Abstract
Preterm birth is a leading cause of infant mortality and can lead to poor life-long health and adverse neurodevelopmental outcomes. The pathophysiologic mechanisms that precede preterm labor remain elusive, and the role that epigenetic phenomena play is largely unstudied. The objective of this study was to assess the association between microRNA (miRNA) expression levels in cervical cells obtained from swabs collected during pregnancy and the length of gestation. We analyzed cervical samples obtained between 16 and 19 weeks of gestation from 53 women in a prospective cohort from Mexico City, and followed them until delivery. Cervical miRNA was extracted and expression was quantified using the NanoString nCounter Analysis System. Linear regression models were used to examine the association between miRNA expression levels and gestational age at delivery, adjusted for maternal age, education, parity, body mass index, smoke exposure, and inflammation assessed on a Papanicolaou smear. We identified 6 miRNAs that were significantly associated with gestational age at the time of delivery, including miR-21, 30e, 142, 148b, 29b, and 223. Notably, per each doubling in miR-21 expression, gestations were 0.9 (95% CI: 0.2–1.5) days shorter on average (P = 0.009). Per each doubling in miR-30e, 142, 148b, 29b, and 223 expression, gestations were shorter by 1.0 to 1.6 days. The predicted targets of the miRNAs were enriched for molecules involved in DNA replication and inflammatory processes. The levels of specific miRNAs in the human cervix during pregnancy are predictive of gestational age at delivery, and should be validated in future studies as potential biomarkers of preterm birth risk.
Abbreviations
| BMI | = | body mass index |
| FDR | = | false discovery rate |
| IPA | = | Ingenuity Pathway Analysis |
| LMP | = | last menstrual period |
| miRNA | = | microRNA |
| mRNA | = | messengerRNA |
| TNF | = | tumor necrosis factor |
| PROGRESS | = | Programming Research in Obesity, GRowth Environment and Social Stress |
Introduction
Preterm birth is a leading cause of infant mortality affecting nearly 15 million (10%) live births worldwide every year.Citation1,2 Infants born preterm, or less than 37 completed weeks (259 d) of gestation, are at increased risk of poor developmental outcomes and adverse life-long health effects.Citation3 In 2005, the estimated annual cost of preterm birth in the US was $26.2 billion from medical expenditures and productivity losses,Citation3 and these figures do not include future medical costs later in life. Of concern, rates of preterm birth are on the rise globally,Citation1,3 while the pathophysiologic mechanisms that underlie preterm birth remain elusive.
Known risk factors associated with preterm birth include demographic, clinical and behavioral factors, including African American race/ethnicity, low socioeconomic status, hypertension, diabetes, infections, prior history of a preterm delivery, infertility treatments, multiple gestations, short cervical length, psychosocial stress, and smoking.Citation3,4 There is a clear need to understand how these factors lead to premature labor induction and preterm birth in order to develop and optimize prevention strategies. While inflammation has been associated with preterm delivery,Citation5,6 how all the aforementioned preterm risk factors lead to inflammation (race for example) and how inflammation leads to cervical dysfunction or uterine contractions remains unclear. Epigenetic biomarkers identified in population-based studies may illuminate biological mechanisms for additional study,Citation7 and, jointly, such studies hold the promise of both improving the identification of women at risk of delivering preterm and highlighting potential therapeutic targets to prevent preterm birth. microRNAs (miRNAs) in particular may represent an epigenetic mechanism that may be both a biomarker of risk, and a mechanism potentially amenable to future interventions.
miRNAs are short, single stranded RNAs that posttranscriptionally regulate gene expression. miRNAs typically regulate gene expression by binding and targeting specific mRNA transcripts for degradation or translational repression. A single miRNA can regulate the expression of hundreds of target mRNAsCitation8; hence, miRNAs serve as an ideal unifying molecular marker to better understand the pathophysiological processes that may regulate labor. miRNAs play a critical role in embryonic development,Citation9 the body's inflammatory response,Citation10 as well as signaling cascades that are vital to labor and delivery.Citation11 For example, the miR-200 family of miRNAs regulates contraction-associated genes.Citation11 To our knowledge, only 2 previous case-control studies have investigated the role of cervical miRNAs in preterm labor and delivery.Citation12,13 Because miRNA expression is tissue-specific, investigating a key tissue for delivery initiation is critical to understand mechanisms and obtain valid biomarkers.Citation7 Additionally, while systemic illness may contribute to preterm birth, local cervical changes are required for preterm labor to occur.
In this study, we assessed the association between miRNA expression in the cervix during the second trimester and the length of gestation in a prospective cohort of 60 pregnant women. We used cervical swabs to collect cells during a Papanicolaou (Pap) smear and analyzed the expression profiles of 800 miRNA using the NanoString nCounter assay. This study aimed to identify miRNAs that were significantly associated with the length of gestation. The findings herein have the potential to enhance the current understanding of the complex molecular systems that govern preterm labor and delivery.
