 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Observations over the last decade suggest that some RNA transcripts, such as non-coding RNAs, function in regulating the transcriptional and epigenetic state of gene expression. DNA methylation appears to be operative in non-coding RNA regulation of gene expression. Interestingly, methylated cytosines undergo deamination to remove the methylation, which if not properly repaired results in the methylated cytosine being recognized by the cell as a thymine. This observation suggests that the process of non-coding RNA-directed epigenetic targeting also has the potential to alter the genomic landscape of the cell by changing cytosines to thymines and ultimately influence the evolution of the cell. This proposed theory of “RNA-mediated gene evolution” might be one possible mechanism of action whereby RNA participates in the natural selective process to drive cellular and possibly organismal evolution.
RNA Functions as Driver of Epigenetic and Transcriptional States
Sequencing of the human genome was one of the greatest milestones in science. Almost immediately following this monumental achievement it became apparent that the vast majority of the genome—roughly 80%—is not involved in generating proteins but rather in producing non-coding transcripts.Citation1-4 These observations have proven perplexing, because it had previously been assumed that the vast majority of the human genome was functionally inert. For years, various notions have been put forth as to the function of these non-protein-coding regions in the human genome; ranging from acting as genomic spacers involved in partitioning genes to structural entities involved in chromosomal integrity and folding. The rationale for these interpretations was the presumption that the majority of this “junk DNA” was not transcriptionally active.Citation5-7 This viewpoint, however, has been radically altered as a result of exciting findings generated from the FantomCitation8 and Encode consortiums,Citation3,4 whereby a paradigm shift has occurred, suggesting that this genomic dark matter—the non-coding or “junk DNA”—is actually transcribed and active.
While it is now evident that the vast majority of the human genome is transcribed into both coding and non-coding transcripts,Citation4,7,8 the function of these long non-coding RNAs (lncRNAs) has until recently remained largely unknown. Studies carried out over the last decade have demonstrated that many non-coding RNAs are functional in the regulation of gene transcription (reviewed inCitation9,10). Mechanistic studies in human cells with both small and long forms of non-coding RNAs have found that these transcripts interact with the proteins involved in epigenetic cellular processes of regulating chromatin states and gene accessibility ().
Figure 1. Non-coding RNA-directed transcriptional silencing. (A and B) Small non-coding RNAs have been observed to target epigenetic silencing complexes to actively transcribed loci, resulting in stable transcriptional gene silencing (reviewed inCitation10,39). This form of silencing involves the action of DNMT3a interacting directly with the small non-coding RNAsCitation20,21,44,45 and directing both histone and DNA methylation to the homology containing loci.Citation17,22 This targeting can ultimately result in stable and heritable epigenetically silent loci.Citation17 (C and D) Long non-coding RNAs that are antisense to particular loci have also been found to interact with DNMT3aCitation19 and modulate transcription.Citation19,46-48
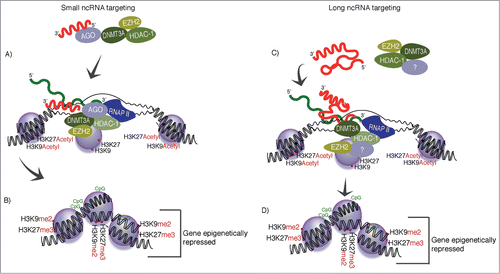
Epigenetics is the study of heritable changes in gene expression caused by mechanisms other than changes in genomic DNA, and focuses predominantly on changes to the chromatin. Chromatin is composed of DNA and proteins, and epigenetics is the modifications to these elements. Changes to the local chromatin architecture can drastically affect the accessibility of a gene to outside influences. Examples abound of epigenetic changes that range from histone modifications to DNA methylation, both of which can result in transcriptional silencing of a particular locus (reviewed inCitation11).
