), axonal injury signals (wnd/MAP3K12/DLK), and move retrogradely even after Drosophila nerve crush, in contrast to perturbed motility of synaptic vesicles. Taken together, our observations propose a previously unknown role for HTT in the retrograde movement of a Rab7-LAMP1-containing signaling LE, with a functional relevance during axonal injury
Results
The retrogradely moving HTT-Rab7 vesicle complex is aided by adaptor proteins Rilpl and Hip1
We previously found that adaptor proteins Rip11 (Rab11-interacting protein) and Hip1, but not milton, nuf (nuclear fallout), or nmo (nemo), likely aid the bi-directional motility of the HTT-Rab4 vesicle complex [Citation10]. To identify adaptor proteins that aid the retrograde motility of the HTT-Rab7 vesicle complex, we performed a candidate screen similar to what we have done previously [Citation10]. Using the pGAL4-62B-SG26-1 driver, which expresses in a small population of motor neurons [Citation39,Citation40], we generated Drosophila larvae simultaneously expressing Rab7-GFP and HTT15Q-mRFP (nonpathogenic form of human HTT) within larval axons and observed co-migrating retrogradely moving vesicles (). Colocalized trajectories (yellow) from merged kymographs revealed that 39% of vesicles containing HTT and Rab7 moved retrogradely while only 14% vesicles moved anterogradely (). Reduction of htt (htt-KO/+, Df(98E2);CG999075 [Citation41], or htt-RNAi [Citation7,Citation9]) with Rab7-GFP or reduction of Rab7 (Rab7EY10675/+ or Rab7d1/+ [null allele [Citation42]]) with HTT-mRFP disrupted Rab7 or HTT motility; significantly decreasing the retrograde populations of Rab7-GFP or HTT-mRFP vesicles (Fig. S1AB). Decreased retrograde motility corresponded to increased distribution of Rab7-GFP at neuromuscular junctions (NMJs) and decreased localization at cell bodies (Fig. S1 CD). Likewise, expression of a dominant negative form of Rab7 (Rab7T22N) also disrupted HTT motility (Fig. S1B). Western blot analysis showed that Rab7 protein levels in homozygous Rab7EY10675/Rab7EY10675 flies were reduced to 28% of WT levels (Fig. S2A), similar to what was previously observed for the Rab7d1 null allele [Citation42]. Further, reductions in either kinesin 1 (Khc8/+) or dynein (Dhc6-10/+) decreased both the anterograde and retrograde motility of either HTT-mRFP or Rab7-GFP-containing vesicles (Fig. S1EF).
Figure 1. The retrogradely moving HTT-Rab7 vesicle is aided by adaptor proteins Hip1 and Rilpl. (A) Schematic diagram of larval nervous system showing the brain and segmental nerves. Red box = imaged area (90 µm). Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs from simultaneous dual-color movies from larvae expressing both HTT-mRFP (red) and Rab7-GFP (green). Co-migrating tracks that contain HTT and Rab7 are seen (yellow arrow). Note that green or red only tracks are also seen (arrowheads). Bar: 5 µm. (B) Quantification of the directional analysis of trajectories containing HTT and Rab7 (yellow). n = 10. (C-D) Representative images from movies and kymographs from larvae expressing either Rab7-GFP or HTT-mRFP alone, with a Hip1 reduction, or with a Rilpl reduction. Bar: 10 µm. (E-F) Directional analysis of Rab7-GFP or HTT-mRFP trajectories compared to Rab7-GFP or HTT-mRFP trajectories with a Hip1 or Rilpl reduction. n = 10. (G) Schematic diagram of homogenate fractionation into the perinuclear supernatant (PNS), vesicle fraction (VF), soluble fraction (SF), and heavy membrane pellet (P1) by ultra-centrifugation and sucrose gradient separation. Representative western blot of an immunoprecipitation of RAB7-containing mouse VF, probed with HTT, DYNC1I1, KIF5C, DCTN1, HIP1, LAMP1, SYT1, and SYP. HTT, DYNC1I1, KIF5C, DCTN1, HIP1, and LAMP1 show presence in the RAB7 IP, while SYT1 and SYP do not. (H) Representative western blot of an immunoprecipitation of HTT-containing mouse VF, probed with RAB7, which shows presence in the HTT IP. (I) Representative western blot of an immunoprecipitation of HTT-containing human iNeuron VF, probed with HTT, DYNC1I1, KIF5C, HIP1, RAB7, LAMP1, and SYT1, which all show presence in the HTT IP. No bands are seen in the negative no antibody control (−crtl). n = 3. Statistical analysis was conducted using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001. Also see Fig. S1 and S2.
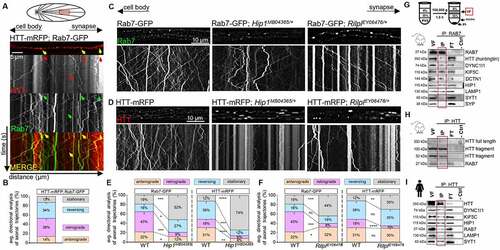
To screen for adaptor proteins that aid the motility of the HTT-Rab7 vesicle complex, we generated larvae expressing Rab7-GFP or HTT-mRFP in the context of reductions in Rab-associated proteins nemo (nmoP1/+) [Citation43,Citation44], Rip11 (Rip11KG02485/+) [Citation45], or Rilpl (RilplEY06476/+) [Citation46,Citation47], Hip1 (Hip1MB04365/+) [Citation48–50] or milton (miltk04704/+). Although HTT moves bi-directionally within larval axons, Drosophila lacks HAP1, but milton shares a HAP1-like N-domain [Citation51–53]. Reduction of Rilpl or Hip1 disrupted Rab7-GFP/HTT-mRFP motility causing Rab7 or HTT blockages, while reductions in milton, nemo, or Rip11 did not (, Fig. S1GH). Reduction of Hip1 significantly decreased both the anterograde and retrograde movement of Rab7-HTT-containing vesicles (), while reduction of Rilpl only decreased the retrograde movement of Rab7 or HTT-containing vesicles (). Biochemical analysis coupled with immunoprecipitation (IP) of mouse brains or iPSC-derived human neurons (iNeurons) showed that RAB7, HTT, HIP1, and motor proteins kinesin 1 and dynein were present on RAB7-containing or HTT-containing membranes (). Homogenized brains/neuron lysates were first fractionated and the PNS (post-nuclear supernatant), VF (vesicle fraction), SF (soluble fraction), and P1 (heavy-membrane pellet) were isolated using sucrose-gradient centrifugation (Fig. S2B), and then HTT or RAB7 was immunoprecipitated from the VF. Both full length and fragmented HTT were seen in the RAB7-VF IP (). Likewise, RAB7 was seen in the HTT-VF IPs (). In addition, HIP1, KIF5C and DYNC1I1 were also present in both the HTT-VF IPs () and the RAB7-VF IP (). Interestingly, SYT1 (synaptotagmin 1) and SYP (synaptophysin) were not observed with the RAB7-VF IP, but LAMP1 was present (), indicating that the HTT-RAB7 vesicle is likely distinct from the putative HTT-RAB4 synaptic vesicle complex we previously isolated [Citation10].
The retrogradely moving HTT-Rab7 vesicle complex is likely a LAMP1/Lamp1-containing late endosome
Because RAB7/Rab7 is seen with retrogradely moving LEs [Citation47], lysosomes [Citation46], and APs [Citation25], and HTT/htt is implicated during autophagy [Citation26,Citation27], we next tested the hypothesis that the HTT-Rab7 vesicle complex is either a LE, lysosome and/or an AP (Fig. S3A). Larvae simultaneously expressing either Rab7-GFP/HTT-mRFP, mCherry-Atg8a/HTT-eGFP, or LAMP1-GFP/HTT-mRFP showed retrogradely co-migrating Rab7-HTT, Atg8a-HTT or LAMP1-HTT containing vesicles (). Colocalized trajectories (yellow) revealed that 25% of vesicles contained HTT and Rab7, 17% of vesicles contained HTT and Atg8a, and 22% of vesicles contained HTT and LAMP1 (). Furthermore, reduction of htt (htt-RNAi) or Rab7 (Rab7EY10675/+) with either mCherry-Atg8a or LAMP1-GFP disrupted Atg8a or LAMP1 motility (). While reduction of htt significantly decreased the retrograde movement of Atg8a-containing vesicles (), reduction of Rab7 decreased both the anterograde and retrograde motility of Atg8a-containing vesicles (). Reduction of htt or Rab7 decreased both the anterograde and retrograde motility of LAMP1-containing vesicles (). Therefore, HTT and Rab7 are likely present together during autophagy and with lysosomes or LEs.
Figure 2. The HTT-Rab7 vesicle complex is likely a LAMP1/Lamp1-containing late endosome and/or autophagosome. (A) Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs from simultaneous dual-color movies from larvae co-expressing either HTT-mRFP and Rab7-GFP, HTT-eGFP and mCherry-Atg8a, HTT-mRFP and LAMP1-GFP. Co-migrating tracks that contain both red and green trajectories are seen (yellow arrow). X axis=distance (µm), Y axis=time (s). Bar: 5 µm. (B) Quantification of the average co-migratory trajectories reveal the percent co-migration between HTT-Rab7, HTT-LAMP1, and HTT-Atg8a. n = 6. (C-D) Representative images from movies and kymographs from larvae expressing mCherry-Atg8a or LAMP1-GFP alone, in the context of htt (htt-RNAi) or Rab7 (Rab7EY10675/+) reduction. Bar: 10 µm. (E-F) Directional analysis of mCherry-Atg8a or LAMP1-GFP trajectories alone compared to mCherry-Atg8a or LAMP1-GFP trajectories with a htt or Rab7 reduction. n = 10. Statistical significance was determined using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001. ****p < 0.00001. Also see Fig. S3.