Results
Characteristics of study participants
Demographics of the 60 Mexican women who were participating in the Programming Research in Obesity, GRowth Environment and Social Stress (PROGRESS) study and consented to a cervical swab during pregnancy are presented in . The average maternal age was 28 y and ranged from 18 to 40. A large proportion of the women had delivered a previous child (57%), had at least 12 y of education (40%), and were normal weight (50%). The majority of women also reported no exposure to tobacco smoke in the home (70%), and had no evidence of inflammation on the Pap smear (56%). Four women (7%) were lost to follow-up. The mean gestational age at delivery of the remaining 56 women was 38 weeks (271 d), and ranged between 34 and 42 weeks (243–298 d). Four women (7%) delivered preterm at less than 37 weeks of gestation.
Table 1. Maternal demographics for 60 pregnant womena participating in the cervical miRNA PROGRESS subcohort, Mexico City
miRNAs associated with gestational age
We identified miRNAs associated with gestational age at delivery using unadjusted linear regression models and calculated FDR q-values (Supplemental Material, Table S1). Eight miRNAs passed our statistical criteria of P < 0.05 and FDR q-value <0.2 in the unadjusted model including miRs 142, 148b, 30e, 4516, 21, 25, 223, and 29b (). These estimates corresponded to an approximately 3-day gestational age difference comparing the 25th to 75th percentiles (interquartile range) of miRNA expression for each miRNA (data not shown).
Table 2. Unadjusted associations (P < 0.05, q < 0.2) of miRNA expression with the length of gestation (n = 53)
Six of the miRNAs including miRs 21, 30e, 142, 148b, 29b, and 223 remained statistically significant (P < 0.05) after adjustment for maternal age, education, pre-pregnancy BMI, parity, smoke exposure inside the home, as well as evidence of inflammation on the Pap smear (). The miRNAs all had higher expression in pregnancies with shorter gestations (). Specifically, a doubling in miR-21 expression levels was associated with a 0.9 day (95% CI: 0.2–1.5) decrease in length of gestation (P = 0.009). Similarly, each doubling of miR-30e, 142, 148b, 29b, and 223 expression was associated with approximately 1.6, 1.3, 1.3, 1.0 and 1.0 day shorter gestational ages.
Table 3. Adjusted associations of miRNA expression with the length of gestation from linear models adjusted for maternal age, parity, education, pre-pregnancy BMI, smoke exposure in the home, and evidence of inflammation on the Pap smear (n = 53)
Figure 1. Heatmap of the top 6 differentially expressed miRNA by gestational age at delivery. Log2 miRNA expression is z-scored, where red indicates higher expression and blue indicates lower expression. Subjects (n = 53) are ordered from shortest to longest gestational age in days on the x-axis, miRNAs are ordered based on Euclidean distance on the y-axis.
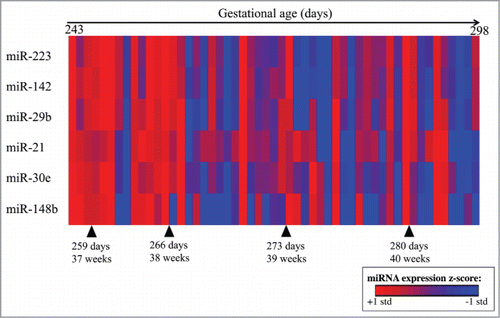
A subanalysis comparison of the preterm (n = 4) and term (n = 25; restricted to 39 and 40 week gestations) deliveries identified 12 miRNAs that were upregulated among mothers who delivered less than 37 weeks gestation (P < 0.05, FDR q-value < 0.2) (Supplemental Material, Table S2). The subanalysis showed increased expression of miR-21, miR-142, miR-30e, miR-148b, and miR-29b among preterm vs. term deliveries, which were also identified in the linear regression models. Additional miRNAs that had significantly increased expressed preterm vs. term subgroups included miR-107, miR-769, miR-29a, miR-548d, miR-15b, miR-93, and miR-590.
In the subgroup of 30 women without evidence of inflammation on their Pap smear, the associations of gestational age and expression of the top 6 miRNA were similar to that of the larger sample, although miR-148b did not reach statistical significance (Supplemental Material, Table S3).
Functional network and pathway analysis of miRNA targeted genes
The 6 miRNAs that passed P < 0.05 in the adjusted model were selected for downstream target prediction and subsequent functional enrichment analysis. We identified 4,733 mRNA that were experimentally observed or predicted targets of the 6 miRNAs. Inclusion of only the experimentally observed downstream mRNA targets resulted in a set of 219 target mRNA, of which 212 were unique (; Supplemental Material Table S4). Notably, miR-30e, which had the largest effect estimate of the significantly upregulated miRNAs associated with shorter gestational age, had 100 experimentally observed downstream mRNA targets. miR-21 and miR-29b were also significantly upregulated with shorter gestations, and had 43 and 58 experimentally observed targets, respectively. miR-142, miR-148b, and miR-223 had 4, 5, and 9 experimentally observed downstream targets, respectively ().