Fascinating studies have found that some phenotypic traits can be passed on to offspring solely by the action of epigenetic changes at particular genetic loci. These observations, termed transgenerational epigenetic inheritance, suggest that our understanding of evolution is incomplete, and that some level of epigenetic heritability is also functional in evolution. Studies carried out in nematode, mouse, and pig model systems all suggest that transgenerational epigenetic inheritance involves non-coding RNAs.Citation12-14 There is also evidence for this phenomenon in humans, suggesting that some level of epigenetic inheritance is operative in the fabric of our existence.Citation15,16 Furthermore, work carried out in human cell culture systems has found that small non-coding RNAs direct epigenetic changes that can be passed on to daughter cells in a long-term heritable manner.Citation17 Collectively, these observations suggest that non-coding RNAs emanating from essentially “junk” DNA, including pseudogenes and natural antisense transcripts (NATs)(reviewed inCitation18), may actually be involved in not only controlling transcription but also in the natural selection of the genome and the cell via transgenerational epigenetic inheritance. Indeed, we are only now realizing how intricately involved both small and long non-coding RNAs are with regards to controlling gene expression in human cells.Citation7
The Theory of “RNA Mediated Gene Evolution”
Based on the emerging realization that some non-coding RNAs are active modulators involved in controlling the transcriptional and epigenetic processes, and that the results of this process appear to be heritable and passed on to offspring, I theorize that an “RNA-mediated form of gene evolution” is endogenous to human cells. This evolutionary process plays out via perturbations to the endogenous non-coding RNA, DNA methylation or DNA deamination pathways. The purported mechanism of action driving this process in human cells is shown schematically (). The notion presented here is that non-coding RNA-associated recruitment of DNA methyltransferase 3a (DNMT3a) and Enhancer of Zeste 2 (EZH2) to targeted loci can result in those cytosines at the target locus undergoing DNA methylationCitation17,19-23 (). Typically, such methylated cytosines (C) are subjected to the endogenous removal and repair process of deamination, whereby APOBEC3A deaminates the methylated cytosine to a thymine (T),Citation24-26 which is subsequently followed by repair with thymine DNA glycosylase (TDG)Citation27 (reviewed inCitation28)(). However, if this process is perturbed by some selective condition, and the deaminated nucleotide not properly repaired, then some methylated cytosines will undergo a hydrolysis reaction resulting in the production of ammonia and the retention of the converted methylated cytosine as a thymine in the DNA sequence. Following DNA replication the cell will recognize this deaminated cytosine as a thymine, which will become fixed in one of the 2 daughter strandsCitation29,30 (). While this cytosine to thymine (CT) conversion has been considered by some to be random, the spontaneous deamination of methylated cytosines has been found to be ∼2-fold faster than non-methylated cytosines,Citation30 suggesting a bias toward cytosine methylation at CpG regions in the deamination process. In light of these eventualities I envision a cellular process involving RNA and in particular non-coding transcripts that under certain conditions—such as increased lncRNA expression or decreased deamination repair—result in the alteration of the nucleotide content at a particular locus (shown schematically ). Note, this entire process involves both RNA and proteins; the RNAs providing target recognition capabilities and the protein components providing the epigenetic modifying and DNA repair components ().
Figure 2. Non-coding RNA-directed gene editing. (A) Antisense long non-coding RNAs function in transCitation19 (and possibly also in cis—not shown) to target epigenetic regulatory complexes to homology-containing loci. (B) This targeting has been found to require DNMT3a and possibly several other protein components known to interact directly with DNMT3a or be involved in epigenetic silencing. (C) The epigenetic silencing is carried out resulting in (D) histone 3 lysine 9 di-methylation (H3K9me2) or histone 3 lysine 27 trimethylation (H3K27me3) and DNA methylation at the homology-containing target loci, chromatin compaction, and, ultimately, gene silencing (reviewed inCitation18,49). (E) The methylated cytosines at particular CpG sites of the RNA-targeted locus undergoes deamination by the actions of APOBEC3A and (F and G) a thymine becomes incorporated in place of the methylated cytosine. (H and I) This thymine is typically repaired by TDG, MBD4, and SMUG, but (J) if this process is disrupted and/or replication occurs before repair of the locus is completed then the thymine may become fixed in the DNA such that (K) those transcripts emanating from the affected locus are altered and the cytosine becomes replaced with a thymine. These changes might inhibit future trans non-coding RNA targeting as well as affect the translated product or promoter activity and/or transcription factor binding at the RNA-targeted deaminated locus.
