Abstract
Background
Artificial intelligences (AIs) are emerging in the field of medical informatics in many areas. They are mostly used for diagnosis support in medical imaging but have potential uses in many other fields of medicine where large datasets are available.
Aim
To develop an artificial intelligence (AI) “ToxNet”, a machine-learning based computer-aided diagnosis (CADx) system, which aims to predict poisons based on patient’s symptoms and metadata from our Poison Control Center (PCC) data. To prove its accuracy and compare it against medical doctors (MDs).
Methods
The CADx system was developed and trained using data from 781,278 calls recorded in our PCC database from 2001 to 2019. All cases were mono-intoxications. Patient symptoms and meta-information (e.g., age group, sex, etiology, toxin point of entry, weekday, etc.) were provided. In the pilot phase, the AI was trained on 10 substances, the AI’s prediction was compared to naïve matching, literature matching, a multi-layer perceptron (MLP), and the graph attention network (GAT). The trained AI’s accuracy was then compared to 10 medical doctors in an individual and in an identical dataset. The dataset was then expanded to 28 substances and the predictions and comparisons repeated.
Results
In the pilot, the prediction performance in a set of 8995 patients with 10 substances was 0.66 ± 0.01 (F1 micro score). Our CADx system was significantly superior to naïve matching, literature matching, MLP, and GAT (p < 0.005). It outperformed our physicians experienced in clinical toxicology in the individual and identical dataset. In the extended dataset, our CADx system was able to predict the correct toxin in a set of 36,033 patients with 28 substances with an overall performance of 0.27 ± 0.01 (F1 micro score), also significantly superior to naïve matching, literature matching, MLP, and GAT. It also outperformed our MDs.
Conclusion
Our AI trained on a large PCC database works well for poison prediction in these experiments. With further research, it might become a valuable aid for physicians in predicting unknown substances and might be the first step into AI use in PCCs.
Background
Artificial intelligence (AI) refers to machines or software that process information and interact with the world as understanding beings [Citation1,Citation2]. In a medical context, they are mostly known for their proficiency in diagnosing radiology images or detecting heart dysrhythmias from wearables [Citation3]. However, due to recent improvements in computation and advancements in algorithms, they offer a myriad of new possibilities, especially datasets which are too vast for humans to deeply conceptualize. Deep learning and Knowledge representation structures offer an especially interesting aspect when it comes to large datasets [Citation1,Citation2,Citation4].
Intoxications are one of the most significant factors of global suffering and death. The abuse of ethanol alone accounted for 99.2 million DALYs (disability-adjusted life-years) in 2016 with an addition of 31.8 million DALYs from other drugs [Citation5]. In case of an intoxication, fast diagnosis and treatment are essential to prevent permanent organ damage or even death [Citation6]. Poison Control centers (PCCs) were established in most countries to provide quick diagnosis and give treatment recommendations to laymen or health professionals with medical doctors (MDs) trained in clinical toxicology staffing the PCCs. Most of the time, the substance responsible for the intoxication is known. However, when this is not the case, the MD working at the PCC must reach a diagnosis solely based on the reported symptoms, without seeing the patient face to face, and give treatment recommendations accordingly.
Current computer-aided diagnosis (CADx) systems in toxicology have not solved these problems so far. Most are rule-based expert systems that are very sensitive towards input variations [Citation7–9]. Chary et al. took the first step in AI technology in toxicology by creating an AI that predicts toxidromes based on clinical findings and showed that probabilistic logic networks can perform medical reasoning comparably to humans in a restricted domain [Citation10].
Aim
First, we wanted to establish our own AI model, called “ToxNet”, using Graph Convolutional Networks (GAT), a relatively new type of neural network model with the task of predicting a toxin from a PCC dataset [Citation11,Citation12]. The aim of the AI was to give an accurate diagnosis proposition based on patient data and symptoms with actual patient data available to the PCC. Therefore, we performed a pilot study testing it on a large dataset with a limited amount of toxin classes. Then we wanted to compare its accuracy with our MDs trained in clinical toxicology. If it was equal or superior to our MDs, we wanted to broaden the spectrum of the AI model to the most common toxins encountered in PCC consultations.