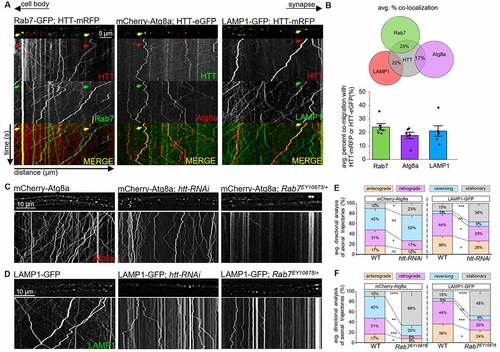
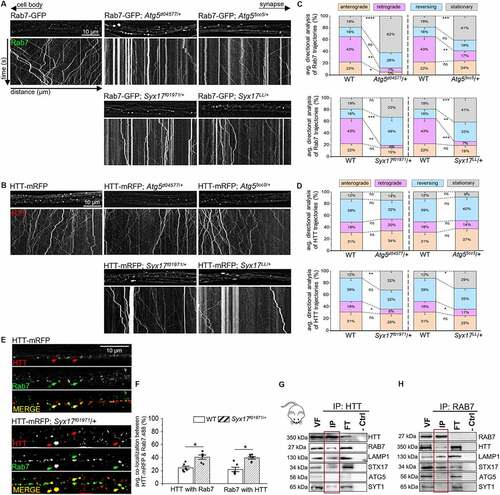
LEs can fuse with lysosomes [Citation54], forming endolysosomes, which can converge onto the autophagic pathway upon AP-lysosome fusion to form ALs (Fig. S3A [Citation23]). Since HTT and Rab7 co-migrated with Atg8a and LAMP1, the retrogradely moving HTT-Rab7 vesicle complex could be a LE, an AP, a lysosome, or all three. To isolate these specific compartments, we focused on Atg5 and Syx17. Atg5 is a key protein involved in the assembly of an AP from a PG [Citation19,Citation55], and is a marker for early autophagic compartments (Fig. S3A), while Syx17 is a SNARE protein identified in Drosophila that aids LE-AP fusion to form amphisomes [Citation23,Citation24] and is a candidate marker for middle autophagic compartments (Fig. S3A). APs fuse with lysosomes to become ALs (Fig. S3A [Citation23,Citation56]). APs can also fuse with LEs to form amphisomes [Citation57,Citation58], which can then fuse with lysosomes (Fig. S3A [Citation23]). Interestingly, Syx17-dependent LE-AP fusion is thought to be required for the retrograde motility of APs within cultured dorsal root ganglion neurons [Citation25]. We found that Atg5, Syx17, and LAMP1 mark discrete compartments in larval axons that do not show colocalization with each other (Fig. S3BE). To further define the HTT-Rab7 retrogradely co-migrating vesicle, we generated larvae expressing either HTT-mRFP or Rab7-GFP in the context of Atg5 reductions (Atg5d04577/+ or Atg55cc5/+) or Syx17 reductions (Syx17f01971/+ or Syx17LL/+). Similar to what was seen previously in Atg55cc5 and Syx17LL null alleles [Citation24,Citation59], western blot analysis showed that Atg5 protein levels in Atg5d04577/+ flies were reduced to 64% and Syx17 protein levels were reduced to 81%= 34KD, 62%= 40KD in Syx17f01971/+ flies compared to WT levels (Fig. S2A). Further, Atg5d04577 mutant flies also showed an ataxic phenotype (Fig. S2C), similar to what was observed in null Atg55cc5 mutant flies [Citation59], while Syx17f01971 was previously verified by PCR [Citation60,Citation61]. Intriguingly, ref(2)P protein levels were significantly increased in Atg5d04577/+, Syx17f01971/+ and Rab7EY10675/Rab7EY10675 flies (Fig. S2D), similar to what was previously observed in Atg55cc5, Syx17LL, and Rab7d1 null mutants [Citation24,Citation42,Citation59], indicating the disruption of degradation in these mutant lines. Reduction of Atg5 significantly decreased the bi-directional motility of Rab7-GFP-containing vesicles (), however the motility of HTT-mRFP containing vesicles was unaffected (). Immunolocalization analysis showed that while Rab7 and Atg5 colocalized (Fig. S3CE), HTT and Atg5 did not (Fig. S3DE). Reduction of Syx17 disrupted the retrograde motility of both HTT-mRFP or Rab7-GFP, with increased populations of stationary/stalled vesicles (), decreased populations of retrograde vesicles (), and an increased frequency of HTT-Rab7 colocalization (), but not HTT-Lamp1 colocalization (Fig. S4C). Note that both HTT and Rab7 colocalize with Syx17 (Fig. S3CDE). Reduction of htt increased the colocalization of Rab7-Syx17 (Fig. S4A) and reduction of Rab7 increased HTT-Syx17 colocalization (Fig. S4B), while the colocalization of Rab7-Lamp1 (Fig. S4A) and HTT-Lamp1 (Fig. S4B) were decreased. Further, STX17 (syntaxin 17) co-immunoprecipitated with HTT-containing vesicles isolated from mouse brain tissue, but not with ATG5 in mice (). STX17 also co-immunoprecipitated with RAB7-containing vesicles (). Therefore, while the retrogradely moving HTT-RAB7/Rab7 vesicle is likely a LE, disruption of LE-AP fusion through Syx17 reduction/loss likely disrupts the fusion of the putative HTT-RAB7/Rab7 LE with APs.
Figure 3. Disrupting autophagosome-late endosome fusion with mutant Syx17 impairs the motility of the HTT-Rab7 vesicle complex. (A-B) Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs from larvae expressing (A) Rab7-GFP or (B) HTT-mRFP alone, with a reduction of Atg5 (Atg5d04577/+ or Atg55cc5/+), or with a reduction of Syx17 (Syx17f01971/+ or Syx17LL/+). X axis=distance (µm), Y axis=time (s). Bar: 10 µm. (C-D) Directional analysis of (C) Rab7-GFP or (D) HTT-mRFP trajectories alone compared to Rab7-GFP or HTT-mRFP trajectories with Atg5 or Syx17 reductions. n = 10. (E) Representative images from larvae expressing HTT-mRFP alone or with a reduction of Syx17 (Syx17f01971/+) that have been immunostained for Rab7. Note the presence of colocalized puncta (yellow) in the merged overlay panels with larger accumulates with Syx17 reduction. Bar: 10 µm. (F) Quantification of colocalization (mander’s correlation) between HTT and Rab7 in larval axons in the context of Syx17 reduction compared to WT. n = 6. (G-H) Representative western blot of an immunoprecipitation of HTT-containing mouse VF (G) or an immunoprecipitation of RAB7-containing mouse VF (H), probed for HTT, RAB7, LAMP1, STX17, ATG5, and SYT1. Note that while RAB7, LAMP1, STX17, and SYT1 are present in the HTT IP, ATG5 is not. While HTT, LAMP1, and STX17 are present in the RAB7 IP, SYT1 and ATG5 are not. No bands are seen in the negative no antibody control (−crtl). Statistical significance was determined using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001. Also see Fig. S2, S3, and S4.
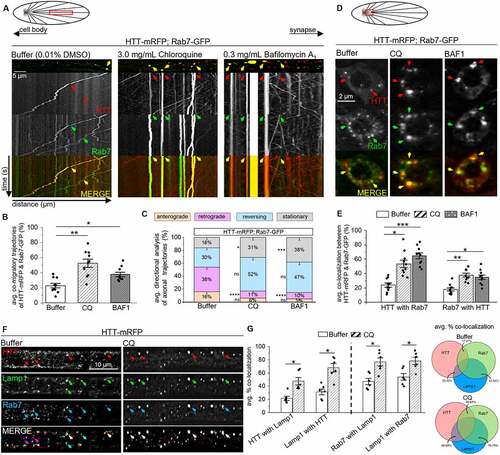
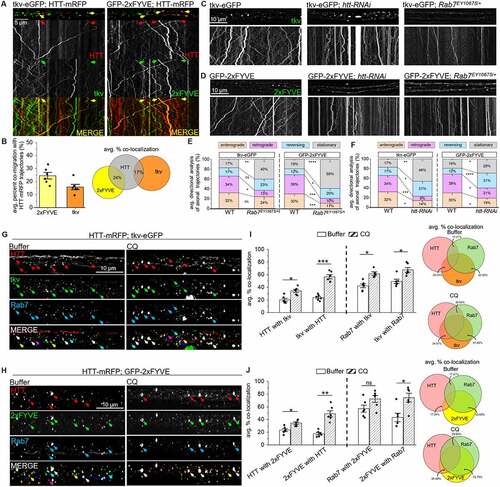
To further test the proposal that HTT-RAB7/Rab7-containing LEs fuse with APs, we next pharmacologically inhibited lysosome acidification and AP-lysosome fusion using chloroquine (CQ) or bafilomycin A1 (BAF1) [Citation28,Citation62–64]. Previous studies show that CQ or BAF1 treatment cause LAMP1 accumulations [Citation65–71]. Indeed, we also found accumulations of LAMP1 within Drosophila larval neurons expressing LAMP1-GFP fed on food laced with CQ [Citation72] or BAF1 [Citation22,Citation70] for 24 h prior to in vivo imaging compared to buffer (Fig. S5A). Accumulations of Rab7 were also seen within CQ or BAF1 treated larvae expressing Rab7-GFP (Fig. S5A). However, CQ or BAF1 treatment had no effect on Syt1-containing synaptic vesicles in larvae expressing Syt1-eGFP (Fig. S5A). Further, larvae simultaneously expressing HTT-mRFP and Rab7-GFP treated with either CQ or BAF1 showed impaired motility of HTT-Rab7-containing vesicles (yellow trajectories, ). CQ or BAF1 treatment significantly increased the co-migration of HTT-mRFP and Rab7-GFP (), but significantly decreased the retrograde motility of HTT-Rab7 vesicles () compared to buffer-treated larvae. While impaired motility of HTT-Rab7 vesicles corresponded to increased levels of colocalization between Rab7-GFP and HTT-mRFP at cell bodies ( S5B), decreased colocalization was observed at NMJs (Fig. S5C), suggesting that perhaps CQ also disrupts endocytosis [Citation73] and the packaging of Rab7 and HTT into LEs/lysosomes at NMJs. Furthermore, we found that HTT-Rab7 vesicles colocalized with Lamp1 in larval axons, which significantly increased upon treatment with CQ (). Taken together, these observations support the proposal that the retrogradely moving HTT-RAB7/Rab7 vesicle complex is likely a LAMP1/Lamp1-containing LE, that can fuse with an amphisome or an AP in a STX17/Syx17-dependent manner.
Figure 4. CQ- and BAF1-mediated inhibition of lysosome acidification and lysosome-autophagosome fusion impairs the motility of HTT-Rab7 vesicles. (A) Schematic diagram of larval nervous system showing the brain and segmental nerves. Red box=imaged area (90 µm). Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs from simultaneous dual-color movies from larvae expressing both HTT-mRFP (red) and Rab7-GFP (green) that have been fed either buffer-, CQ-, or BAF1-laced food. Co-migrating tracks that contain HTT and Rab7 are seen (yellow arrow). X axis=distance (µm), Y axis=time (s). Bar: 5 µm. (B) Quantification of co-migratory (yellow) trajectories (%) between HTT and Rab7 in buffer-, CQ-, and BAF1-treated larvae. (C) Quantification of directional analysis of yellow (HTT and Rab7) trajectories (%) in buffer-, CQ-, and BAF1-treated larvae. (D) Schematic diagram of larval nervous system showing the brain and segmental nerves. Red box=imaged area. Representative images from simultaneous dual-color images of neuron cell bodies from larvae co-expressing HTT-mRFP (red) and Rab7-GFP (green) that have been fed either buffer-, CQ-, or BAF1-laced food. Colocalized puncta that contain HTT and Rab7 are seen (yellow arrows). Bar: 2 µm. (E) Quantification of percent colocalization (mander’s) between HTT and Rab7%) in nerves of buffer-, CQ-, and BAF1-treated larvae. n = 10. (F) Representative images of segmental nerves from larvae expressing HTT-mRFP (red) that have been simultaneously immunostained with Lamp1 (green) and Rab7 (blue) following treatment with either buffer or CQ. Note that the merged panel of buffer-treated larvae reveals heterogenous overlays containing either HTT-Rab7 (purple arrows), Rab7-Lamp1 (teal arrows), or HTT-Rab7-Lamp1 (white arrows), while the merged panel of CQ-treated larvae show more overlays containing HTT-Rab7-Lamp1 (white arrows) Bar: 10 µm. (G) Quantification of colocalization (mander’s) between HTT-Rab7, HTT-Lamp1, and Rab7-Lamp1%) in nerves of buffer-, CQ-, or BAF1-treated larvae. Data represented as overlaid histogram-dot plots (left) as well as proportional Venn diagrams (right). n = 6. Statistical significance was determined using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001. Also see Fig. S5. .