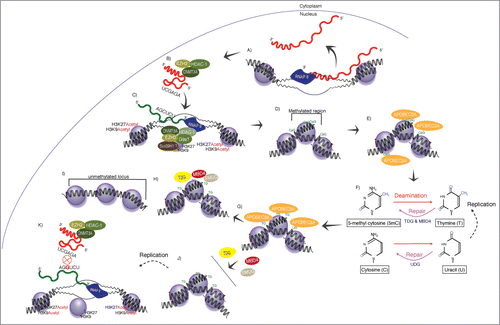
Conditions for such occurrences in the genome could be: (1) the result of robust non-coding RNA targeting of a particular locus leading to increased or overt DNA methylation or; (2) when DNA replication occurs without proper repair to epigenetically modified nucleotides. Such conditions could lead to some methylated cytosines undergoing accidental deamination resulting in stable CT transitions in the genomic code. The result of such C
T transitions are essentially gene editing at non-coding RNA targeted sites (). This mechanism of action is perhaps an inherent aspect of using DNA cytosine methylation to regulate gene expression. Indeed, the methylation of cytosine in CpG residues is recognized as an important avenue of epigenetic regulation,Citation31 cytosine to thymine changes represent the most common single nucleotide mutation found;Citation32,33 furthermore, this mutation is also found to be involved in the evolution of transcription factor binding sites.Citation34 With regard to human cells, it is difficult to resist the temptation to speculate that the methylation of CpG residues acts as a selective force driving the evolution of the genome and also in human disease.Citation33 While such a pathway () is surmised here to exist, there is limited direct experimental validation for this notion that RNA acts to shape the genomic content of the genome. However, there are several observations over the last decade that suggest that non-coding RNAs function as mediators of selection involved in the evolution of the cell, and there are clear experimental avenues to testing this theory (see below).
Experimental Validation of the Theory of “RNA-Mediated Gene Evolution”
Experimental observations in human cells suggest that methylated cytosines are repaired by the cell using a process of deamination, where APOBEC3A deaminates the methylated cytosine to a thymine,Citation24-26 which is subsequently followed by repair with TDG. Other glycosylase proteins may also be involved in this repair pathway, including methyl-CpG (mCpG) binding domain protein 4 (MBD4),Citation35,36 SMUG,Citation37 and uracil DNA glycosylase (UDG),Citation27 which can repair those unmodified cytosines that have been deaminated by APOBEC3A to a uracil (reviewed inCitation38)(). It is noteworthy that UDG has also been observed to interact directly with HDAC-1, which interestingly appears to be a required component in non-coding RNA directed epigenetic regulation (reviewed inCitation10,39)(). This observation also suggests an intersection between both pathways of interest: RNA-directed epigenetic gene silencing and repair of deaminated cytosines.
Collectively, data generated over the last decade suggest that: (1) non-coding RNAs direct epigenetic gene silencing and DNA methylation to those CpGs in the RNA targeted locusCitation17,22,40 (); (2) methylated cytosines that do not undergo proper deamination and repair can become incorporated into the genome as thymines (); and (3) these 2 processes have overlapping protein components, namely DNMT3a and HDAC-1. This overlap in proteins provides for a starting point to experimentally validate the theory of RNA-directed gene evolution. One experimental approach is to use a stable and inducible cell lines expressing smallCitation17 or long non-coding RNACitation19 to induce RNA-targeted epigenetic silencing (as shown in ) in the absence of the endogenous deamination repair pathway (). This works by suppressing APOBEC3A, TDG, SMUG, UDG, and MBD4 via RNA interference or CRISPR/Cas9.Citation41 These cultures could then be assessed for the genesis of CT transitions using either a surveyor assay,Citation42 a quantitative RT-PCR small nucleotide polymorphism assay, or deep-sequencing of the RNA-targeted locus in the various cultures.
Conclusions
Although the notions presented here have yet to be experimentally substantiated, one can envision a model whereby RNA-driven genetic variation, followed by selection, could work (). One possible area in which such an evolutionary mechanism could be operative is in the evolution of transcription factor binding sites.Citation34 Cytosine-to-thymine transitions seem to be involved in the evolution of transcription factor binding sites; moreover, promoter elements—where transcription factor binding sites are ubiquitous—are often bona fide targets for antisense lncRNAs (Citation19 and reviewed inCitation43).