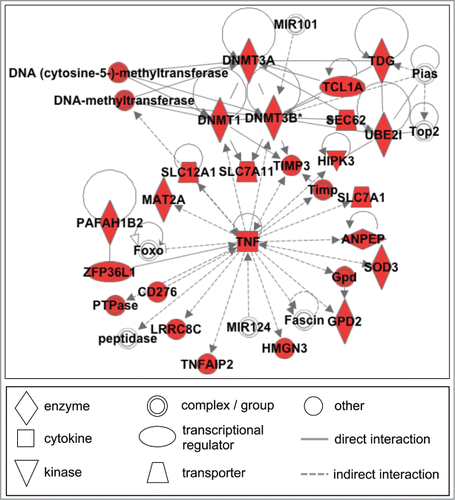
To understand the biological functions of the mRNA targets, we performed 3 types of enrichment analysis on the 212 unique mRNA targets that included i) molecular network mapping, ii) canonical pathway enrichment, and iii) physiological system function enrichment. To understand how these molecules interact within a cell, we mapped the mRNA targets to molecular networks. The most significantly enriched network of interacting molecules was associated with DNA replication, recombination and repair, as well as amino acid and nucleic acid metabolism (P = 1 × 10–41) (). Tumor necrosis factor (TNF) and several DNA methyltransferases are identified as key nodes in this regulatory network. Other networks were enriched for cancer, gastrointestinal, and hepatic disease (P = 1 × 10–31), as well as cell death and survival, inflammatory response, and cellular development (P = 1 × 10–29).
Figure 2. The most significantly enriched network of gestational age-associated molecules was enriched for cellular functions including DNA replication, recombination and repair, amino acid and nucleic acid metabolism (P = 1 × 10–41). Colors represent molecular targets of the miRNAs (red symbols) or associated proteins (clear symbols). TNF and several DNA methyltransferases are identified as key nodes in this regulatory network.
The top 3 canonical pathways identified included hepatic fibrosis P = 5 × 10–11), molecular mechanisms of cancer (P = 5 × 10–10), and aryl hydrocarbon receptor signaling (P = 4 × 10–9). Notably, the most significantly enriched physiological system functions were organismal survival (P < 2 × 10–8), cardiovascular system development and function (P < 3 × 10–6), organismal development (P < 3 × 10–6), and tissue morphology and development (P < 4 × 10–6).
Discussion
In this prospective cohort study of pregnant women, we identified distinct miRNAs measured in cervical samples during pregnancy that are associated with the subsequent gestational age of offspring. We also identified differentially expressed miRNAs with respect to preterm vs. term birth in a subset of women.
The six miRNAs highlighted in this study include miR-21, miR-30e, miR-142, miR-148b, miR-29b, and miR-223. All of the miRNAs had higher expression with shorter gestations, and miR-21, miR-142, and miR-29b have not been previously identified in association with gestational age or delivery. Three of the 6 identified miRNAs (miR-30e, miR-142, and miR-223), were identified in 2 previous studies of cervical miRNAs in labor and delivery.Citation12,13 In the only previous prospective study of preterm birth and cervical miRNA expression, which consisted of 20 women in a nested prospective case-control study that measured expression twice during pregnancy (samples were obtained at 20–23 and 24–27 weeks gestation),Citation12 miR-148b, and miR-30e were upregulated in women who delivered preterm vs. those who did not. In addition, miR-223 was previously found to be upregulated in biopsied cervical tissue of laboring vs. non-laboring women.Citation13 In agreement, our study identified increased levels of miR-148b, miR-30e, and miR-223 were associated with shorter gestational age. Notably, while miR-21, miR-142, and miR-29b have not been previously associated with pregnancy outcomes and markers from cervical cells, their downstream mRNA targets are involved in cancer, inflammatory response, and apoptosis,Citation14-16 and have a biologically plausible role in preterm birth.
Each of the 6 identified miRNAs are involved in innate or adaptive immune responseCitation16-20; we speculate that increased expression of the miRNAs associated with shorter gestations may be evidence of a miRNA-mediated response that contributes to preterm birth. Notably, network analysis of the 212 mRNA targets of the 6 significantly upregulated miRNAs identified tumor necrosis factor (TNF) as a central node of a network that was enriched for DNA replication. TNF-induced inflammatory response is associated with preterm birth,Citation21,22 and levels of TNF are increased in the amniotic fluid from cases of preterm labor 22. Several DNA methyltransferases were also identified within the network, which suggests that additional layers of epigenetic control are involved. This is supported by the observations that miR-142 is directly repressed by DNA methylation,Citation23 and miR-29b and miR-148 target and repress DNA methyltransferases.Citation24,25 Ultimately, initiation of labor and delivery is likely regulated by a concerted effort of miRNAs as well as other regulatory molecules. We have shown in a previous study that epigenetic markers, such as long-interspersed nuclear element-1 (LINE-1) and prostaglandin E receptor-2 (PTGER2) methylation in the cervix were associated with gestational age.Citation26 Future studies will aim to perform gene expression analysis of mRNA targets and incorporate other epigenetic markers such as DNA methylation.