Evidence for a retrogradely moving HTT-Rab7 signaling late endosome during axonal injury
Rab7 has been implicated in facilitating the retrograde axonal movement of long-distance signaling components [Citation12,Citation16,Citation36,Citation74]. To examine whether the HTT-Rab7-containing LE co-migrates with long-distance signaling components, we evaluated the retrograde BMP-signaling receptors tkv and wit [Citation75–77]. We also examined phosphatidylinositol-3-phosphate (PtdIns3P) phospholipids (FYVE), which are predominantly found within EEs, LEs, or lysosomes and perform key functions during signaling and membrane trafficking [Citation78–82]. Drosophila larval axons expressing the type-I receptor tkv-eGFP showed strong colocalization (80%) with the type-II receptor wit, which are known to form a complex upon ligand binding (Fig. S2E). Simultaneous expression of HTT-mRFP and tkv-eGFP showed co-migrating HTT-tkv vesicles (), with 17% of vesicles containing both HTT and tkv (). Further, immunolocalization showed that 20–24% of HTT colocalized with tkv while 42–50% of Rab7 colocalized with tkv vesicles (). Reduction of htt (htt-RNAi) or reduction of Rab7 (Rab7EY10675/+) impaired the motility of tkv-eGFP (), while reduction of Rab7 significantly decreased only the retrograde motility of tkv-eGFP (), and reduction of htt significantly decreased both the anterograde and retrograde motility of tkv-eGFP (). Further in Drosophila larval axons simultaneously expressing HTT-mRFP and GFP-2xFYVE, which probes for PtdIns3Ps, 24% of vesicles showed co-migration (yellow trajectories, ). Immunolocalization showed that 17–24% of HTT-2xFYVE or 2xFYVE-HTT vesicles were colocalized, while 43–58% of Rab7-2xFYVE or 2xFYVE-Rab7 vesicles were colocalized (, S2B). Moreover, reduction of htt or Rab7 with GFP-2xFYVE significantly impaired 2xFYVE motility (), decreasing both the anterograde and retrograde movement of GFP-2xFYVE (). Intriguingly, treatment of larval axons with CQ increased the colocalization of HTT-Rab7 vesicles with tkv or 2xFYVE, suggesting that inhibition of lysosome acidification and/or AP-lysosome fusion () perturbs the motility of the HTT-Rab7-tkv-wit-2xFYVE-containing vesicle complex. Taken together, these observations support the proposal that the retrogradely moving HTT-Rab7-containing LE is likely a long-distance signaling endosome.
Figure 5. The HTT-Rab7 vesicle complex co-transports with long-distance signaling components. (A) Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs from simultaneous dual-color movies from larvae co-expressing HTT-mRFP with either tkv-eGFP or GFP-2xfyve (PtdIns3ps). Co-migrating tracks that contain both red and green trajectories are seen (yellow arrow). X axis=distance (µm), Y axis=time (s). Bar: 5 µm. (B) Quantification of the average co-migratory trajectories reveal the percent co-migration (%) between HTT-tkv and HTT-2xfyve. n = 6. (C-D) Representative images from movies and kymographs from larvae expressing tkv-eGFP or GFP-2xfyve alone, in the context of htt reduction (htt-RNAi), or a genetic reduction of Rab7 (Rab7EY10675/+). Bar: 10 µm. (E-F) Directional analysis of tkv-eGFP or GFP-2xfyve trajectories alone compared to tkv-eGFP or GFP-2xfyve trajectories with a htt or Rab7 reduction. n = 10. (G-H) Representative images of segmental nerves from larvae co-expressing HTT-mRFP (red) and either (G) tkv-eGFP (green) or (H) GFP-2xfyve (green) that have been immunostained with Rab7 (blue) following treatment with buffer or CQ. Note that the merged panel of buffer-treated larvae reveals heterogenous overlays containing either HTT-Rab7 (purple arrows), Rab7-tkv or Rab7-2xfyve (teal arrows), HTT-tkv or HTT-2xfyve (yellow arrows) or HTT-Rab7-tkv and HTT-Rab7-2xfyve (white arrows), while the merged panel of CQ-treated larvae show more overlays containing HTT-Rab7-tkv and HTT-Rab7-2xfyve (white arrows) Bar: 10 µm. (I-J) Quantification of colocalization (mander’s, %) between (I) HTT-Rab7, HTT-tkv, and tkv-Rab7 or (J) HTT-Rab7, HTT-2xfyve, and 2xfyve-Rab7 in nerves of buffer-, CQ-, and BAF1-treated larvae. Data represented as overlaid histogram-dot plots (left) as well as proportional Venn diagrams (right). n = 6. Statistical significance was determined using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001. Also see Fig. S2.
Work shows that Rab7 colocalizes with MAP3K12/DLK, a long-distance signaling component which is activated after axon injury to support axon regeneration and neuron survival [Citation33,Citation36]. Moreover, HTT is implicated in the reprogramming of the regenerative transcriptome following axonal injury in mouse corticospinal neurons [Citation37]. To test the hypothesis that the HTT-Rab7-containing LE moves retrogradely and can be activated by axonal injury, we first evaluated wallenda (wnd) a conserved MAPKKK, homologous to MAP3K12/DLK which functions as an upstream mediator of injury [Citation30]. Larvae expressing either Rab7-GFP or HTT-mRFP in the context of endogenous wnd tagged with mCherry or GFP (wnd-mCherry or wnd-GFP [Citation83,Citation84]) showed wnd retrogradely co-migrating with HTT or Rab7-containing vesicles (Fig. S6E). Intriguingly, similar to what was observed with tkv (), treatment with CQ disrupted HTT-wnd or Rab7-wnd vesicle motility, suggesting that wnd is also present with the HTT-Rab7-tkv-wit-containing vesicle.
To directly test the hypothesis that the HTT-Rab7-containing LE moves retrogradely after axonal injury we crushed Drosophila larval nerves expressing GFP using the pucGAL4 driver ([Citation30,Citation85,Citation86], Fig. S6B). In these larvae, puc (puckered) regulatory sequences control the expression of GAL4, which will in turn express GFP after injury, and increased GFP expression will be seen in injured neuron cell bodies compared to non-injured neurons. Indeed, significant increases in GFP were observed in motor neuron cell bodies 16–24 h after nerve crush compared to 0–12 h (, S6B). Next, larvae expressing HTT-mRFP or Rab7-GFP were imaged at three axonal segments (, S6A, far-proximal to crush site-red box, proximal to the crush site-blue box, distal to crush site-green box) 16 h after nerve crush. As a control, larvae expressing Syt1-eGFP were imaged 16 h after nerve crush. Interestingly, while Syt1-eGFP vesicles were stalled/immobile in all three axonal segments imaged (far-proximal to crush site (red box), proximal to crush site (blue box), and distal to crush site (green box)), HTT-mRFP or Rab7-GFP vesicles showed motility at the far-proximal axonal segment to the crush site (red) and at the proximal crush site (blue box), but not at the distal axonal segment to the crush site (green box, ). Quantification of the anterograde:retrograde flux ratio revealed a significant retrogradely biased motility for both HTT-mRFP or Rab7-GFP vesicles (). Note that an increase in Rab7 and HTT localization is seen in cell bodies after nerve crush compared to uninjured controls (Fig. S6D). Moreover, Drosophila larval axons simultaneously expressing HTT-mRFP or Rab7-GFP imaged 16 h after nerve crush also showed an increase in the retrogradely co-migrating HTT-Rab7 vesicle population proximal to the crush site (red box, ), indicating that axonal injury does not perturb the retrograde motility of the HTT-Rab7 LE but rather stimulates its motility.
Figure 6. Axonal injury encourages enhanced retrograde movement of HTT-vesicles: evidence for a HTT-Rab7 signaling late endosome. (A) Schematic diagram of larval nervous system showing the brain. Red box=imaged area. Representative images from simultaneous dual-color images of neuron cell bodies from larvae expressing UAS-eGFP under the pucGAL4E69 driver that have been immunostained with elav at 0, 12, 16, 18, or 24 h after being subjected to larval nerve crush in the presence of fly food laced with buffer or CQ. The presence of eGFP in motor neuron cell bodies corresponds to increased expression of puckered (puc). Bar: 2 µm. (B) Quantification of the intensity ratio of GFP:elav normalized to the uninjured-buffer condition at motor neuron cell bodies at t24 of uninjured and injured/crushed larvae in buffer- or CQ-laced fly food. n = 5. (C) Representative line plot of the GFP:elav intensity ratio at motor neuron cell bodies of larvae subjected to nerve crush in either buffer- or CQ-laced fly food at 0, 12, 16, 18, or 24 h. n = 5. (D) Schematic diagram of larval nervous system showing the brain and segmental nerves. Red box=imaged area (90 µm). Arrows depict location of the cell bodies (retrograde) and synapses (anterograde). Representative images and kymographs of movies from larvae expressing Syt1-eGFP, Rab7-GFP, or HTT-mRFP that have been either left uninjured or injured/crushed for 16 h prior to in vivo imaging. Injured larvae were imaged at the far-proximal to crush site (red), proximal to crush site (blue), and distal to crush site (green). X axis=distance (µm), Y axis=time (s). Bar: 5 µm. (E) Representative images and kymographs from movies of larvae expressing HTT-mRFP (red) or Rab7-GFP (green) that have been injured for 16 h in CQ-laced fly food prior to in vivo imaging at the far-proximal to crush site (red) or the proximal to crush site (blue). (F) Quantification of the avg. anterograde:retrograde flux ratio per larvae (AU) of HTT-mRFP or Rab7-GFP trajectories. A retrograde bias is depicted as less than zero (left on x-axis), while an anterograde bias is depicted as greater than zero (right on x-axis). n = 5. (G) Representative images and kymographs from simultaneous dual-color movies from larvae expressing HTT-mRFP (red) and Rab7-GFP (green) that have been either left uninjured, injured for 16 h in buffer-laced fly food, or injured for 16 h in CQ-laced fly food prior to in vivo imaging at the far-proximal to crush site (red). Co-migrating tracks that contain HTT and Rab7 are seen (yellow arrow). X-axis=distance (µm), Y-axis=time (s). Bar: 5 µm. (H) Quantification of directional analysis (%) of HTT-Rab7 (yellow) trajectories of uninjured larvae compared to buffer-treated injured larvae and/or CQ-treated injured larvae. n = 5. (I) Representative western blot of an immunoprecipitation of HTT-containing human iNeuron VF, probed with HTT, RAB7, BDNF, and MAPK8/JNK, which all show presence in the HTT VF-IP. No bands are seen in the negative no antibody control (−crtl). n = 3. Statistical significance was determined using the two-sample two-sided Student’s t-test. Data represented as mean±sem. ns=p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001. Also see Fig. S5 and S6.
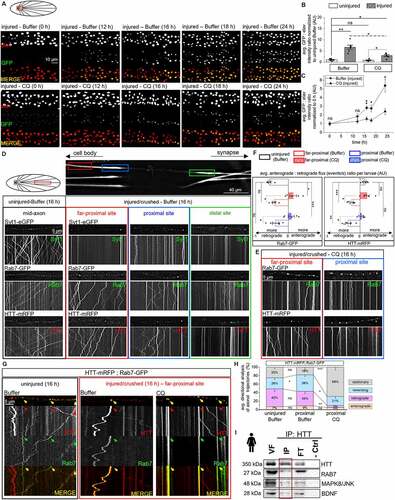
We next postulated that if the retrogradely moving HTT-Rab7 LE carries signaling components to the cell body after axonal injury then CQ treatment should disrupt this motility. Indeed, CQ treatment perturbed the retrograde motility of HTT-mRFP or Rab7-GFP containing vesicles (, S5A), with significant decreases in pucGAL4-driven GFP expression in motor neuron cell bodies 16–24 h after nerve-crush (, Fig. S6C). Quantification analysis of the anterograde:retrograde flux ratio revealed a significant anterogradely biased motility for both HTT-mRFP or Rab7-GFP vesicles with CQ treatment after nerve crush (). Note that Rab7-GFP and HTT-mRFP localization to cell bodies was also decreased with CQ treatment after nerve crush (Fig. S6D). Further, larvae co-expressing HTT-mRFP and Rab7-GFP with CQ treatment showed decreased co-migrating retrograde populations with increased stalling and reversing populations () after nerve crush. Additionally, biochemical analysis of HTT VF-IPs from human iNeurons revealed, in addition to RAB7, the presence of long-distance signaling components BDNF (brain-derived neurotrophic factor), which promotes axonal regrowth after sciatic nerve crush [Citation87], and MAPK8/JNK (), which is activated [Citation88,Citation89] and transported retrogradely following injury [Citation29,Citation32] to promote regeneration. Therefore, taken together, the retrogradely moving HTT-RAB7/Rab7-LAMP1-containing LE we isolated is likely a signaling endosome containing wnd/MAP3K12/DLK, BDNF, MAPK8/JNK, and/or the BMP-receptors tkv and wit which are carried to the cell body after axonal injury (Fig. S6F).