If all this was successful, we wanted to create an easy interface in the form of a web App. With the App, it should be possible to easily enter cases and calculate a poison prediction. Furthermore, it should be possible to import cases from our PCC database and calculate the prediction. The AI and web App are meant to be used by the clinicians working in the PCC to support them in their clinical decision-making.
Methods
This project was a collaboration between the Division of Clinical Toxicology and Poison Control Centre Munich, Department of Internal Medicine II, TUM School of Medicine, and Computer Aided Medical Procedures, TUM Department of Informatics, both Technical University of Munich. Ethical approval was obtained by our ethics commission (650/20 S-KH). Anonymized patient data was collected by the Division of Clinical Toxicology and Poison Control Centre Munich, the AI was programmed by the TUM Department of Informatic, and the web App was programmed as a student project.
The overall dataset included patient data from 1 January 2002 through 9 September 2020 with a total of 781,278 cases. All eligible cases were mono-intoxications. Patient symptoms, meta-information (e.g., age group, sex, etiology, toxin point of entry, weekday, etc.), and responsible toxin were provided by the PCC database. Laboratory or clinical examinations like ECGs were not included. All caller origins (hospitals, general practitioners, emergency medical services (EMS), members of the public, etc.) were included. The included substances were chosen from our region’s most relevant and common intoxications including medication and drugs of abuse [Citation13]. For the pilot study, the 10 most common substance classes with one representative substance from each class were chosen (e.g., citalopram for antidepressants of the SSRI type). For the extended cohort, 28 substance classes with all substances of that class were included (e.g., antidepressants of the SSRI type).
The AI uses two approaches: a literature matching branch and the graph convolutional network (GCN). For the literature matching branch, intoxication symptoms for each substance were extracted from IBM Micromedex® (Truven Health Analytics, IBM Watson Health Company, Ann Arbor, MI, USA) and our own poison control knowledge database. Using the GCN, we incorporated meta information and population context into the prediction process. The prediction was optimized using inductive graph attention networks (GAT). This parallel approach stabilizes the output and was called ToxNet. A sequential approach was called ToxNet(S). The technical details of the AI used for the pilot study have been published at the MICCAI conference 2020 [Citation14] ().
Figure 1. Schematic architecture of ToxNet. The symptom vectors are processed in the graph-based GAT layers and the literature-matching network in parallel. The upper branch is the literature branch, and the lower branch is the GAT branch. GAT: graph attention layer; FC: fully connected layer; GT: ground truth; S: symptom vector; G: graph containing symptoms and metadata; M: number of patients.
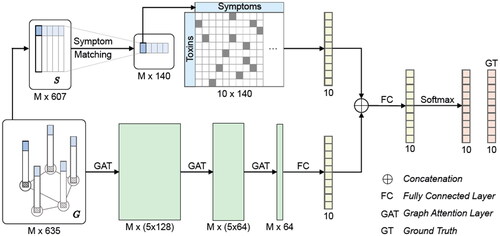
All experiments use a 10-fold cross-validation, where each fold contains 10% of the data as test data, the remaining 90% are further split into 80% training and 20% validation. The AI was tested against different benchmarks like naïve matching, a decision tree approach, and a multilayer perceptron (MLP), but also against different approaches within the AI: the literature matching and the GAT.
The MLP is a fully connected class of feedforward artificial neural network and consists of three layers of nodes, an input layer, in our case three hidden layers, and an output layer [Citation15]. Since MLPs are established as machine learning solutions, we used a standard MLP as a benchmark for our AI.
For the literature matching, a list of commonly occurring symptoms for every toxin class was created. These were encoded in a binary symptom vector for every poison. We designed a specific symptom-matching layer that learns a mapping of the patient symptom vectors to the literature symptoms. This concept results in an interpretable transfer function. With a second transformation, the resulting transformation maps the patient symptoms onto the poison classes with the explicit usage of literature information [Citation14].
ToxNet uses a GAT, which is a leading representative of a GCN. It is a novel neural network architecture that operates on graph-structured data, leveraging masked self-attentional layers to address the shortcomings of prior methods based on graph convolutions or their approximations [Citation16].