Comparison of the preterm to term gestations showed significant upregulation of 12 miRNAs. Notably, miR-30e, miR-148b, and miR-93 (as part of the miR-106b cluster) were previously identified as upregulated among cases of preterm birth compared to matched controls by Elovitz et al.Citation12 The subanalysis of preterm vs. term gestations was limited by sample size and we were unable to adjust for covariates. When results from this subanalysis are compared to the continuous gestational age analysis, miR-21, miR-30e, miR-142, miR-148b, and miR-29b were statistically significant in both models. This indicates that in this study, miR-223 was distinctly identified when gestational age was examined as a continuous vs. dichotomous (term vs. preterm) outcome. Even though the sample size is limited, this may be an important consideration in future studies since using gestational age as a continuous variable in a regression model increases statistical power.
While our results replicate several findings from previous studies, differences in the miRNAs identified in this study may originate from the type of quantification platform used, the study design (cohort vs. case-control), time of sample collection during pregnancy, and differences in the source population. Our source population was largely a healthy group of pregnant women in Mexico City, whereas in the most comparable study to ours Elovitz et al. used a population in Philadelphia at “high risk” of preterm or using 17-α-hydroxyprogesterone caproate, which could independently affect miRNA expression.
Our study has several strengths. First, we prospectively enrolled women during pregnancy who were participating in an ongoing population-based cohort study with carefully collected covariate data. Given our cohort design, we were able to adjust for potential confounding variables and analyzed gestational age as a continuous outcome. While residual confounding may limit our study, in general, the effect estimates were remarkably unchanged after covariate adjustment indicating little confounding (with the exception of miR-4516 estimates, for which adjustment for inflammation noted on the Pap smear attenuated the effect estimate). Although the cells obtained through cervical swabs were too few to perform cell sorting, the clinical pathologist provided a qualitative assessment of the inflammatory cells noted on the Pap smear thereby allowing our analysis to adjust for cell type. We note that the prevalence of Pap smear inflammation in this study (44%) is similar to findings from a previous study of pregnant women in the US (51%).Citation27 The present study is the largest study to date (n = 60) to examine the association between cervical miRNA and gestational age. We sampled the cervix during the second trimester; this was comparable to the first of 2 sampling time points performed by Elovitz et al..Citation12 Reassuringly, Elovitz et al. reported that miRNA profiles between the 2 time points did not differ substantially. Taken together this suggests that some of the miRNAs associated with gestational age may be stable biomarkers in the second trimester. The prospective study design allowed us to collect cervical samples prior to the onset of the outcome as opposed to a cross sectional study design as performed by Hassan et al. of laboring vs. not laboring women. This distinction is important as the outcome (labor) may cause changes in cervical cells that are independent of the upstream variables that are truly causative.
Our study also has several limitations. The PROGRESS cohort is a racially/ethnically homogeneous population, which may limit external generalizability, although results may likely be better generalized to Mexican Americans. While rates of preterm birth are lower in Mexico than the US (7.3% vs. 12%),Citation1 preterm birth rates are markedly higher (12.8%) among Hispanic women living in the US near the US-Mexico border region compared to the national Mexican rate.Citation28 The measure of smoke exposure used in this study was ascertained using a questionnaire rather than direct measurement of cotinine biomarkers, and thus could potentially result in exposure misclassification. We sampled the cervix just once mid-pregnancy and thus cannot identify from this study the best time window to predict preterm delivery with miRNA biomarkers. Pregnancy dating was done using maternal report of last menstrual period (LMP) at the time of study enrollment. In a cohort enrolled in an identical fashion in this setting, we validated LMP dating on a subset of 98 women with ultrasound and found a correlation of 0.89 (P < 0.0001) (data not shown). Given the limitations of LMP-estimated gestational age,Citation29,30 and that ultrasound-based methods are not standard practice in Mexico's public hospitals, we further improved upon the estimated gestational ages by checking agreement with the Capurro method physical examination at the time of birth. In this study, the 3 infants for whom the Capurro method was subsequently applied were each reassigned a longer gestational age. We note that ascertainment of the LMP was done prior to delivery and in that regard to any misclassification bias of gestational age by LMP should be non-differential with respect to preterm delivery and levels of miRNA expression. Likewise, although measured at delivery any bias in the Capurro assignment of gestational age is non-differential with respect to miRNA expression. Non-differential misclassification bias typically drives effect estimates toward the null value and would lead to underestimation of the true effect size but not a false positive result. Because we were concerned that the pregnancies whose gestational lengths were reassigned might nonetheless introduce bias, we reanalyzed our data excluding these subjects and the results were similar and remained statistically significant with the exception of miR-223 (P = 0.07) (data not shown). An additional limitation of our study was the use of cervical swabs, which will collect a heterogeneous cell mixture that, in addition to cervical epithelial cells, includes leukocytes and other cell types. A subanalysis of participants without evidence of inflammation on their Pap smears revealed similar results. Women without inflammation would be likely to have a more homogeneous cell population collected. Additionally, with just a single cervical swab, we did not have sufficient yield to perform additional analyses to validate our findings using an alternate platform. However, the NanoString platform uses a quantitative assay that has been validated in other studies.Citation31 We did not collect information on fathers and thus our study cannot address any effects of sperm or other paternal factors on cervical miRNA expression.