Discussion
Despite more than 350 binding partners for HTT identified across a wide-range of cellular processes, including trafficking, the molecular mechanisms by which HTT and its binding partners function remain elusive. Here we provide evidence for a retrogradely moving HTT-Rab7 vesicular complex using Drosophila genetics, in vivo imaging in Drosophila larval axons coupled with a custom particle tracking analysis and pharmacological inhibitors. Specifically, we found that the retrogradely moving HTT-RAB7/Rab7 vesicle is likely a LAMP1/Lamp1-containing LE that can traffic long-distance signaling components such as the BMP-receptors tkv and wit, neurotrophic factor BDNF and axonal damage response components wnd/MAP3K12/DLK and MAPK8/JNK following axonal injury. Taken together, our observations unravel a previously unknown role for HTT in the retrograde movement of a RAB7-LAMP1-containing signaling LE (Fig. S6F), which has functional relevance during axonal injury.
The biological relevance of the putative retrogradely moving HTT-RAB7-LAMP1-containing late endosome
Rab7 is an established LE marker [Citation11]. Several studies also show that Rab7 has a substantial role in the maturation of APs and LEs, by aiding in their retrograde motility and by participating in fusion with lysosomes [Citation11,Citation90–92]. Further, HTT facilitates the retrograde axonal movement of Atg8a/LC3-containing autophagic vesicles ( [Citation27]). However, Atg8a/LC3 is used as a marker for autophagy and this is problematic because Atg8a/LC3 is present in multiple autophagy compartments from PGs to ALs [Citation28] as well as during LE-AP fusion in distal axons [Citation25]. Therefore, knowledge of the exact location of where HTT enters the autophagy pathway is ambiguous. To overcome these issues, we probed HTT and Rab7 with markers specific for PGs, APs, amphisomes and lysosomes together with genetic mutants and pharmacological inhibitors to identify the cellular compartment where HTT and Rab7 merge. While HTT co-migrates with retrogradely moving Rab7-LAMP1 vesicles, reduction/loss of LE-AP fusion with mutant Syx17 or by pharmacological inhibition using CQ/BAF1 impaired the retrograde movement of axonal HTT-Rab7-LAMP1-containing vesicles. In contrast, reduction/loss of Atg5-dependent PG assembly to APs had no effect (). Despite HTT being implicated as a molecular scaffold during induction and cargo loading of elongating PGs [Citation93] through associations with ULK1 [Citation26], the lack of colocalization between HTT and Atg5 (Fig. S3) within axons and the failure to disrupt HTT motility with loss/reduction of Atg5 () suggests that HTT likely has no role at the PG within axons. Therefore, our observations suggest that the only location that HTT and Rab7 can enter the autophagy pathway is when HTT-Rab7 containing LEs fuse with Atg8a/LC3-containing APs (Fig. S3A). It is also possible that the HTT-Rab7 LE also contains LAMP1/Lamp1 since recent work shows that LAMP1 may not only mark degradative lysosomes [Citation14]. Therefore, it is clear that Atg8a/LC3 and LAMP1/Lamp1 cannot be used to conclusively label APs and lysosomes.
The presence of HTT with Rab7 in the endocytic pathway is further supported by our finding that loss/reduction of Hip1, a membrane bound component of clathrin-mediated endocytosis and endosomal trafficking [Citation50,Citation94], disrupted the motility of both HTT and Rab7 (, S1). Upon endocytic internalization, cargo marked to be degraded remains at EEs, which mature to LEs prior to long distance retrograde movement within axons to the cell body for lysosome-mediated degradation. In fact, pharmacological inhibition of lysosome acidification and lysosome-AP-LE fusion fusion impaired the retrograde co-migration of the putative HTT-Rab7-LAMP1 vesicle complex (). Similar to work that HTT facilitates kinesin 1 [Citation95] and dynein-mediated axonal motility [Citation96], and that HTT moves bidirectionally by interacting with kinesin 1 and dynein [Citation7], one potential model for HTT is to function as a molecular scaffold to coordinate dynein-mediated retrograde movement of RAB7/Rab7-LAMP1/Lamp1-containing LEs. Furthermore, while the Rab7-interacting protein FYCO1 (FYVE And coiled-coil domain autophagy adaptor 1) could mediate kinesin 1-mediated movement of LEs [Citation97], we observed that loss/reduction of Rilpl disrupted the retrograde movement of axonal HTT-Rab7 vesicles (, S1). Indeed, RAB7-interacting/accessory proteins RILP [Citation46,Citation47], ORP1 L [Citation98,Citation99], and PLEKHM1 [Citation100,Citation101] were shown to function during dynein-mediated movement of LEs. Therefore, we propose that a retrogradely moving axonal HTT-RAB7/Rab7-LAMP1 LE exists in vivo and is likely involved in autophagy through LE-AP fusion (Fig. S3A).
A novel role for HTT during axonal injury
RAB7 is implicated in the retrograde trafficking of neurotrophin signaling receptors NGF (nerve growth factor)-activated TRKA [Citation102,Citation103], and BDNF-activated TRKB [Citation12] within a LE, while a RAB7 mutant causing Charcot-Marie-Tooth disease showed dysregulated TRKA-NGF retrograde signaling resulting in axonal degeneration [Citation15,Citation104]. Work has shown that a majority of the retrograde TRKA signaling endosomes are ultra-structurally and molecularly defined multivesicular bodies [Citation16]. HTT is also implicated in the retrograde trafficking of TRKB-BDNF signals [Citation105,Citation106], with defective TRKB signaling seen in HD [Citation107,Citation108]. In fact, we also found BDNF with HTT-associated vesicles isolated from WT human iNeurons (), which was decreased in HD iNeurons (data not shown). Additionally, the BMP-receptors tkv and wit co-migrated with HTT and Rab7 (, S2), with pathogenic HTT disrupting tkv-containing vesicle motility [Citation109] causing synaptic growth defects [Citation76]. Therefore, the putative retrogradely moving HTT-RAB7-LAMP1 containing endosome we isolated here likely traffics long distance signaling components such as NGF and BDNF (Fig. S6F).
Further, a role for Rab7 in mediating the retrograde transport of injury signals has also been proposed. In response to axonal injury, transcriptional reprogramming is thought to occur via several protective signaling pathways, such as the MAPK8/JNK pathway [Citation110,Citation111] and DLK [Citation30,Citation33,Citation34,Citation112,Citation113]. DLK was shown to move retrogradely with Rab7-containing LEs following palmitoylation of DLK [Citation36]. We also found the axonal damage response signal wnd co-migrating with HTT and Rab7 (Fig. S6E). Additionally, work has suggested that retrograde signaling following axonal injury is mediated by vesicle transport complexes, such as the syd/JIP3-containing endosome that carries phosphorylated- MAPK8/JNK to neuronal cell bodies [Citation29,Citation35] and mass spectrometry of isolated syd/JIP3-associated vesicles showed the presence of Rab7/RAB7 [Citation35]. Macrophage-derived NADPH oxidase 2 complexes were shown to incorporate into RAB7-positive endosomes after injury in dorsal root ganglion neurons, influencing PtdIns3K signaling to stimulate neurite outgrowth and axon regeneration after sciatic nerve lesion [Citation114]. Moreover, the RAB7 mutant associated with Charcot-Marie-Tooth showed reduced protein synthesis in the axon with compromised axon integrity after initial axon outgrowth [Citation115]. A non-canonical BMP-signaling pathway has been suggested to mediate retrograde injury signaling from spinal cord injury sites [Citation116]. Additionally, HTT was recently proposed to influence the reprogramming transcriptome after spinal cord injury with deletion of HTT attenuating regeneration of mouse corticospinal axons [Citation37]. Further, loss/reduction of HTT displayed an altered proteomic profiling network in axotomized retinal ganglion cells, suggesting a role for HTT as an injury-response signaling hub [Citation38]. Taken together, the HTT-RAB7/Rab7-containing signaling endosome we identified likely has an important role during axonal injury (Fig. S6F). Indeed, we found that the retrograde motility of the HTT-RAB7/Rab7-LAMP1 LE was not perturbed but stimulated in Drosophila larval nerve crush experiments, which activates retrograde injury signaling reported by puc, a MAPK8/JNK-phosphatase, compared to uninjured larvae (, S6). Therefore, our observations identify a previously unknown role for HTT during axonal injury: to facilitate the movement of retrograde signals, such as BMP-receptors (tkv and wit), MAPK8/JNK, BDNF and the canonical damage signal wnd/DLK within a RAB7/Rab7-LAMP1/Lamp1 signaling LE to the cell body for transcriptional reprogramming of neurons. Future work will investigate a more direct mechanism by which HTT helps promote axonal regeneration following injury.