For each approach, the F1 micro and macro score were calculated and tested for superiority, p < 0.05 was considered significant. The F1 score was chosen as a measure of our model’s accuracy since the dataset is imbalanced. An F1 score of 1.0 is the best possible result, and 0.0 is the worst possible result. The Wilcoxon signed-rank test was used to test for significant differences within the F1 scores.
The results were validated against 10 MDs with different experiences in clinical toxicology (ranging from fully trained clinical toxicologists to MDs in their training in clinical toxicology after at least 6 months of training). Each MD classified 50 selected cases out of the overall dataset in the pilot study, where 25 cases within the dataset were unique and 25 cases were identical to other MD’s cases to test for intervariability (overall 275 cases, 250 cases in the individual set and 25 cases in the intervariability study). In the extended cohort, 10 MDs with different experiences in clinical toxicology received each 50 unique and 50 identical cases out of the overall dataset (overall 550 cases, 500 cases in the individual set, and 50 cases in the intervariability study). The MDs received the same cases as the AI which were extracted from our PCC and consisted of patient symptoms and meta-information (age group, sex, etiology, toxin point of entry, weekday, etc.). They then needed to select the most likely poison responsible for that individual case. The F1 scores of their performance was then measured and compared with the AI’s. All available data was the data at the time of the PCC contact, MDs had no access to the whole PCC case, only the refined data.
Results
Pilot study
We included 8995 cases with the following 10 different substances and substance classes in the pilot study [Citation14]: ramipril representing ACE-inhibitors/sartans, acetaminophen, citalopram representing antidepressants (SSRI), diazepam representing benzodiazepines, bisoprolol representing beta-blockers, amlodipine representing calcium channel antagonists (dihydropyridine type), cocaine, ethanol, ibuprofen representing NSAIDs (excluding acetyl salicylic acid (ASA) and acetaminophen) and heroin representing opiates (pure µ-agonists) ().
Table 1. Included substances in pilot and extended cohort with number of cases with respective F1 score of ToxNet.
Sixty-five percent of the calls originated from hospitals, 17% from members of the public, 12% from EMS, and 4% from general practitioners.
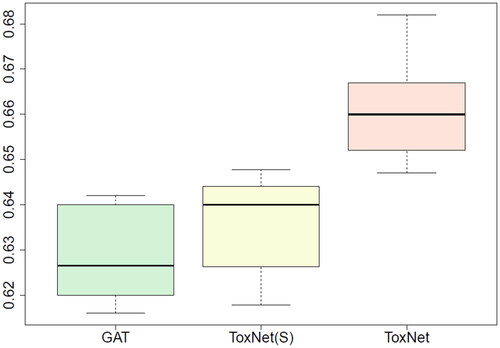
Our AI “ToxNet” was able to predict the correct toxin with a performance of 0.66 ± 0.01 (F1 micro score) and was significantly superior (all p < 0.005) to all benchmark methods: it was superior to naïve matching (0.20 ± 0.01 F1 micro score), the decision tree approach (0.25 ± 0.02) and the MLP (0.54 ± 0.02). It was also superior to its own components by themselves, the literature matching branch (0.47 ± 0.01) and the GAT alone (0.63 ± 0.01). The best performance was obtained using a parallel setting of ToxNet which is slightly superior to a sequential setting ().
Figure 2. Comparison of ToxNet in different settings compared with the GAT branch over 10-fold cross-validation in the pilot cohort, 8995 cases. GAT: graph attention network; ToxNet(S): ToxNet in sequential setting; ToxNet: ToxNet in parallel setting.
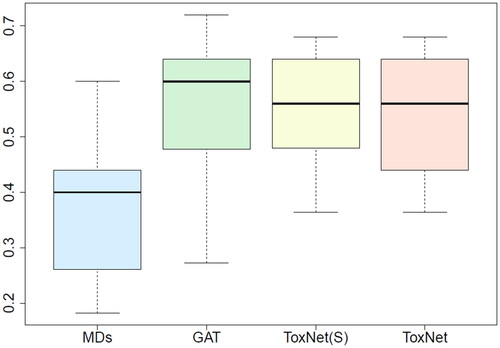
shows a box plot of the performance of the 10 MDs compared with ToxNet in different settings and the GAT alone. Each MD classified 25 cases in one out of ten individual sets, resulting in 250 rated cases in total. All three graph-based approaches [GAT, ToxNet(S), and ToxNet] rated the 250 cases and outperformed the MDs ().