Paradoxically, the ability of this system () to constitutively target and specifically control a locus is limited. For instance, the greater frequency of cytosine to thymine changes occurring at a particular RNA-targeted locus could result in an overall loss of sequence complementarity between the effector RNA and its targeted locus-specific transcript (). This eventuality could lead to a loss in the capability of the RNA to target a particular locus, while simultaneously permitting the targeted transcript to fold into a different conformation. As such, changes in the non-coding RNA transcript might lead to a loss of its targeting abilities and/or its susceptibility to different interactions with other lncRNAs and/or proteins (). Such a notion would play out with trans targeted RNAs while a different scenario might play out in the cis functional RNAs, possibly resulting in more stabilized RNA:RNA interacting regions of the genome. Indeed, the possibilities become infinitely complex, an eventuality that may be advantageous in the evolution of biological systems. No doubt as data emerge in this exciting area of research, so will additional layers of regulation of the central dogma of molecular biology and our understanding of the role of RNA in basic evolutionary processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This project is funded by 1P01AI099783–01 to KVM and Australian Research Council Future Fellowship FT130100572.
References
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr., Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 2012; 22:1646-57; PMID:22955977; http://dx.doi.org/10.1101/gr.134767.111
- St Laurent G, 3rd, Shtokalo D, Tackett M, Yang Z, Eremina T, Wahlestedt C, Inchima SU, Seilheimer B, McCaffrey TA, Kapranov P. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics 2012; 13:504; PMID:23006825; http://dx.doi.org/10.1186/1471-2164-13-504
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome browser: update 2012. Nucleic Acids Res 2012; 40:D912-7; PMID:22075998; http://dx.doi.org/10.1093/nar/gkr1012
- Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012; 337:1159, 61; PMID:22955811; http://dx.doi.org/10.1126/science.337.6099.1159
- Ohno S. So much "junk" DNA in our genome. Brookhaven Sym Biol 1972; 23:366-70; PMID:5065367
- Ohno S, Yomo T. The grammatical rule for all DNA: junk and coding sequences. Electrophoresis 1991; 12:103-8; PMID:2040257; http://dx.doi.org/10.1002/elps.1150120203
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet 2014; 15:423-37; PMID:24776770; http://dx.doi.org/10.1038/nrg3722
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science 2005; 309:1559-63; PMID:16141072; http://dx.doi.org/10.1126/science.1112014
- Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther 2010; 12:214-22; PMID:20373265
- Turner AM, Morris KV. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques 2010; 48:ix-xvi; PMID:20569216; http://dx.doi.org/10.2144/000113442
- Allis CD, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2007.
- Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 2011; 147:1248-56; PMID:22119442; http://dx.doi.org/10.1016/j.cell.2011.10.042
- Braunschweig M, Jagannathan V, Gutzwiller A, Bee G. Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS ONE 2012; 7:e30583; PMID:22359544; http://dx.doi.org/10.1371/journal.pone.0030583
- Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry 2010; 68:408-15; PMID:20673872; http://dx.doi.org/10.1016/j.biopsych.2010.05.036
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 2008; 105:17046-9; PMID:18955703; http://dx.doi.org/10.1073/pnas.0806560105
- Barry G. Lamarckian evolution explains human brain evolution and psychiatric disorders. Front Neurosci 2013; 7:224; PMID:24324395; http://dx.doi.org/10.3389/fnins.2013.00224
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res 2009; 37:2984-95; PMID:19304753; http://dx.doi.org/10.1093/nar/gkp127
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 2009; 4:296-301; PMID:19633414; http://dx.doi.org/10.4161/epi.4.5.9282
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 2013; 20:440-6; PMID:23435381; http://dx.doi.org/10.1038/nsmb.2516
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 2006; 12:256-62; PMID:16373483; http://dx.doi.org/10.1261/rna.2235106