Our selection of miRNAs was informed by the NanoString platform. The NanoString platform analyzed 800 human and viral miRNA at the time of this study. Many of these targets (n = 726) were detectable in fewer than 60% of cervical samples, and thus were not included in our analyses. This may have been conservative as lack of expression may be due to non/low expression and not necessarily poor sample quality. As such, comparison of quantitative miRNA technologies has shown NanoString to have less sensitive probe detection than other methodologies.Citation32
Identifying novel biomarkers for pregnancies at risk of preterm delivery remains a critically important goal. Current prevention strategies for preterm birth include screening for prior spontaneous preterm birth and shortened cervix mid-pregnancy. Healthcare providers then offer various therapies depending on a woman's individual risk including progesterone, cerclage and/or pessary placement.Citation33 However, using this strategy, the prediction of preterm birth remains inexact. Improvements in preterm birth prediction promises to have 2 major impacts on patient care. First, new biomarkers with improved prediction ability could help to better target these effective, but invasive, therapies to individuals who are most likely to benefit. Second, novel interventions could be designed to interrupt the cascade of molecular events that occur to trigger labor inappropriately and produce preterm birth. Our findings suggest that miRNAs represent promising biomarkers for identifying women at risk of preterm delivery.
In conclusion, we identified a set of 6 miRNAs with increased expression in the cervix mid-pregnancy that are associated with the subsequent shorter length of gestation. This study highlights miR-21, miR-142, and miR-29b as novel miRNAs associated with gestational age, and replicated findings from other investigators that miR-148b, miR-30e, and miR-223 are cervical miRNA predictors of preterm birth. If larger studies from other populations and mechanistic studies confirm our findings, cervical miRNAs and their downstream targets hold promise as preventive screening tools and potential therapeutic interventions for preterm birth.
Materials and Methods
Study design
This study was conducted on a subcohort of 60 Mexican women ages 18 to 40 y participating in the PROGRESS birth cohort in Mexico City, and who consented to a cervical swab during pregnancy. Details of enrollment for the parent cohort are published elsewhere.Citation34 Briefly, women in their second trimester were recruited between 2007 and 2011 through the Mexican social security system (Instituto Mexicano del Seguro Social). The parent cohort consists of 1,054 mothers. Because this study was conceived after the majority of recruitment had occurred only the last 100 women enrolled in the parent cohort were approached. Eighty enrollees provided written informed consent for an obstetrician to obtain a cervical swab mid-pregnancy (16–19 weeks gestation) for miRNA analysis. Women were offered a free Pap smear in return for their participation, which is the standard of care during pregnancy in the US, but is not routinely offered in Mexico. The IRBs of the participating institutions approved this study. For funding reasons, 60 (80%) of the 80 samples collected were randomly selected for miRNA profiling analysis. Four women were lost to follow-up and 56 of these women continued in the study through delivery of their infants. Fifty-three women had complete covariate data and were included in the final models.
Participant data collection
Demographics and birth outcomes were collected as part of the parent study including maternal age, parity, self reported pre-pregnancy body mass index (BMI), and years of education. Staff conducted in-person surveys, which included a question about household smoke exposure. Household smoke exposure was dichotomized as yes/no based on the mother's report that at least one household member smoked. All participants reported that they did not smoke during pregnancy. A histopathological assessment for evidence of inflammation on the Pap smear served as a proxy for shifts in the cell type mixture. The cervical swab sample collection contains mostly cervical epithelial cells (ectocervical and endocervical cells), but also leukocytes, which can be categorically quantified by a trained histopathologist. We dichotomized Pap smear inflammation as yes/no based on the histopathologist's blinded interpretation of the smear. Gestational age was calculated in units of “days” starting with the maternally reported LMP to the date of delivery. The Capurro method (i.e., an infant physical exam) was used as a secondary confirmatory estimate of gestational age, as it is a well-established 6-variable physical assessment that can be used to estimate gestational age.Citation35 In instances where the gestational age estimated from the LMP differed by more than 3 weeks from the Capurro method, the Capurro method-derived estimate was considered more accurate in this study (n = 3). To convert the Capurro method estimates from weeks into days, the gestational age in weeks was multiplied by 7 plus 3.5 d to convert to days mid-week. We performed a sensitivity analysis to ensure our results were unchanged when excluding these pregnancies.