Figure 3. Comparison of MDs’ performance with ToxNet over 10 different sets evaluated by one MD each set (25 cases per set) in the pilot cohort, overall 250 cases. MDs: medical doctors; GAT: graph attention network; ToxNet(S): ToxNet in sequential setting; ToxNet: ToxNet in parallel setting.
We also performed a detailed inter-variability study on the 25 cases evaluated by all doctors. Except for one case (Acetaminophen), every intoxication case correctly classified by most MDs was also correctly classified by ToxNet. Furthermore, for eight cases, where only half of the MDs or less correctly predicted the intoxication, our method still succeeded. Even compared to the two best MDs, who correctly classified 12 cases, our method overall resulted in 15 correct poison predictions. Six cases were wrongly classified by all doctors and our method [one ACE inhibitor, one antidepressant (SSRI), one calcium channel antagonist (dihydropyridine type), one cocaine, and two NSAID cases] ().
Figure 4. Clinician inter-variability and comparison with ToxNet. Poison classes are ordered alphabetically, each group separated with a white spacing in the pilot cohort, 25 cases.

In the pilot cohort, ToxNet was especially good at predicting ethanol (0.90 ± 0.01 F1 score) and opiates (0.91 ± 0.02) while it had difficulties with ACE-inhibitors (0.23 ± 0.03), acetaminophen (0.23 ± 0.03) and calcium channel antagonists (0.21 ± 0.01) ().
Extended cohort
For the extended cohort, the initial list was expanded to the whole substance class and the following further substance classes, resulting in 28 substance classes and 36,033 cases: ACE-inhibitors/sartans, acetaminophen, ASA, antidepressants (SSRI, SSNRI, and tricyclic), benzodiazepines, beta blockers, calcium channel antagonists (dihydropyridine type and class IV according to the Vaughan Williams classification), cocaine, cetirizine/loratadine, diphenhydramine/doxylamine/dimenhydrinate, gamma-hydroxybutyrate (GHB)/gamma-butyrolactone (GBL), ethanol, levetiracetam, lithium, methylphenidate, NSAIDs (excluding acetyl salicylic acid (ASA) and acetaminophen), neuroleptics (atypical and typical), opiates (pure µ-agonists and buprenorphine), pregabalin/gabapentin, stimulants, tetra-hydro cannabinol (THC), valproic acid and Z-substances ().
Sixty-eight percent of the calls originated from hospitals, 15% from EMS, 12% from members of the public, and 4% from general practitioners.
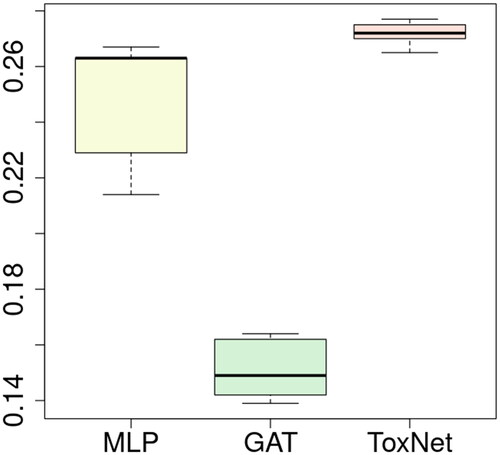
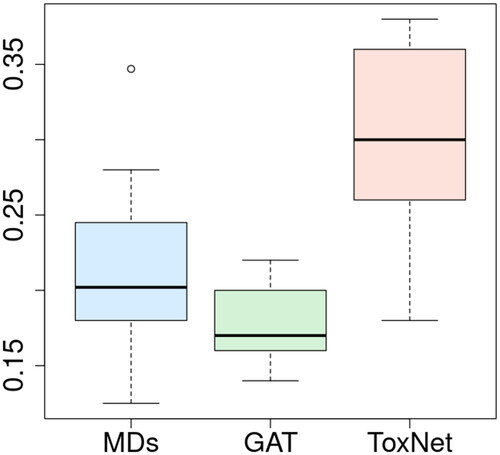
ToxNet was able to predict the correct toxin with an overall performance of 0.27 ± 0.01 (F1 micro score) and was significantly superior (all p < 0.005) to all benchmark methods: it was superior to naïve matching (0.05 F1 micro score) and the MLP (0.25 ± 0.02). It was also superior to its own components by themselves, the literature matching (0.21 ± 0.01), and the GAT alone (0.15 ± 0.01). Since the parallel setting of ToxNet was superior to the pilot setting, the sequential setting was not tested anymore. Notably, the GAT had a poor performance in this dataset ().