Materials and methods
Drosophila genetics
Neuronal drivers Appl-GAL4 (pan neuronal) and pGAL4-62B-SG26-1 (8 motor neurons) were used for neuronal expression of transgenic lines [Citation39,Citation40,Citation117]. For dual-color imaging, males from UAS-Rab7-GFP, UASp-YFP.rab7T22N, UAS-Syt1-eGFP, UAS-LAMP1-GFP, UAS-GFP-myc-2xFYVE, UAS-tkv-eGFP, MiMIC-wnd-mCherry, MiMIC-wnd-GFP, or UAS-mCherry-Atg8a (BDSC) were crossed to Appl-GAL4;T(2:3),CyO,TM6B,Tb/Pin88K virgin females. The chromosome carrying T(2:3),CyO,TM6B,Tb is referred to as B3 and carries the dominant markers, Hu, Tb and CyO. The larval Tb (Tubby) marker is used to select larvae of interest. Appl-GAL4/Y;UAS-/Cyo,TM6B males were then crossed with UAS-HTT15Q-mRFP (gift from Troy Littleton [Citation118]), UAS-HTTex1-25Q-eGFP (gift from Norbert Perrimon [Citation119]), or UAS-Rab7-GFP (BDSC) virgins. Additionally, UAS-Rab7-GFP males were crossed to pGAL4-62B SG26–1;T(2:3),CyO,TM6B,Tb/Pin88K virgin females and progeny that were pGAL4-62B-SG26-1/Y,CyO,TM6B,Tb/UAS-Rab7-GFP males were then crossed to UAS-HTT15Q-mRFP virgin females. For genetic interaction analysis, males from miltk04704/CyO [Citation52,Citation53], RilplEY06476 [Citation120], Hip1MB04365 [Citation121], Rip11KG02485/FM7c [Citation122], nmoP1/TM6 [Citation44,Citation123], Khc8/cyO (null allele [Citation124]), Dhc6-10/TM6B (null allele [Citation125]), Atg5d04577 (BDSC), Atg55cc5/FM7 (null allele [Citation59], BDSC), Syx17f01971/TM6B (BDSC [Citation60,Citation61]), Syx17LL/TM6 (null allele [Citation24], a gift from Gábor Juhász) Rab7EY10675 (BDSC [Citation42]), Rab7d1/TM6 (null allele [Citation42], a gift from Gábor Juhász), Df(98E2);CG9990 (htt-KO/+, [Citation41], gift from Sheng Zhang), or UAS-htt-rnai [Citation7] were crossed to Appl-GAL4;T(2:3),CyO,TM6B,Tb/Pin88K virgin females. Males were then crossed to UAS-HTT15Q-mRFP or UAS-Rab7-GFP. Note that Appl-GAL4 (× chromosome) and UAS-HTT15Q-mRFP (3rd chromosome) are on the same chromosomes as Atg5 (× chromosome), Syx17, or Rab7 (both on the 3rd chromosome) mutants. Western blot analysis showed that rab7 was reduced to 28% of WT levels in homozygous Rab7EY10675/Rab7EY10675 and Atg5 was reduced to 64% of WT levels in heterozygous Atg5d04577/+ (Fig. S2A). Additionally, significant increases in ref(2)P protein levels were seen in homozygous Rab7EY10675/Rab7EY10675, heterozygous Atg5d04577/+, heterozygous Syx17f01971/+, and the HTT lines htt-rnai and htt-KO/+ (Fig. S2D), similar to what was previously seen [Citation24,Citation26,Citation42,Citation59]. Rab7EY10675 showed an enhanced ectopic midline crossing phenotype [Citation126]. Syx17f01971 showed an Atg8-rnai phenotype of small perinuclear dots [Citation24]. Similar to Atg55cc5 [Citation59], Atg5d04577 flies showed an ataxic phenotype (Fig. S2C). Progeny that were Appl-GAL4/Y;Cyo,TM6B males were then crossed to UAS-Rab7-GFP, UAS-LAMP1-GFP, UAS-GFP-myc-2xFYVE, UAS-tkv-eGFP, or UAS-mCherry-Atg8a, or UAS-HTT15Q-mRFP virgin females. Conversely, Rip11KG02485/FM7c or UAS-htt-RNAi virgins were crossed to Appl-GAL4/Y;UAS-/Cyo,TM6b males. X/Y; +/+; pucGAL4E69/TM6B (BDSC) males were crossed with UAS-eGFP (BDSC) virgin females and female progeny were selected for larval nerve crush confirmation assays. UAS-Rab7-GFP, UAS-Syt1-eGFP, UAS-LAMP1-GFP, UAS-GFP-myc-2xFYVE, UAS-HTT15Q -mRFP, or UAS-eGFP males were crossed to Appl-GAL4 or pGAL4-62B-SG26-1 virgin females. In all cases non-tubby female 3rd instar larvae were dissected for in vivo imaging or immunohistochemistry. Sibling tubby larvae were evaluated as controls. Reciprocal crossings were also done to confirm observations. A comprehensive list of Drosophila lines used for this study is organized in .
Table 1. Key resource table.
Human iPSC cultures
The following cell lines were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: iPSCs from WT (polyQ = 25, 26y, female) patients were purchased from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research (GM23279*C) and iPSCs from WT (polyQ = 17, 48y, female) patients were purchased from the NINDS Human Cell Repository (ND38555).
Mouse brains
CBA/CaJ background mouse brains (gift from Matthew Xu-Friedman, The State University of New York at Buffalo) were dissected, halved, then stored on dry ice and used immediately or stored at −80°C for future use.
In vivo imaging and analysis of vesicle motility within whole-mount larval axons
Larvae were dissected and immediately imaged under physiological conditions as previously detailed [Citation9]. Non-tubby, female larvae were dissected and imaged under physiological conditions in dissection buffer. Motility was visualized in the red (568 nm), green (488 nm), or both channels (568/488 nm using a dual-view beam splitter attachment) within larval segmental nerves using a Nikon TE-2000 microscope (Nikon, Melville, NY, USA). From each larva, four sets of movies at an imaging window frame size of 90 μm at 150 frames were taken from the mid-region of the larva at an exposure of 500 ms using a Cool Snap HQ cooled CCD camera (Photometrics, Tucson, AZ, USA) and the Metamorph imaging system (Molecular Devices, Sunnyvale, CA, USA). Kymographs were generated in Metamorph using the kymograph stack tool. From a total of 10 larvae a set of 40 movies were imaged for each genotype at a spatial resolution of 0.126 μm/pixel. The four movies, each lasting a 1.25 min span a total time of 5 min. Because most of the vesicles take <1 min to move they will have moved out of the 90 μm imaging window by the end of the first movie since each time frame for each movie lasts 1.25 min. Movies were analyzed using a MATLAB-based particle tracker program as previously described [Citation129]. Briefly, vesicle trajectories were analyzed to obtain the overall distribution of cargo populations (directional analysis) and individual vesicle movement behaviors (velocities, pause frequencies/durations, run lengths). Duration-weighted segmental velocity evaluates the average velocity behavior that vesicles exhibit per time spent moving. Individual vesicles were automatically categorized as either anterograde, retrograde, reversing, or stationary. Reversing refers to a vesicle that has at least one switch event between anterograde and retrograde motility. Both anterograde or retrogradely moving mitochondria show a net movement in the respective direction without pausing or reversing.
Simultaneous dual-view in vivo imaging
A narrow single-band GFP/DsRED (488/568 nm) filter cassette was added to the Cool Snap HQ camera. Movies were taken in dual-view mode using the split view software in Metamorph at 150 frames from the mid-region of the larva at an exposure of 500 ms to simultaneously image RFP and GFP tagged vesicles. The Cool Snap HQ camera dual-view mode was aligned using Metamorph software (Split-View settings) before each imaging session. Movies were split by wavelength and each kymograph for each split movie was created, merged and analyzed for co-migration. Trajectories of vesicles with colocalized tracks were identified from kymographs using Metamorph software. For each fluorescence channel, a kymograph was generated using Metamorph as previously done [Citation10]. Briefly, after selecting the first channel, all frames within the time-lapse image sequence of this channel were added together to produce a summed image. Then a polyline was generated on an axon. After generation of the polyline, the kymograph was created using Metamorph software. The polyline was then copied from one fluorescent channel to the other and used to create a kymograph for the other fluorescent channel. To identify vesicles with colocalized signals from both channels, the kymographs were colored in red and green, respectively, and combined into a single RGB kymograph. Non-stationary vesicles with colocalized signals were identified and counted by their yellow color in the combined kymograph. Note that to differentiate meaningful colocalization we evaluated the colocalization of the entire trajectory of a moving particle during the entire time frame of the movie. Therefore, only particles containing the same trajectory in both the red and green channels would show colocalization in yellow when merged and spurious colocalization observed in one-time frame would be avoided. The total number of colocalized full trajectories for 20 kymographs across five larvae was counted for each genotype. Pearson’s and Mander’s coefficients were obtained using Coloc2 in ImageJ. Briefly, kymographs from Drosophila larval segmental nerves expressing two fluorophores were separated into red and green channels using Metamorph and analyzed using Coloc2 in ImageJ.
Immunohistochemistry of larval axons for quantification of accumulations or colocalization
3rd instar larvae were dissected and fixed in 8% paraformaldehyde, washed with PBT (phosphate buffered saline [PBS; 0.15 M NaCl and 10 mM Na2HPO4, pH 7.5] supplemented with 0.1% Tween-20 [Pierce, 28,352]) and incubated overnight with antibodies against Atg5 (1:100; Sigma-Aldrich, A0856), Syx17 (1:100; gift from Gábor Juhász, Eötvös Loránd University & Eötvös Loránd Research Network), Lamp1 (1:100; Abcam, ab30687), Syt1 (1:100; Phosphosolutions, 1975-STG), elav (1:100; DSHB [Developmental Studies Hybridoma Bank], elav-9F8A9), p-MAPK8/JNK (1:500; Cell Signaling Technology,), wit (1:100; DSHB, 23C7 anti-wit), and/or Rab7 (1:100; DHSB, Rab7). Larvae were incubated in secondary antibodies (1:100; Alexa Fluor 568 anti-mouse [Invitrogen, A11004], Alexa Fluor 488 anti-mouse [Invitrogen, A11001], Alexa Fluor 647 anti-mouse [Invitrogen, A21235], Alexa Fluor 568 anti-rabbit [Invitrogen, A11011], Alexa Fluor anti-rabbit 488 [Invitrogen, A11008], and/or Alexa Fluor 647 anti-rabbit [Invitrogen, A21244]), and mounted using Vectashield mounting medium (Vector Laboratories, NC9265087). Images of segmental nerves were collected using a Nikon Eclipse TE 2000 U microscope at 40X using the 40X objective or 90X using the 60X objective with 1.5X gain (Nikon, Melville, NY, USA) alongside the FITC (488 nm), TxRED (568 nm), both FITC/TxRED (568/488 nm using a dual-view beam splitter attachment) and/or Cy5 (647 nm) filters. Quantitative analysis on axonal blockages was carried out by collecting six confocal optical images from middle segmental larval nerves. For each genotype, at least six confocal optical images across five larvae were imaged and the number of axonal blockages was measured using NIH ImageJ software as previously done [Citation7,Citation117]. Quantitative analysis on colocalization of compartments in larval nerves reported with simultaneous dual-view imaging was carried out by collecting six confocal optical images from middle segmental larval nerves. For each genotype, at least six confocal optical images across five larvae were imaged. The total number of colocalized particles in larval nerves from six confocal images across five larvae was counted for each genotype. Mander’s and Pearson’s coefficients were obtained using Coloc2 in image J. Fixed imaged of larval nerves taken at 90X using the FITC, TxRED, and Cy5 filters were merged into a single RGB image to analyze colocalization noted as triple colocalization (white puncta) or double colocalization (yellow, magenta, or cyan puncta)/
Larval feeding
For chemical feeding experiments, 3rd instar larvae expressing UAS-HTT15Q-mRFP, UAS-rab7-GFP, UAS-LAMP1-GFP, UAS-Syt1-eGFP, UAS-GFP-myc-2xFYVE, or UAS-eGFP were grown with fly flood (24 g agar, 22 g yeast, 164 g cornmeal, 12 g soy flour, and 200 mL molasses, and 5 L H2O) containing buffer (0.01% DMSO,), 3 mg/mL chloroquine (Cayman Chemical, 14,194) dissolved in buffer/0.01% DMSO, 0.3 mg/mL bafilomycin A1 (Cayman Chemical, 11,038) dissolved in buffer (0.01% DMSO), 1.0 µg/mL rapamycin (Cayman Chemical, 13,346) dissolved in buffer/0.01% DMSO, or 10 µm paclitaxel (Millipore Sigma, 58-055-65 MG [Citation130]) dissolved in buffer/0.01% DMSO for 12, 16, 18, or 24 h. Larvae were dissected and imaged for in vivo motility, described above. Alternatively, larvae were dissected for immunohistochemistry, described above. For dual-view imaging, larvae simultaneously expressing UAS-HTT15Q-mRFP with either UAS-Rab7-GFP, UAS-LAMP1-GFP, UAS-tkv-eGFP, or UAS-GFP-myc-2xFYVE were subjected to either fly food feeding condition and dissected and imaged for dual-view imaging, described above. Alternatively, larvae were dissected for immunohistochemistry, described above.