Cervical miRNA collection and extraction
Cervical cells were collected in a method similar to a standard Pap smear protocol, wherein a cotton swab was used to collect cells from the endocervix of the external os. The sample was immediately placed in RNALater (Qiagen, Valencia, CA) and the specimen was frozen at -80°C until subsequent analysis. Total RNA was extracted using the Exiqon miRCURY kit (Exiqon, Woburn, MA) according to the manufacturer's protocol. A cleanup step was then performed using an Amicon Ultra 0.5 mL clean up kit (EMD Millipore, Billerica, MA). miRNA were quantified using a NanoPhotometer P-300 (Implent GmbH, Westlake Village, CA).
NanoString nCounter assay for miRNA expression
miRNA expression was assessed using the NanoString nCounter system (NanoString Technologies, Seattle, WA). This method enables multiplexed direct digital counting of miRNA molecules.Citation36 This method measured a total of 800 probes that were available for analysis at the time of this study, and included both endogenous human-associated miRNAs as well as viral miRNAs that are expressed in human cells.Citation37-39 We performed a feasibility pilot with 10 of the original samples, including 2 technical replicates. Technical replicates showed strong correlation and an additional figure including these preliminary samples is provided in Supplemental Material (Supplemental Material, Figure S1).
The raw count data from the 60 samples were normalized using the NanoStringNorm R package.Citation40 Data were background-corrected by subtracting the mean of the 6 negative controls included on the platform, and normalized using the geometric mean of the 10 probes with the lowest coefficients of variation—which were used to calculate a scaling factor as suggested by the package guidelines. A priori, we required that probes be detectable in at least 60% of the samples. This resulted in 74 probes that were included in the analyses. Individual probes with expression levels below the LOD were assigned a nominal value of 1. Note that the proportion of samples below detect for each miRNA is reported in Supplemental Table S1. The distributions of miRNA expression and model residuals showed that our selection of a linear model was appropriate for these data.
Statistical analysis
To examine the association between miRNA expression levels and gestational age, we used linear regression models. We used separate models to estimate the mean difference in gestational age at delivery (expressed in days) associated with a doubling of expression (log2 unit increase) of each miRNA. We chose covariates a priori and included maternal age, education, pre-pregnancy BMI, parity, smoke exposure inside the home, as well as evidence of inflammation on the Pap smear. Gestational age at delivery was estimated in days, and smoke exposure and inflammation were dichotomized as described above. Maternal education was categorized as having less than high school, high school, or greater than high school education. Pre-pregnancy BMI was categorized as normal (BMI 18.5–25 kg/m2), overweight (BMI 25–30 kg/m2), and obese (≥30 kg/m2). Both adjusted and unadjusted regression models were performed to examine the association between each of the 74 probes’ log2-transformed miRNA levels with gestational age. P-values and Storey's false discovery rate (FDR) q-values were calculated to estimate significanceCitation41; P < 0.05 was considered statistically significant. miRNAs with P < 0.05 and FDR q-values <0.2 in the unadjusted model were retained for downstream target prediction and pathways enrichment analyses. To exclude the influence of confounders, we dropped from these subsequent analyses any miRNAs with P ≥ 0.05 in the adjusted model. A heatmap to visualize the top 6 differentially expressed miRNAs over gestational age was prepared using Partek Genomics Suite 6.6 (St Louis, MO). Hierarchical cluster analysis of z-scored expression using Euclidean distance as a measure was used to identify the relationships between miRNAs.Citation42
To compare our findings to those of a previous study,Citation12 we compared a subset of the 4 mothers who delivered preterm (<37 weeks gestation) to a control group of mothers (n = 25) who delivered at term (completed 39 or 40 weeks gestation). We used Cochran t-tests for unequal variances to compare the means of each of the 74 miRNA between the preterm and term groups; P <0.05 and FDR q-value <0.2 was considered statistically significant. To address concerns about cell mixture, we also re-analyzed the associations between the miRNAs we identified and the length of gestation among women without Pap smear inflammation (n = 30).
Prediction of miRNA targets
To predict downstream mRNA targets, the set of up-regulated miRNAs which passed P-value < 0.05 in the adjusted model (and FDR q-value < 0.2 in the unadjusted model) were uploaded into the Ingenuity Pathway Analysis (IPA) tool (Ingenuity® Systems, Redwood City, CA). Putative miRNA-mRNA relationships were identified using the IPA microRNA Target Filter, based on a knowledgebase of predicted and experimentally observed relationships. We stringently selected for only the experimentally observed miRNA-mRNA relationships, and the resulting target gene list was analyzed for functional network and pathway analysis.