Figure 5. Comparison of ToxNet with MLP and the GAT branch over 10-fold cross-validation in the extended cohort, 36,033 cases. MLP: multi-layer perceptron; GAT: graph attention network; ToxNet: ToxNet in parallel setting.
shows a box plot of the performance of the 10 MDs compared with ToxNet and the GAT alone. Here, each MD classified 50 cases in one out of ten individual sets, resulting in 500 rated cases in total. ToxNet still outperforms the MDs, the GAT alone had a poorer performance ().
Figure 6. Comparison of MDs’ performance with ToxNet over 10 different sets evaluated by one MD each set (50 cases per set) in the extended cohort, overall 500 cases. MDs: medical doctors; GAT: graph attention network; ToxNet: ToxNet in parallel setting.
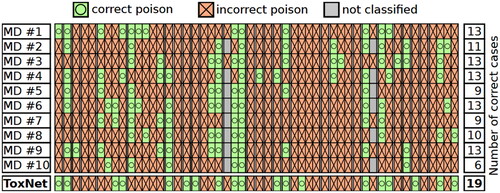
We repeated the detailed inter-variability study on 50 cases evaluated by all doctors. Except for one case (tricyclic antidepressant), every intoxication case correctly classified by most MDs was also correctly classified by ToxNet. For 12 cases, where only half of the MDs or less correctly predicted the intoxication, our method still succeeded. Compared to the five best-performing MDs, who correctly classified 13 cases, our method overall resulted in 19 correct poison predictions. 15 cases were wrongly classified by all doctors and our method [one antidepressant (SSRI), one antidepressant (tricyclic), one acetylic salicylic acid, one beta blocker, one Cetirizine/Loratadine, one cocaine, one Diphenhydramine/Doxylamine/Dimenhydrinate, one GHB/GBL, three Methylphenidate, one neuroleptic (atypical), one neuroleptic (typical), one NSAID, one opiate (Buprenorphine), one Valproic acid, and one Z-substance case] ().
Figure 7. Clinician inter-variability and comparison with ToxNet. Poison classes are ordered alphabetically, each group separated with a white spacing in the extended cohort, 50 cases.
In the extended cohort, ToxNet was especially good at predicting ethanol (0.70 ± 0.02 F1 score), opiates (0.48 ± 0.02), THC (0.48 ± 0.04), lithium (0.45 ± 0.05), GHB (0.45 ± 0.03), and stimulants (0.44 ± 0.03). It had difficulties with substances that appeared rarely like class IV calcium-channel blockers (0.07 ± 0.03), cetirizine/loratadine (0.07 ± 0.01), levetiracetam (0.01 ± 0.01), valproic acid (0.01 ± 0.02), and buprenorphine (0.05 ± 0.00). Substances with unspecific symptoms like NSAIDS (0.05 ± 0.01) and Pregabalin/Gabapentin (0.04 ± 0.01) were also hard to predict ().
Discussion
While AI models might play an important role in radiology, pathology, ophthalmology, and dermatology, and while some applications have already been shown in those fields, the use of an AI in a PCC context has not been shown so far [Citation17]. In our study, we demonstrated that AIs might become a potentially helpful tool in diagnosing intoxications from a large dataset in the future when presented with a specific task.
All PCCs have their unique dataset with different regional intoxications and AIs are especially good at analyzing meta-data and large datasets. Our AI could well be trained on another PCC and give region-specific results. AIs also have the advantage over new MDs, that the AI’s training has been completed before it is put into use. Therefore, AIs are capable to produce consistent results from the start. However, AIs don’t critically question cases and might miss important details, the input of the data still relies on the MDs and the history they take—critical interpretation of the results is also necessary.