Larval nerve crush
Larval nerve crush was performed as done [Citation30]. Larvae were anesthetized with CO2 gas for 1 min prior to pinching larval segmental nerves through the ventral cuticle for 5 s with Dumont #5 Inox forceps (Fine Science Tools, 11,252–20). Successful injury events were recorded by posterior paralysis of larvae with actively moving mouthpieces. Observational recording of successful injury was confirmed by Phase-contrast imaging and HRP-staining (HRP-FITC [Jackson Immuno Research Labs, 123-095-021] or HRP-TxRED [Jackson Immuno Research Labs, 123-585-021] at 1:50) of nerve crush site of dissected larvae as well as through reporting of increased GFP at motor neuron cell bodies in larvae expressing pucGAL4E69; UAS-eGFP, which was validated by p-MAPK8/JNK (Cell Signaling Technology, 9251) immunostaining of neuron cell bodies (data not shown). For incubations, larvae were either immediately dissected for t0 results, or transferred to fresh fly-food plates containing either Buffer/0.01% DMSO, 10 µm paclitaxel dissolved in Buffer/0.01% DMSO, or 3 mg/mL CQ dissolved in Buffer/0.01% DMSO. These larvae were further incubated for 12, 16, 18, or 24 h at 29ºC prior to dissection for immunohistochemistry or in vivo imaging as described above.
Drosophila geotaxis assay
Negative geotaxis assays were performed as previously done [Citation59,Citation131,Citation132]. WT and Atg5d04577 flies were reared for 4 weeks at 25°C and 60% humidity. 20 healthy flies (10 males and 10 females) were then selected and transferred into a negative geotaxis cylinder. Flies were at rest for 5 min after transfer and then tapped to induce negative geotaxis. Fly movements were captured by images taken 3 s post-induction. Fly climbing speed and distance traveled after negative geotaxis induction were calculated using ImageJ and analyzed.
Human iPSC neuronal differentiation
Human iPSCs were grown and expanded on corning matrigel (Thermofisher Scientific, CB40230A) using E8 iPSC media (Invitrogen, A1517001) as previously done [Citation10]. Pluripotency was analyzed using an antibody against OCT-3/4 (1:200; SCBT [Santa Cruz Biotechnology], sc-5279) and Hoechst (Thermofisher Scientific, PI62249) was used as a nuclear staining as detailed below. After 4 passages iPSCs were differentiated into NPCs (neuronal precursors) using PSC neural induction media (Invitrogen, A1647801) and published protocols (publication #MAN0008031). NPCs were identified using an antibody against Nestin (1:200; SCBT, sc -23,927) and then differentiated into mature iNeurons using neurobasal media supplemented with 1× B27 (Invitrogen, 17,504–044) and 2 mM glutamine (ThermoFisher Scientific, 25,030,081). Differentiated neurons, identified using antibodies against MAP2 (1:100, BD Biosciences, BDB556326), TUBB3 (1:100; Biolegend, 801,201) and SYP (1:100, EMD Millipore, MAB5258) exhibited an extensive neurite network after 21 days at which time they were used for biochemical experiments as previously done [Citation10].
Preparation of protein extracts from fly brains, mouse brains, and human iNeurons
Mouse or fly brains were homogenized in homogenization buffer (10 mM HEPES, pH 7.4, 100 mM K acetate, 150 mM sucrose, 5 mM EGTA, 3 mM Mg acetate, 1 mM DTT) containing a cocktail of protease inhibitors (Pierce, PIA32965) and phosphatase inhibitors (Pierce, PI88667) with 5 mM EDTA. Neuronal extracts from Fly brains were centrifuged at 1000 g for 15 min and the supernatant (PNS) was used as detailed below for western blotting. Mouse brain extracts were then centrifuged at 1000 g for 15 min at 4°C. The PNS was then used for sucrose gradient fractionation analysis as detailed below or for western blotting. iNeurons were manually removed from 6-well or 12-well plates using ice-cold homogenization buffer (10 mM HEPES, pH 7.4, 100 mM K acetate, 150 mM sucrose, 5 mM EGTA, 3 mM Mg acetate, 1 mM DTT) containing a cocktail of protease inhibitors (Pierce, PIA32965) and 5 mM EDTA, blended for 30 s on ice using a motorized pestle, and then quickly snap-frozen in liquid nitrogen. Neuronal extracts were then centrifuged at 1000 g for 15 min and the supernatant (PNS) was used for sucrose gradient fractionation analysis as detailed below or for western blotting.
Sucrose gradient fractionations
PNS samples human iNeuron extracts or mouse brains were further fractionated into soluble fractions (SF), heavy membrane pellet (P1), and vesicle fractions (VF) by sucrose gradient ultra-centrifugations using lysis buffer (4 mM HEPES, 320 mM sucrose, pH 7.4) containing a phosphatase inhibitor (Pierce, PI88667) and protease inhibitor cocktail (Pierce, PIA32965) as previously done [Citation10,Citation133,Citation134]. Briefly, 300 ul of PNS was combined with 300 ul 62% sucrose and layered onto a sucrose gradient (35% and 8% sucrose) and centrifuged at 50,000 g for 90 min. The VF (35/8 layer), SF, and P1 were removed and used in western blot analysis. 100 ul of lysis buffer was used to dissolve P1.
Co-immunoprecipitation analysis
1000 ug of total protein from the mouse VF or the iNeuron VF were incubated with a mouse monoclonal antibody to RAB7 (1:1000, SCBT, sc -13,156) or a mouse monoclonal antibody to HTT (1:500, EMD Millipore, MAB2166) overnight at 4°C. Protein homogenates were then incubated with protein A/G magnetic beads (Pierce, PI88802) with rotation for 90 min and eluted with elution buffer (0.1 M glycine, pH 2.0). Eluents were then analyzed by western blot analysis as detailed below. Alternatively, Co-IP was performed on protein homogenates using the crosslinking co-immunoprecipitation kit (Pierce, 26,147) with 0.25 mM DSS (Pierce, 21,655) for 30 min before incubating antibody-bound magnetic beads with VFs.
SDS-PAGE and western blot analysis
Fly brains, mouse brains, or human iNeuron fractions were separated by SDS-PAGE. Samples were denatured in NuPage LDS sample buffer (Invitrogen, NP008) with 4 mM β-mercaptoethanol (Sigma-Aldrich, M6250) and run on 4–12% Bis-Tris gels (Invitrogen, NP0322BOX) which were then transferred to nitrocellulose membranes (Cytiva, 10,119–996). Blots were blocked using TBST (tris buffered saline [TBS; 150 mM NaCl and 20 mM Tris-base, pH = 7.5] supplemented with 0.1% Tween-20 [Pierce, 28,352]) with 5% BSA (Lampire, 50-413-345) for 60 min at 25°C and incubated with primary antibodies: SYT1 (1:1000; Phosphosolutions, 1975-STG), ref(2)P (1:500; gift from Gábor Juhász, Eötvös Loránd University & Eötvös Loránd Research Network), RAB7 (1:1000; SCBT, sc -13,156), Rab7 (1:100; DSHB, Rab7), Syx17 (1:500; gift from Gábor Juhász, Eötvös Loránd University & Eötvös Loránd Research Network), ATG5 (1:1000; Sigma-Aldrich, A0856), Lamp1 (1:1000; Abcam, ab30687), RTN1 (1:1000; SCBT, sc -23,880), SYP (1:1000, Thermofisher Scientific, MAB5258), Cyt-c-p (1:1000; SCBT, sc -13,156), HIP1 (1:1000, Novus biological, NB300–203), DCTN1 (1:500; Abcam, ab96004), KIF5C (1:500; gift from Lawrence Goldstein, University of California, San Diego), DYNC1I1 (1:1000, Abcam, ab23905), ACTA1/Actin (1:1000; ThermoFisher Scientific, MA5– 32,479), TUBA4A (1:2000, Abcam, ab7291), MAPK8/JNK (1:1000, Cell Signaling Technology, 9252), BDNF #1 (1:250; DSHB, BDNF-#1), BDNF #9 (1:250; DSHB, BDNF-#9), HTT rabbit polyclonal (1:1000, Abcam, ab45169), and HTT mouse monoclonal (1:1000; EMD Millipore, MAB2166) for 16 h at 4°C. Blots were then incubated with appropriate secondary HRP-conjugated anti-mouse (1:1000; ThermoFisher Scientific, 32,430) or anti-rabbit (1:1000; ThermoFisher Scientific, 32,460) secondary antibodies and imaged using a ChemiDoc MP imaging system (Bio-Rad Laboratories, Hercules, CA, USA) with ECL (Pierce, PI32106) or diluted West Femto (1:5 in TBS [detailed above]; Pierce, 34,095). Images from 3 blots were quantified using ImageJ.
Statistical analysis
The statistical analysis used for each experiment is indicated in each figure legend. First power and sample size (n) calculations were performed on Minitab18 for each experimental paradigm: comparing 2 means from 2 samples, with two-sided equality to identify the sample size that corresponds to a power of 0.9 with α = 0.01.For each experiment, a stringent significance threshold of p < 0.01 (99% confidence) was used as detailed in [Citation9]. Based on the power analysis, quantifications were performed across 5, 6, or 10 larvae. The n-value refers to the number of larvae or the number of immunoblots from 3 independent experiments. Individual data points for each analysis were averaged for each n and compared.
To select the appropriate statistical test, data distributions for each transport dynamic analyzed were first checked for normality using the nortest package of R: the Lilliefors test and Anderson – Darling test as previously detailed [Citation9,Citation40,Citation129]. Statistical significance of normal distributions was calculated by one-way ANOVA/post-hoc analysis to reduce Type I error, followed by two-sample two-tailed Student’s t-tests to test to compare individual groups in Excel and Minitab18. Statistical analysis reported in figures report results from Student’s t-tests, as results from ANOVA/post-hoc and Student’s t-tests were consistent. Statistical significance of normal distributions was calculated by a two-sample two-tailed Student’s t-test and/or ANOVA while the non-normal segmental velocity distributions were compared using the non-parametric Wilcoxon – Mann–Whitney rank sum test in Excel and Minitab18. Unless otherwise specified, the data compared was found to be normally distributed. For in vivo motility quantifications, each larva (total of 4 movies, > 500 vesicles) were pooled, then the averaged before performing statistical analysis. Therefore, statistical analysis was performed on global quantifications from each larva rather than individual vesicle dynamics (n = 10 larvae, 4 movies per larvae). Overlaid dot plots were constructed for all figures using OriginLab/OriginPro to represent mean±SEM and biological models were generated in BioRender.
Data availability
All data generated or analyzed during this study are included in this published article or in supplementary information. Raw data is available from the corresponding author upon request.
Supplemental Material
Download MS Word (6.6 MB)Acknowledgment
We thank the members of the Gunawardena lab for constructive discussions and insight. We thank Dr. Troy Littleton for the UAS-HTT15Q-mRFP line, Dr. Norbert Perrimon for the UAS-HTTex1-25Q-eGFP line, Dr. Sheng Zhang for the htt-KO line, Dr. Matthew Xu-Friedman for mouse brains, the Coriell Cell Repository and the NINDS Repository for the WT human iPSC lines, Dr. Gábor Juhász for the Syx17LL and Rab7d1 fly lines as well as the Syx17 and the ref(2)P antibodies, Dr. Lawrence Goldstein for the KIF5C antibody, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank. SG thanks Priyantha Karunaratne for constant support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15548627.2022.2119351
Additional information
Funding
References
- Nasir J, Floresco SB, O’Kusky JR, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995 Jun 2 81;(5)811–823. DOI:10.1016/0092-8674(95)90542-1
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003 Aug;28(8):425–433.