Functional pathway and network enrichment analysis of miRNA transcripts
Functional analysis was carried out to identify molecular networks and biological functions significantly associated with the mRNA target gene set. Analysis of i) molecular network mapping, ii) canonical pathway enrichment, and iii) physiological system function enrichment was performed using the IPA tool. The IPA proprietary database curates gene-phenotype associations, molecular interactions, regulatory events, and chemical knowledge to provide a global molecular network. Related networks were algorithmically constructed based on connectivity. Statistical significance of each biological function was calculated using Fisher's exact text with an α set at 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Table_S4.xls
Download MS Excel (115 KB)Table_S2.xlsx
Download MS Excel (15.3 KB)Table_S1.xlsx
Download MS Excel (61.6 KB)Supplemental_Material.docx
Download MS Word (285.2 KB)Acknowledgments
We thank Erroll Reuckert at NanoString Technologies for his assistance with the miRNA expression profiling and analysis. We thank ABC Hospital in Mexico City for providing facilities during data collection. We also thank Lorena Pantano and Shannon Ho Sui of the Bioinformatics Core at the Harvard School of Public Health for their assistance with diagnostic informatics analyses.
Funding
This work was supported in part by Pilot Project funding from the HSPH-NIEHS Center for Environmental Health (ES000002) and NIH/NIEHS: K23ES022242, K99ES023450, P42 ES016454, P30ES23515, R01ES013744, R01ES020268, R01ES021357, the Klarman Scholars Program at Beth Israel Deaconess Medical Center, and the Harvard Catalyst D-MaPS Program.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 2012; 379:2162-72; PMID:22682464; http://dx.doi.org/10.1016/S0140-6736(12)60820-4
- Beck S, Wojdyla D, Say L, Betran AP, Merialdi M, Requejo JH, Rubens C, Menon R, Van Look PF. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ 2010; 88:31-8; PMID:20428351; http://dx.doi.org/10.2471/BLT.08.062554
- IOM. In: Behrman RE, Butler AS, eds. Preterm Birth: Causes, Consequences, and Prevention. Washington (DC), 2007; http://www.ncbi.nlm.nih.gov/pubmed/20669423.
- Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet 2008; 371:75-84; PMID:18177778; http://dx.doi.org/10.1016/S0140-6736(08)60074-4
- Trivedi S, Joachim M, McElrath T, Kliman HJ, Allred EN, Fichorova RN, Onderdonk A, Heitor F, Chaychi L, Leviton A, et al. Fetal-placental inflammation, but not adrenal activation, is associated with extreme preterm delivery. Am J Obstet Gynecol 2012; 206:236 e1-8; PMID:22264652; http://dx.doi.org/10.1016/j.ajog.2011.12.004
- Hecht JL, Fichorova RN, Tang VF, Allred EN, McElrath TF, Leviton A. Relationship Between Neonatal Blood Protein Concentrations and Placenta Histologic Characteristics in Extremely Low GA Newborns. Pediatr Res 2011; 69:68-73; PMID:20921924; http://dx.doi.org/10.1203/PDR.0b013e3181fed334
- Bakulski KM, Fallin MD. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen 2014; 55:171-83; PMID:24449392; http://dx.doi.org/10.1002/em.21850
- Freedman JE, Tanriverdi K. Defining miRNA targets: balancing simplicity with complexity. Circulation 2013; 127:2075-7; PMID:23625958; http://dx.doi.org/10.1161/CIRCULATIONAHA.113.003058
- Collignon J. miRNA in embryonic development: the taming of Nodal signaling. Dev Cell 2007; 13:458-60; PMID:17925220; http://dx.doi.org/10.1016/j.devcel.2007.09.012
- Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol 2008; 8:120-30; PMID:18204468; http://dx.doi.org/10.1038/nri2252
- Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A 2010; 107:20828-33; PMID:21079000; http://dx.doi.org/10.1073/pnas.1008301107
- Elovitz MA, Brown AG, Anton L, Gilstrop M, Heiser L, Bastek J. Distinct cervical microRNA profiles are present in women destined to have a preterm birth. Am J Obstet Gynecol 2014; 210:221 e1-11; PMID:24565431
- Hassan SS, Romero R, Pineles B, Tarca AL, Montenegro D, Erez O, Mittal P, Kusanovic JP, Mazaki-Tovi S, Espinoza J, et al. MicroRNA expression profiling of the human uterine cervix after term labor and delivery. Am J Obstet Gynecol 2010; 202:80 e1-8; PMID:AMBIGUOUS; http://dx.doi.org/10.1016/j.ajog.2009.08.016
- Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer 2011; 30:371-80; PMID:21627859; http://dx.doi.org/10.5732/cjc.011.10132
- Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 2012; 44:237-44; PMID:22214600; http://dx.doi.org/10.1152/physiolgenomics.00141.2011
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PloS One 2007; 2:e1020; PMID:17925868; http://dx.doi.org/10.1371/journal.pone.0001020