Chary et al. created an AI predicting toxidromes based on clinical findings [Citation10]. This approach has the advantage that the AIs reasoning is understandable for users, however, it only predicts toxidromes. With our approach, we wanted to be able to give an accurate diagnosis proposition, since toxidromes might be helpful, but are not specific to one toxic etiology [Citation18]. We also based our AI on real PCC patient data and not artificial synthetic cases to get a representation of cases that is as realistic as possible. One of the main issues will probably be for MDs to use results created by an AI without knowing how the result was created [Citation19]. One advantage of ToxNet is that it uses the GAT technology as well as the literature-based approach. Especially in the extended cohort, the GAT approach alone did not perform well, but the AI’s overall results were stabilized by the literature branch.
In our dataset, we also included calls from origins other than hospitals, like EMS, general practitioners, or members of the public. This might influence the accuracy of the patient history or symptoms: However, our aim was to create a dataset with as much data as possible and as close to reality as possible. On the other hand, we expected that our PCC staff would ask sufficiently detailed questions to get a good description of a patient’s symptom even from a medical layperson.
The AI had rather solid results; however, it also misclassified many poisonings. One issue was the quality of documentation and history taking, another issue was the time of presentation. Often, only the main symptom was reported to the PCC like, e.g., drowsiness, since no structured medical examination was asked on the phone. Other times, the patient presented before developing symptoms. This also made the prediction for the MDs so difficult. Furthermore, in the extended cohort, some substance classes have overlapping syndromes or toxidromes, like antidepressants of the different types, benzodiazepines, GHB/GBL, and Z-substance. Twenty-eight substance classes are also about the limit of substances this experimental setup can identify. With a larger amount of substance classes, other ranking methods would be needed, this could be looked at in other studies.
For this study, we also only included mono-intoxications to get a more distinct differentiation between patient symptoms. In real life, most intoxications are mixed ingestions—which our AI cannot predict currently. Therefore, the proposed diagnosis of the AI should only be used as a suggestion and still needs critical interpretation by the PCC physician. Since the AI is trained on a PCC database, it would also only be useful for the PCC it has been trained for and not for other users like hospital physicians, EMS, or members of the public. Since poison prediction is only a small part of the daily PCC consultations, our AI will not remove the need for PCCs—however, AIs could be a vital part of a PCC’s IT infrastructure in the future.
The main effort in programming and training the AI is data collection, preparation, and modelling of the algorithm. One big effort for medical toxicology research is data preparation out of medical records [Citation20]. The advantage of PCC documentation is that this documentation is already structured in most cases. However, there are cases with insufficient documentation quality (e.g., only a single generic symptom) which indicate intrinsic challenges from medical data which make it hard to predict a toxin. A reduced level of consciousness was often one of the only symptoms.
Translating the algorithm into an easy-to-use web App is another small, but important, step. If this is done well, it will increase the acceptance in daily use. After finishing the pilot study, a web App was developed by informatics students in our collaboration. It provides a browser-based interface within our hospital intranet. Here, MDs can enter new cases and predict the intoxication. They can also export a case from our PCC database, import that files into the App, and start the prediction algorithm. The web App also shows disclaimers that results are “ongoing scientific research”. ToxNet may well add to the many upcoming Applications of AI in healthcare, although it is far from productive use [Citation17]. Importantly, the regular usage of the web App leads to better and more structured documentation of cases, and increases the labeled data pool gradually over time, e.g., for prospective studies.
While these technologies hold promise for increasing productivity and improving outcomes in the future, it must be remembered that they, just like their human creators, are not infallible [Citation17]. AI’s perform specific tasks, for which they are programmed but their results should not be blindly adopted. Therefore, it is necessary to evaluate and implement AIs with a critical eye.
Limitation
Our study has several limitations. Firstly, it was a pilot study on retrospective cases and focusses on mono-intoxications. Therefore, documentation quality for some cases was poor and toxin prediction was performed on limited data. This might also explain the performance of our MDs. Secondly, the toxin which was considered the “true” toxin was the one reported by the caller without later confirmation. Furthermore, all available symptoms were recorded, even though they might not have been caused by the intoxication. Finally, ToxNet gives no reasoning for its decision which could make it hard for physicians to believe the AI. To increase trust, future work could incorporate methods for explainable AI (XAI) into our ToxNet model and web app system [Citation21,Citation22].