- Ross JP, Suetake I, Tajima S, Molloy PL. Recombinant mammalian DNA methyltransferase activity on model transcriptional gene silencing short RNA-DNA heteroduplex substrates. Biochem J 2010; 432:323-32; PMID:20846120; http://dx.doi.org/10.1042/BJ20100579
- Turner AM, De La Cruz J, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol Ther 2009; 17:360-8; PMID:19066594; http://dx.doi.org/10.1038/mt.2008.268
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439:871-4; PMID:16357870; http://dx.doi.org/10.1038/nature04431
- Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res 2012; 40:9206-17; PMID:22798497; http://dx.doi.org/10.1093/nar/gks685
- Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem 2012; 287:34801-8; PMID:22896697; http://dx.doi.org/10.1074/jbc.M112.385161
- Suspene R, Aynaud MM, Vartanian JP, Wain-Hobson S. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PLoS ONE 2013; 8:e63461; PMID:23840298; http://dx.doi.org/10.1371/journal.pone.0063461
- Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res 2012; 40:10203-14; PMID:22962365; http://dx.doi.org/10.1093/nar/gks845
- Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma 2012; 121:1-20; PMID:22048164; http://dx.doi.org/10.1007/s00412-011-0347-4
- Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res 1999; 437:11-20; PMID:10425387; http://dx.doi.org/10.1016/S1383-5742(99)00065-4
- Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res 1994; 22:972-6; PMID:8152929; http://dx.doi.org/10.1093/nar/22.6.972
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6-21; PMID:11782440; http://dx.doi.org/10.1101/gad.947102
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem 1996; 271:12767-74; PMID:8662714; http://dx.doi.org/10.1074/jbc.271.48.30986
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature 2013; 500:415-21; PMID:23945592; http://dx.doi.org/10.1038/nature12477
- Zemojtel T, Kielbasa SM, Arndt PF, Behrens S, Bourque G, Vingron M. CpG deamination creates transcription factor-binding sites with high efficiency. Genome biology and evolution 2011; 3:1304-11; PMID:22016335; http://dx.doi.org/10.1093/gbe/evr107
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012; 40:4841-9; PMID:22362737; http://dx.doi.org/10.1093/nar/gks155
- Kondo E, Gu Z, Horii A, Fukushige S. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol Cell Biol 2005; 25:4388-96; PMID:15899845; http://dx.doi.org/10.1128/MCB.25.11.4388-4396.2005
- Kemmerich K, Dingler FA, Rada C, Neuberger MS. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res 2012; 40:6016-25; PMID:22447450; http://dx.doi.org/10.1093/nar/gks259
- Budworth H, McMurray CT. Bidirectional transcription of trinucleotide repeats: roles for excision repair. DNA repair 2013; 12:672-84; PMID:23669397; http://dx.doi.org/10.1016/j.dnarep.2013.04.019
- Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides 2009; 19:299-306.PMID:19943804; http://dx.doi.org/10.1089/oli.2009.0212
- Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum Gene Ther 2012; 23:473-83; PMID:22122263; http://dx.doi.org/10.1089/hum.2011.165
- Weinberg MS, Morris KV. A new world order: tailored gene targeting and regulation using CRISPR. Mol Ther 2014; 22:893; PMID:24787973; http://dx.doi.org/10.1038/mt.2014.54
- Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques 2004; 36:702-7; PMID:15088388
- Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long noncoding RNAs; sequence, structure, function. Biochim Biophys Acta 2013; PMID:24184936; http://dx.doi.org/10.1016/j.bbagen.2013.10.035
- Holz-Schietinger C, Reich NO. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res 2012; 40:8550-7; PMID:22730298; http://dx.doi.org/10.1093/nar/gks537
- Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem 2004; 279:49479-87; PMID:15342650; http://dx.doi.org/10.1074/jbc.M409070200
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008; 451:202-6; PMID:18185590; http://dx.doi.org/10.1038/nature06468
- Hawkins PG, Morris KV. Transcriptional regulation of oct4 by a long non-coding RNA antisense to oct4-pseudogene 5. Transcription 2010; 1:165-75; PMID:21151833; http://dx.doi.org/10.4161/trns.1.3.13332
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 2008; 4:e1000258; PMID:19008947; http://dx.doi.org/10.1371/journal.pgen.1000258
- Morris KV. Non-coding RNAs, epigenetic memory, and the passage of information to progeny. RNA Biol 2009; 6:242-7; PMID:19305164; http://dx.doi.org/10.4161/rna.6.3.8353