- Goehler H, Lalowski M, Stelzl U, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol Cell. 2004 Sep 24 15;(6)853–865. DOI:10.1016/j.molcel.2004.09.016
- Kaltenbach LS, Romero E, Becklin RR, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007 May 11 3;(5)e82. DOI:10.1371/journal.pgen.0030082
- Culver BP, Savas JN, Park SK, et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J Biol Chem. 2012 Jun 22 287;(26)21599–21614. DOI:10.1074/jbc.M112.359307
- Ratovitski T, Chighladze E, Arbez N, et al. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle. 2012 May 15 11;(10)2006–2021. DOI:10.4161/cc.20423
- Gunawardena S, Her LS, Brusch RG, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003 Sep 25 40;(1)25–40. DOI:10.1016/S0896-6273(03)00594-4
- Power D, Srinivasan S, Gunawardena S. In-Vivo evidence for the disruption of Rab11 vesicle transport by loss of huntingtin. Neuroreport. 2012 Nov 14;23(16):970–977. DOI:10.1097/WNR.0b013e328359d990
- White JAs2nd, Anderson E, Zimmerman K, et al. Huntingtin differentially regulates the axonal transport of a sub-set of Rab-containing vesicles in vivo. Hum Mol Genet. 2015 Dec 20 24;(25)7182–7195. DOI:10.1093/hmg/ddv415
- White JAs2nd, Krzystek TJ, Hoffmar-Glennon H, et al. Excess Rab4 rescues synaptic and behavioral dysfunction caused by defective HTT-Rab4 axonal transport in Huntington’s disease. Acta Neuropathol Commun. 2020 Jul 1 8;(1)97. DOI:10.1186/s40478-020-00964-z
- Chavrier P, Parton RG, Hauri HP, et al. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990 Jul 27 62;(2)317–329. DOI:10.1016/0092-8674(90)90369-P
- Deinhardt K, Salinas S, Verastegui C, et al. Rab5 and Rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron. 2006 Oct 19 52;(2)293–305. DOI:10.1016/j.neuron.2006.08.018
- Cai Q, Lu L, Tian JH, et al. Snapin-Regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010 Oct 6 68;(1)73–86. DOI:10.1016/j.neuron.2010.09.022
- Cheng XT, Xie YX, Zhou B, et al. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol. 2018 Sep 3 217;(9)3127–3139. DOI:10.1083/jcb.201711083
- Zhang K, Fishel Ben Kenan R, Osakada Y, et al. Defective axonal transport of Rab7 GTPase results in dysregulated trophic signaling. J Neurosci. 2013 Apr 24 33;(17)7451–7462. DOI:10.1523/JNEUROSCI.4322-12.2013
- Ye M, Lehigh KM, Ginty DD. Multivesicular bodies mediate long-range retrograde NGF-TrkA signaling. Elife. 2018 Jan 30; 7: DOI:10.7554/eLife.33012
- Vv K, Maday S. Neuronal endosomes to lysosomes: a journey to the soma. J Cell Biol. 2018 Sep 3;217(9):2977–2979. DOI:10.1083/jcb.201806139
- Yap CC, Digilio L, McMahon LP, et al. Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J Cell Biol. 2018 Sep 3 217;(9)3141–3159. DOI:10.1083/jcb.201711039
- Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001 Feb 19 152;(4)657–668. DOI:10.1083/jcb.152.4.657
- Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014 Jul 14;30(1):71–85. DOI:10.1016/j.devcel.2014.06.001
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012 Feb 20;196(4):407–417. DOI:10.1083/jcb.201106120
- Maday S, Holzbaur EL. Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci. 2016 Jun 1;36(22):5933–5945. DOI:10.1523/JNEUROSCI.4401-15.2016
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012 Dec 7;151(6):1256–1269. DOI:10.1016/j.cell.2012.11.001
- Takáts S, Nagy P, Varga Á, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013 May 13 201;(4)531–539. DOI:10.1083/jcb.201211160
- Cheng XT, Zhou B, Lin MY, et al. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015 May 11 209;(3)377–386. DOI:10.1083/jcb.201412046
- Rui YN, Xu Z, Patel B, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015 Mar;17(3):262–275. DOI:10.1038/ncb3101
- Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014 Jan 22;34(4):1293–1305. DOI:10.1523/JNEUROSCI.1870-13.2014
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222.
- Cavalli V, Kujala P, Klumperman J, et al. Sunday Driver links axonal transport to damage signaling. J Cell Biol. 2005 Feb 28 168;(5)775–787. DOI:10.1083/jcb.200410136
- Xiong X, Wang X, Ewanek R, et al. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010 Oct 4 191;(1)211–223. DOI:10.1083/jcb.201006039
- Bisby MA, Bulger VT. Reversal of axonal transport at a nerve crush. J Neurochem. 1977 Aug;29(2):313–320.
- Lindwall C, Kanje M. Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol Cell Neurosci. 2005 Jun;29(2):269–282.
- Shin JE, Cho Y, Beirowski B, et al. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012 Jun 21 74;(6)1015–1022. DOI:10.1016/j.neuron.2012.04.028
- Shin JE, Ha H, Kim YK, et al. DLK regulates a distinctive transcriptional regeneration program after peripheral nerve injury. Neurobiol Dis. 2019 Jul;127:178–192.
- Abe N, Almenar-Queralt A, Lillo C, et al. Sunday driver interacts with two distinct classes of axonal organelles. J Biol Chem. 2009 Dec 11 284;(50)34628–34639. DOI:10.1074/jbc.M109.035022
- Holland SM, Collura KM, Ketschek A, et al. Palmitoylation controls DLK localization, interactions and activity to ensure effective axonal injury signaling. Proc Natl Acad Sci U S A. 2016 Jan 19 113;(3)763–768. DOI:10.1073/pnas.1514123113
- Poplawski GHD, Kawaguchi R, Van Niekerk E, et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature. 2020 May;581(7806):77–82. DOI:10.1038/s41586-020-2200-5
- Belin S, Nawabi H, Wang C, et al. Injury-Induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron. 2015 May 20 86;(4)1000–1014. DOI:10.1016/j.neuron.2015.03.060
- Shyamala BV, Chopra A. Drosophila melanogaster chemosensory and muscle development: identification and properties of a novel allele ofscalloped and of a new locus, SG18.1, in a Gal4 enhancer trap screen. J Genet. 1999 08 01;78(2):87.
- Gunawardena S, Yang G, Goldstein LS. Presenilin controls kinesin-1 and dynein function during APP-vesicle transport in vivo. Hum Mol Genet. 2013 Oct 1;22(19):3828–3843. DOI:10.1093/hmg/ddt237
- Zhang S, Feany MB, Saraswati S, et al. Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington’s disease model. Dis Model Mech. 2009 May-Jun;2(5–6):247–266. DOI:10.1242/dmm.000653
- Hegedűs K, Takáts S, Boda A, et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell. 2016 Oct 15 27;(20)3132–3142. DOI:10.1091/mbc.e16-03-0205
- Sahlender DA, Roberts RC, Arden SD, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005 Apr 25 169;(2)285–295. DOI:10.1083/jcb.200501162
- Merino C, Penney J, González M, et al. Nemo kinase interacts with Mad to coordinate synaptic growth at the Drosophila neuromuscular junction. J Cell Biol. 2009 May 18 185;(4)713–725. DOI:10.1083/jcb.200809127
- Schonteich E, Wilson GM, Burden J, et al. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J Cell Sci. 2008 Nov 15 121;(Pt 22)3824–3833. DOI:10.1242/jcs.032441
- Jordens I, Fernandez-Borja M, Marsman M, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001 Oct 30 11;(21)1680–1685. DOI:10.1016/S0960-9822(01)00531-0
- Johansson M, Rocha N, Zwart W, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007 Feb 12 176;(4)459–471. DOI:10.1083/jcb.200606077
- Metzler M, Legendre-Guillemin V, Gan L, et al. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001 Oct 19 276;(42)39271–39276. DOI:10.1074/jbc.C100401200
- Hyun TS, Li L, Oravecz-Wilson KI, et al. Hip1-Related mutant mice grow and develop normally but have accelerated spinal abnormalities and dwarfism in the absence of HIP1. Mol Cell Biol. 2004 May;24(10):4329–4340. DOI:10.1128/MCB.24.10.4329-4340.2004
- Legendre-Guillemin V, Metzler M, Lemaire JF, et al. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005 Feb 18 280;(7)6101–6108. DOI:10.1074/jbc.M408430200
- Li SH, Gutekunst CA, Hersch SM, et al. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998 Feb 15 18;(4)1261–1269. DOI:10.1523/JNEUROSCI.18-04-01261.1998
- Stowers RS, Megeath LJ, Górska-Andrzejak J, et al. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002 Dec 19 36;(6)1063–1077. DOI:10.1016/S0896-6273(02)01094-2
- Glater EE, Megeath LJ, Stowers RS, et al. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006 May 22 173;(4)545–557. DOI:10.1083/jcb.200601067
- Mullock BM, Bright NA, Fearon CW, et al. Fusion of lysosomes with late endosomes produces a hybrid organelle of intermediate density and is NSF dependent. J Cell Biol. 1998 Feb 9 140;(3)591–601. DOI:10.1083/jcb.140.3.591
- Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998 Sep 24 395;(6700)395–398. DOI:10.1038/26506
- Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015 Apr 23 520;(7548)563–566. DOI:10.1038/nature14147
- Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988 Feb 29;151(1):40–47. DOI:10.1016/0006-291X(88)90556-6
- Berg TO, Fengsrud M, Strømhaug PE, et al. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998 Aug 21 273;(34)21883–21892. DOI:10.1074/jbc.273.34.21883
- Kim M, Sandford E, Gatica D, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016 Jan 26;5: DOI:10.7554/eLife.12245
- Gilsohn E, Volk T. Slowdown promotes muscle integrity by modulating integrin-mediated adhesion at the myotendinous junction. Development. 2010 Mar;137(5):785–794.
- Younger MA, Müller M, Tong A, et al. A presynaptic ENaC channel drives homeostatic plasticity. Neuron. 2013 Sep 18 79;(6)1183–1196. DOI:10.1016/j.neuron.2013.06.048
- Ahlberg J, Glaumann H. Uptake–microautophagy–and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp Mol Pathol. 1985 Feb;42(1):78–88.
- Yamamoto A, Tagawa Y, Yoshimori T, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998 Feb;23(1):33–42. DOI:10.1247/csf.23.33
- Juhász G. Interpretation of bafilomycin, pH neutralizing or protease inhibitor treatments in autophagic flux experiments: novel considerations. Autophagy. 2012 Dec;8(12):1875–1876.
- Fass E, Shvets E, Degani I, et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006 Nov 24 281;(47)36303–36316. DOI:10.1074/jbc.M607031200
- Fedele AO, Proud CG. Chloroquine and bafilomycin a mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci Rep. 2020 Apr 30;40(4): DOI:10.1042/BSR20200905
- Guiney SJ, Adlard PA, Lei P, et al. Fibrillar α-synuclein toxicity depends on functional lysosomes. J Biol Chem. 2020 Dec 18 295;(51)17497–17513. DOI:10.1074/jbc.RA120.013428
- Mauthe M, Orhon I, Rocchi C, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. DOI:10.1080/15548627.2018.1474314
- Mauvezin C, Nagy P, Juhász G, et al. Autophagosome-Lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015 May 11;6:7007. DOI:10.1038/ncomms8007
- Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11(8):1437–1438.
- Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 2013 Nov;11(11):e1001708.