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 2007; 449:919-22; PMID:17943132; http://dx.doi.org/10.1038/nature06205
- Ruan Q, Wang P, Wang T, Qi J, Wei M, Wang S, Fan T, Johnson D, Wan X, Shi W, et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis 2014; 5:e1095; PMID:24577093; http://dx.doi.org/10.1038/cddis.2014.47
- O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 2010; 10:111-22; PMID:20098459; http://dx.doi.org/10.1038/nri2708
- Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol 2010; 185:7244-51; PMID:21068402; http://dx.doi.org/10.4049/jimmunol.1001573
- Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol 2014; 11(6):571-81; PMID:24954221
- Romero R, Mazor M, Sepulveda W, Avila C, Copeland D, Williams J. Tumor necrosis factor in preterm and term labor. Am J Obstet Gynecol 1992; 166:1576-87; PMID:1595815; http://dx.doi.org/10.1016/0002-9378(92)91636-O
- Skarn M, Baroy T, Stratford EW, Myklebost O. Epigenetic regulation and functional characterization of microRNA-142 in mesenchymal cells. PloS one 2013; 8:e79231; PMID:24236112; http://dx.doi.org/10.1371/journal.pone.0079231
- Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci 2013; 14:14647-58; PMID:23857059; http://dx.doi.org/10.3390/ijms140714647
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA 2008; 14:872-7; PMID:18367714; http://dx.doi.org/10.1261/rna.972008
- Burris HH, Baccarelli AA, Motta V, Byun HM, Just AC, Mercado-Garcia A, Schwartz J, Svensson K, Téllez-Rojo MM, Wright RO, et al. Association between length of gestation and cervical DNA methylation of PTGER2 and LINE 1-HS. Epigenetics 2014; 9:1083-91; PMID:24827772; http://dx.doi.org/10.4161/epi.29170
- Blake RL, Jr., Gay JW, Brown S, Smith W. Does evidence of inflammation on Papanicolaou smears of pregnant women predict preterm labor and delivery? J Am Board Fam Pract 1992; 5:555-63; PMID:1462789
- McDonald JA, Mojarro O, Sutton PD, Ventura SJ. A binational overview of reproductive health outcomes among US Hispanic and Mexican women in the border region. Prev Chronic Dis 2013; 10:E137; PMID:23948338; http://dx.doi.org/10.5888/pcd10.130019
- Alexander GR, Tompkins ME, Petersen DJ, Hulsey TC, Mor J. Discordance between LMP-based and clinically estimated gestational age: implications for research, programs, and policy. Public Health Rep 1995; 110:395-402; PMID:7638326
- Hall MH, Carr-Hill RA, Fraser C, Campbell D, Samphier ML. The extent and antecedents of uncertain gestation. Br J Obstet Gynaecol 1985; 92:445-51; PMID:3994927; http://dx.doi.org/10.1111/j.1471-0528.1985.tb01347.x
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D'Andrade P, DeMayo M, Dennis L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat methods 2014; 11:809-15; PMID:24973947; http://dx.doi.org/10.1038/nmeth.3014
- Knutsen E, Fiskaa T, Ursvik A, Jorgensen TE, Perander M, Lund E, Seternes OM, Johansen SD, Andreassen M. Performance comparison of digital microRNA profiling technologies applied on human breast cancer cell lines. PloS one 2013; 8:e75813; PMID:24116077; http://dx.doi.org/10.1371/journal.pone.0075813
- Practice bulletin no. 130: prediction and prevention of preterm birth. Obstet Gynecol 2012; 120:964-73; PMID:22996126; http://dx.doi.org/10.1097/AOG.0b013e3182723b1b
- Burris HH, Braun JM, Byun HM, Tarantini L, Mercado A, Wright RJ, Schnaas L, Baccarelli AA, Wright RO, Tellez-Rojo MM. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics 2013; 5:271-81; PMID:23750643; http://dx.doi.org/10.2217/epi.13.24
- Capurro H, Konichezky S, Fonseca D, Caldeyro-Barcia R. A simplified method for diagnosis of gestational age in the newborn infant. J Pediatr 1978; 93:120-2; PMID:650322; http://dx.doi.org/10.1016/S0022-3476(78)80621-0
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008; 26:317-25; PMID:18278033; http://dx.doi.org/10.1038/nbt1385
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science 2007; 317:376-81; PMID:17641203; http://dx.doi.org/10.1126/science.1140956
- Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog 2010; 6:e1000787; PMID:20195463; http://dx.doi.org/10.1371/journal.ppat.1000787
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology 2011; 411:325-43; PMID:21277611; http://dx.doi.org/10.1016/j.virol.2011.01.002
- Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012; 28:1546-8; PMID:22513995; http://dx.doi.org/10.1093/bioinformatics/bts188
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003; 100:9440-5; PMID:12883005; http://dx.doi.org/10.1073/pnas.1530509100
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998; 95:14863-8; PMID:9843981; http://dx.doi.org/10.1073/pnas.95.25.14863