Conclusion
The AI in our experiment, trained on a large PCC database, produces relatively accurate and stable results, especially in the pilot cohort with 10 substance classes. In this experimental context, its results and prediction performance are superior to our MDs’. It also works on an increased number of 28 substance classes, although it loses its prediction performance when expanding the number of substance classes. It performs especially well in common intoxications but has difficulty with rare substances or substances with unspecific symptoms.
It, therefore, needs to be seen as the first step into AI use in PCCs. Further research is needed until a functioning AI might come into full use which could support physicians in the future for the very specific task of predicting an unknown substance.
Ethical approval
Ethical approval was obtained by our ethics commission (650/20 S-KH).
References
- Chary MA, Manini AF, Boyer EW, et al. The role and promise of artificial intelligence in medical toxicology. J Med Toxicol. 2020;16(4):458–464.
- Malik P, Pathania M, Rathaur VK. Overview of artificial intelligence in medicine. J Fam Med Prim Care. 2019;8(7):2328–2331.
- European Society of Radiology. Impact of artificial intelligence on radiology: a EuroAIM survey among members of the European Society of Radiology. Insights Imaging. 2019;10(1):105.
- Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–1358.
- GBD 2016 Alcohol and Drug Use Collaborators. The global burden of disease attributable to alcohol and drug use in 195 countries and territories, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Psychiatry. 2018;5(12):987–1012.
- Kulling P, Persson H. Role of the intensive care unit in the management of the poisoned patient. Med Toxicol. 1986;1(5):375–386.
- Batista-Navarro RTB, Bandojo DA, Gatapia MAJK, et al. ESP: an expert system for poisoning diagnosis and management. Inform Health Soc Care. 2010;35(2):53–63.
- Long JB, Zhang Y, Busic V, et al. Antidote application. Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. ACM Press; 2017. p. 442–448.
- Darmoni SJ, Massari P, Droy JM, et al. SETH: an expert system for the management on acute drug poisoning in adults. Comput Methods Programs Biomed. 1994;43(3–4):171–176.
- Chary M, Boyer EW, Burns MM. Diagnosis of acute poisoning using explainable artificial intelligence. Comput Biol Med. 2021;134:104469.
- Kipf TN, Welling M. Semi-supervised classifcation with graph convolutional networks. 5th International Conference on Learning Representations, ICLR 2017 – Conference Track Proceedings International Conference on Learning Representations. ICLR; 2016.
- Defferrard M, Bresson X, Vandergheynst P. Convolutional neural networks on graphs with fast localized spectral filtering. Adv Neural Inf Process Syst. 2016;29:3844–3852.
- Tschirdewahn J, Eyer F. [Diagnostics and treatment of selected clinically relevant, acute drug intoxications]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2019;62(11):1313–1323.
- Burwinkel H, Keicher M, Bani-Harouni D, et al. Decision support for intoxication prediction using graph convolutional networks. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020: Springer Link; 2020. p. 633–642.
- [cited 2022 Oct]. Available from: https://en.wikipedia.org/wiki/Multilayer_perceptron
- Veličković P, Cucurull G, Casanova A, et al. Graph attention networks; 2017 [cited 2017 Oct 1]. Available from: https://ui.adsabs.harvard.edu/abs/2017arXiv171010903V
- He J, Baxter SL, Xu J, et al. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25(1):30–36.
- Mégarbane B. Toxidrome-based approach to common poisonings. Asia Pac J Med Toxicol. 2014;3(1):2–12.
- Naylor CD. On the prospects for a (deep) learning health care system. JAMA. 2018;320(11):1099–1100.
- Larsen J, Mycyk MB, Thompson TM. Reviewing the record: medical record reviews for medical toxicology research. J Med Toxicol. 2018;14(3):179–181.
- Ying R, Bourgeois D, You J, et al. GNNExplainer: generating explanations for graph neural networks. Adv Neural Inf Process Syst. 2019;32:9240–9251.
- Tjoa E, Guan C. A survey on explainable artificial intelligence (XAI): toward medical XAI. IEEE Trans Neural Netw Learn Syst. 2021;32(11):4793–4813.