- Nagy P, Varga A, Pircs K, et al. Myc-Driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 2013;9(8):e1003664. DOI:10.1371/journal.pgen.1003664
- Chen PM, Gombart ZJ, Chen JW. Chloroquine treatment of ARPE-19 cells leads to lysosome dilation and intracellular lipid accumulation: possible implications of lysosomal dysfunction in macular degeneration. Cell Biosci. 2011 Mar 8;1(1):10. DOI:10.1186/2045-3701-1-10
- Andres-Alonso M, Ammar MR, Butnaru I, et al. SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals. Nat Commun. 2019 Nov 29 10;(1)5448. DOI:10.1038/s41467-019-13224-z
- Smith RB, Machamer JB, Kim NC, et al. Relay of retrograde synaptogenic signals through axonal transport of BMP receptors. J Cell Sci. 2012 Aug 15 125;(Pt 16)3752–3764. DOI:10.1242/jcs.094292
- Kang MJ, Hansen TJ, Mickiewicz M, et al. Disruption of axonal transport perturbs bone morphogenetic protein (Bmp)–signaling and contributes to synaptic abnormalities in two neurodegenerative diseases. PLoS One. 2014;9(8):e104617. DOI:10.1371/journal.pone.0104617
- Liao EH, Gray L, Tsurudome K, et al. Kinesin Khc-73/KIF13B modulates retrograde BMP signaling by influencing endosomal dynamics at the Drosophila neuromuscular junction. PLoS Genet. 2018 Jan;14(1):e1007184. DOI:10.1371/journal.pgen.1007184
- Simonsen A, Wurmser AE, Emr SD, et al. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001 Aug;13(4):485–492. DOI:10.1016/S0955-0674(00)00240-4
- Raiborg C, Wenzel EM, Pedersen NM, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015 Apr 9 520;(7546)234–238. DOI:10.1038/nature14359
- Jaber N, Mohd-Naim N, Wang Z, et al. Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J Cell Sci. 2016 Dec 1 129;(23)4424–4435. DOI:10.1242/jcs.192260
- Schink KO, Tan KW, Stenmark H. Phosphoinositides in Control of Membrane Dynamics. Annu Rev Cell Dev Biol. 2016 Oct 6;32(1):143–171. DOI:10.1146/annurev-cellbio-111315-125349
- Wallroth A, Haucke V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J Biol Chem. 2018 Feb 2;293(5):1526–1535. DOI:10.1074/jbc.R117.000629
- Li J, Zhang YV, Asghari Adib E, et al. Restraint of presynaptic protein levels by Wnd/DLK signaling mediates synaptic defects associated with the kinesin-3 motor Unc-104. Elife. 2017 Sep 19;6: DOI:10.7554/eLife.24271
- Venken KJ, Schulze KL, Haelterman NA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011 Sep;8(9):737–743. DOI:10.1038/nmeth.1662
- Adachi-Yamada T. Puckered-GAL4 driving in JNK-active cells. Genesis. 2002 Sep-Oct;34(1–2):19–22.
- Stone MC, Albertson RM, Chen L, et al. Dendrite injury triggers DLK-independent regeneration. Cell Rep. 2014 Jan 30 6;(2)247–253. DOI:10.1016/j.celrep.2013.12.022
- Zheng J, Sun J, Lu X, et al. BDNF promotes the axonal regrowth after sciatic nerve crush through intrinsic neuronal capability upregulation and distal portion protection. Neurosci Lett. 2016 May 16;621:1–8. DOI:10.1016/j.neulet.2016.04.006
- Kenney AM, Kocsis JD. Peripheral axotomy induces long-term c-Jun amino-terminal kinase-1 activation and activator protein-1 binding activity by c-Jun and junD in adult rat dorsal root ganglia in vivo. J Neurosci. 1998 Feb 15;18(4):1318–1328. DOI:10.1523/JNEUROSCI.18-04-01318.1998
- Lindwall C, Dahlin L, Lundborg G, et al. Inhibition of c-Jun phosphorylation reduces axonal outgrowth of adult rat nodose ganglia and dorsal root ganglia sensory neurons. Mol Cell Neurosci. 2004 Nov;27(3):267–279. DOI:10.1016/j.mcn.2004.07.001
- Mukhopadhyay A, Funato K, Stahl PD. Rab7 regulates transport from early to late endocytic compartments in Xenopus oocytes. J Biol Chem. 1997 May 16;272(20):13055–13059. DOI:10.1074/jbc.272.20.13055
- Rink J, Ghigo E, Kalaidzidis Y, et al. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005 Sep 9 122;(5)735–749. DOI:10.1016/j.cell.2005.06.043
- Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009 May 1;284(18):12110–12124. DOI:10.1074/jbc.M809277200
- Martin DD, Heit RJ, Yap MC, et al. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum Mol Genet. 2014 Jun 15 23;(12)3166–3179. DOI:10.1093/hmg/ddu027
- Engqvist-Goldstein AE, Warren RA, Kessels MM, et al. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001 Sep 17 154;(6)1209–1223. DOI:10.1083/jcb.200106089
- McGuire JR, Rong J, Li SH, et al. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006 Feb 10 281;(6)3552–3559. DOI:10.1074/jbc.M509806200
- Caviston JP, Ross JL, Antony SM, et al. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007 Jun 12 104;(24)10045–10050. DOI:10.1073/pnas.0610628104
- Pankiv S, Alemu EA, Brech A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010 Jan 25 188;(2)253–269. DOI:10.1083/jcb.200907015
- Johansson M, Lehto M, Tanhuanpää K, et al. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005 Dec;16(12):5480–5492. DOI:10.1091/mbc.e05-03-0189
- Ma X, Liu K, Li J, et al. A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning. J Biol Chem. 2018 Sep 7 293;(36)14155–14164. DOI:10.1074/jbc.RA118.001854
- McEwan DG, Popovic D, Gubas A, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015 Jan 8 57;(1)39–54. DOI:10.1016/j.molcel.2014.11.006
- Marwaha R, Arya SB, Jagga D, et al. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J Cell Biol. 2017 Apr 3 216;(4)1051–1070. DOI:10.1083/jcb.201607085
- Saxena S, Bucci C, Weis J, et al. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci. 2005 Nov 23 25;(47)10930–10940. DOI:10.1523/JNEUROSCI.2029-05.2005
- Saxena S, Howe CL, Cosgaya JM, et al. Differential endocytic sorting of p75ntr and TrkA in response to NGF: a role for late endosomes in TrkA trafficking. Mol Cell Neurosci. 2005 Mar;28(3):571–587. DOI:10.1016/j.mcn.2004.11.011
- BasuRay S, Mukherjee S, Romero E, et al. Rab7 mutants associated with Charcot-Marie-Tooth disease exhibit enhanced NGF-stimulated signaling. PLoS One. 2010 Dec 9 5;(12)e15351. DOI:10.1371/journal.pone.0015351
- Gauthier LR, Charrin BC, Borrell-Pagès M, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004 Jul 9 118;(1)127–138. DOI:10.1016/j.cell.2004.06.018
- Liot G, Zala D, Pla P, et al. Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J Neurosci. 2013 Apr 10 33;(15)6298–6309. DOI:10.1523/JNEUROSCI.2033-12.2013
- Ginés S, Bosch M, Marco S, et al. Reduced expression of the TrkB receptor in Huntington’s disease mouse models and in human brain. Eur J Neurosci. 2006 Feb;23(3):649–658. DOI:10.1111/j.1460-9568.2006.04590.x
- Ginés S, Paoletti P, Alberch J. Impaired TrkB-mediated ERK1/2 activation in huntington disease knock-in striatal cells involves reduced p52/p46 Shc expression. J Biol Chem. 2010 Jul 9;285(28):21537–21548. DOI:10.1074/jbc.M109.084202
- Hansen T, Thant C, White JA, et al. Excess active P13K rescues huntingtin-mediated neuronal cell death but has no effect on axonal transport defects. Apoptosis. 2019 Apr;24(3–4):341–358. DOI:10.1007/s10495-019-01520-4
- Fernandes KA, Harder JM, Fornarola LB, et al. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol Dis. 2012 May;46(2):393–401. DOI:10.1016/j.nbd.2012.02.003
- Hao Y, Waller TJ, Nye DM, et al. DegeneratiOn of injured axons and dendrites requires restraint of a protective JNK signaling pathway by the transmembrane protein raw. J Neurosci. 2019 Oct 23 39;(43)8457–8470. DOI:10.1523/JNEUROSCI.0016-19.2019
- Hammarlund M, Nix P, Hauth L, et al. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009 Feb 6 323;(5915)802–806. DOI:10.1126/science.1165527
- Yan D, Wu Z, Chisholm AD, et al. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009 Sep 4 138;(5)1005–1018. DOI:10.1016/j.cell.2009.06.023
- Hervera A, De Virgiliis F, Palmisano I, et al. Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat Cell Biol. 2018 Mar;20(3):307–319. DOI:10.1038/s41556-018-0039-x
- Cioni JM, Lin JQ, Holtermann AV, et al. LaTe endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019 Jan 10 176;(1–2)56–72.e15. DOI:10.1016/j.cell.2018.11.030
- Setoguchi T, Yone K, Matsuoka E, et al. Traumatic injury-induced BMP7 expression in the adult rat spinal cord. Brain Res. 2001 Dec 7 921;(1–2)219–225. DOI:10.1016/S0006-8993(01)03123-7
- Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001 Nov 8;32(3):389–401. DOI:10.1016/S0896-6273(01)00496-2
- Weiss KR, Kimura Y, Lee WC, et al. Huntingtin aggregation kinetics and their pathological roLe in a DROSOPHILA HUNTINGTON’S disease model. Genetics. 2012 Feb;190(2):581–600. DOI:10.1534/genetics.111.133710
- Zhang S, Binari R, Zhou R, et al. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010 Apr;184(4):1165–1179. DOI:10.1534/genetics.109.112516
- Stofanko M, Kwon SY, Badenhorst P. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics. 2008 Sep;180(1):253–267.
- Bellen HJ, Levis RW, Liao G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004 Jun;167(2):761–781. DOI:10.1534/genetics.104.026427
- Li BX, Satoh AK, Df R, et al. Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol. 2007 May 21;177(4):659–669. DOI:10.1083/jcb.200610157
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994 Jul 15;78(1):125–136. DOI:10.1016/0092-8674(94)90579-7
- Saxton WM, Hicks J, Goldstein LS, et al. Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell. 1991 Mar 22 64;(6)1093–1102. DOI:10.1016/0092-8674(91)90264-Y
- Gepner J, Li M, Ludmann S, et al. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996 Mar;142(3):865–878. DOI:10.1093/genetics/142.3.865
- Chance RK, Bashaw GJ. Slit-DepenDent endocytic trafficking of the robo receptor is required for son of sevenless recruitment and midline axon repulsion. PLoS Genet. 2015 Sep;11(9):e1005402.
- Pircs K, Nagy P, Varga A, et al. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 2012;7(8):e44214. DOI:10.1371/journal.pone.0044214
- Xia C, Rahman A, Yang Z, et al. Chromosomal localization reveals three kinesin heavy chain genes in mouse. Genomics. 1998 Sep 1 52;(2)209–213. DOI:10.1006/geno.1998.5427
- Reis GF, Yang G, Szpankowski L, et al. Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol Biol Cell. 2012 May;23(9):1700–1714. DOI:10.1091/mbc.e11-11-0938
- Hamoudi Z, Khuong TM, Cole T, et al. A fruit fly model for studying paclitaxel-induced peripheral neuropathy and hyperalgesia. F1000res. 2018;7:99.
- Kim M, Park HL, Park HW, et al. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy. 2013 Aug;9(8):1201–1213. DOI:10.4161/auto.24811
- Kim M, Semple I, Kim B, et al. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 2015;11(8):1358–1372. DOI:10.1080/15548627.2015.1063766
- Dolma K, Iacobucci GJ, Hong Zheng K, et al. Presenilin influences glycogen synthase kinase-3 β (GSK-3β) for kinesin-1 and dynein function during axonal transport. Hum Mol Genet. 2014 Mar 1 23;(5)1121–1133. DOI:10.1093/hmg/ddt505
- Banerjee R, Rudloff Z, Naylor C, et al. The presenilin loop region is essential for glycogen synthase kinase 3 β (GSK3β) mediated functions on motor proteins during axonal transport. Hum Mol Genet. 2018 Sep 1 27;(17)2986–3001. DOI:10.1093/hmg/ddy190
