Abstract
Alterations of epigenetic marks are linked to normal development and cellular differentiation as well as to the progression of common chronic diseases. The plasticity of these marks provides potential for disease therapies and prevention strategies. Macro- and micro-nutrients have been shown to modulate disease risk in part via effects on the epigenome. The essential micronutrient selenium affects human health outcomes, e.g., cancers, cardiovascular and autoimmune diseases, via selenoproteins and through a range of biologically active dietary selenocompounds and metabolism products thereof. This review provides an assessment of the current literature regarding epigenetic effects of dietary and synthetic selenocompounds, which include the modulation of marks and editors of epigenetic information and interference with one-carbon metabolism, which provides the methyl donor for DNA methylation. The relevance of a selenium-epigenome interaction for human health is discussed, and we also indicate where future studies will be helpful to gain a deeper understanding of epigenetic effects elicited by selenium.
Introduction
Epigenetics describes mitotically stable chromatin-based mechanisms that modulate gene expression without altering the genomic DNA sequence. These mechanisms include modifications to DNA [methylation of cytosine to 5-methylcytosine (5 mC), and 5mC oxidation products] and to histones (acetylation, methylation and many others),Citation1 which interfere with chromosomal packaging and the binding of trans-acting factors. The recent advancements in DNA/RNA sequencing technology have made it possible to study epigenetic marks on a genome-wide level, gaining insight into the so-called epigenome. Multi-centered consortia, namely DEEP (Deutsches Epigenom-Programm, German Epigenome Program, www.deutsches-epigenom-programm.de) and IHEC (International Human Epigenome Consortium, www.ihec-epigenomes.org) are currently deciphering high-resolution epigenome maps of healthy and diseased tissues/cell types to obtain standard and disease-specific epigenome profiles. According to current knowledge, epigenomes show plasticity throughout an organism's lifetime,Citation2,3 during cellular differentiation and in response to manifold external stimuli. Changes to the epigenome are also associated with the onset and progression of cancerCitation4,5 and other complex diseases like autoimmune diseases,Citation6 inflammatory bowel diseases,Citation7 type 2 diabetes,Citation8 and cardiovascular disease.Citation9 Causality of most of these associations remains unknown, but in light of the principal alterability of epigenetic marks—in contrast to the largely stable DNA setup of a cell—, targeting the epigenome may provide a promising strategy in disease therapy and prevention. Major determinants of risk and progression of the above-mentioned common diseases are—besides genetic predispositions—environmental factors like lifestyle and diet. Dietary patterns, specific (micro) nutrients and secondary plant compounds have been found to alter epigenetic marks,Citation10,11 and evidence is increasing that the modulation of health outcomes by food components is (at least in part) mediated by their epigenetic effects. While also macronutrients (e.g., employed as experimental high-fat, high-protein, or calorie-restricted diets) have been shown to modify epigenetic marks,Citation11 mechanistic explanations for these observations are difficult to derive due to multiple confounding effects that are elicited by macronutrients and due to composition variability. Therefore, the majority of studies have assessed epigenetic effects in response to intervention with specific micronutrients and secondary plant compounds. In this regard, selenium (Se), an essential trace element that exists in a range of biologically active compounds, is a particularly interesting micronutrient. It has been found to modify epigenetic marks in studies employing cell systems and animals, and in a limited number of human studies, too. The importance of Se for maintaining optimal health is based on the biological functions that are exerted by 2 main groups of Se species: members of the selenoprotein family, which are encoded by 25 genes in humans and contain cotranslationally inserted selenocysteine,Citation12 and, second, a non-selenoprotein pool of low-molecular-weight selenium compounds contained in the diet or derived from Se metabolism. Well-characterized selenoproteins, such as glutathione peroxidases (GPx), thioredoxin reductases (TrxR), and iodothyronine deiodinases (DIO) are oxidoreductases and act as antioxidant enzymes, regulators of redox-sensitive signaling pathways and of thyroid hormone metabolism. Other selenoproteins facilitate selenium transport (selenoprotein P), selenoprotein biosynthesis (selenophosphate synthetase 2), and are involved in maintaining endoplasmic reticulum homeostasis (e.g., selenoproteins S, K, and 15-kDa selenoprotein).Citation13 The importance of selenoproteins for murine development and health has been demonstrated in transgenic mice with singleCitation14,15 or totalCitation16 selenoprotein depletion. Moreover, humans with genetically impaired selenoprotein biosynthesis suffer from a severe multisystem disorder.Citation17 Biological outcomes of murine and human Se supplementation studies (e.g., disease incidences, effects on the transcriptome, epigenome, and on Se status,Citation18,19(and references therein) are known to be affected not only by Se dosage, but also by its chemical form and the pre-supplemental Se status. We will therefore briefly present an overview of Se requirements and the metabolism of dietary Se compounds. Thereafter, we summarize the current knowledge of the effects of naturally occurring and synthetic Se compounds on epigenetic marks and editors and discuss their possible relevance for health and disease. An aim is to approach the biochemical basis of the Se–epigenome interaction, and to this end we focus on studies that have shown direct and indirect effects of Se compounds on the epigenetic machinery (e.g., DNA methyl transferases, histone modifying enzymes, one-carbon metabolism). These questions cannot fully be answered by the current knowledge, and we therefore indicate which studies may be useful to broaden our understanding of the Se-epigenome interaction in the future.
Selenium Requirements and Metabolism of Dietary Se Compounds
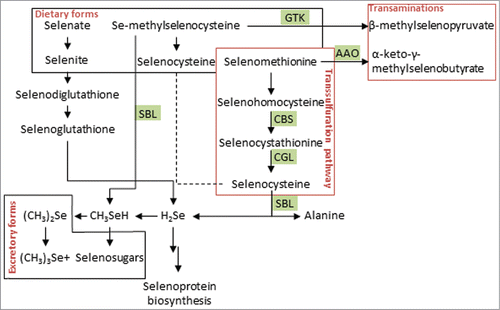
Overt Se deficiency is relatively rare in Western countries. A clinical condition that is associated with Se deficiency is Keshan Disease, which occurred in a Chinese province where affected people had total daily Se intakes of ≤15 μg.Citation13 The tolerable upper intake level of Se has been defined by the World Health Organization as 400 μg per day,Citation20 and the majority of health bodies recommends Se intake levels in the range from 55 to 70 μg per day.Citation19 These recommendations are often based on total Se intakes required to optimize activity/expression of GPx3 and SeP in plasma, 2 commonly used biomarkers of Se status. Optimization of plasma GPx3 activity is achieved at 40–47 μg Se/day,Citation21 whereas SeP requires ∼105 μg Se/day.Citation22 The plasma Se concentration that coincides with SeP optimization (124 μg Se/l) is in a range that is associated with decreased mortality risk and prevention of several cancers.Citation19 Supplementation beyond these Se levels appears to confer no additional benefit but may instead increase the risk of type 2 diabetes.Citation23 Major dietary selenocompounds are the amino acids selenomethionine (SelMet), selenocysteine, Se-methylselenocysteine, and the anions selenite and selenate (for a listing of Se compounds and quantities in food seeCitation19). Se compounds are metabolized through different routes (displayed in ) to hydrogen selenide. Hydrogen selenide is methylated to excretory forms (dimethylselenide, trimethylselenonium, Se-methyl-N-acetylselenohexosamine) or phosphorylated to selenophosphate, used as precursor of the amino acid selenocysteine (Sec), which is cotranslationally inserted from a Sec-specific tRNA (Sec-tRNA[Ser]Sec) into selenoproteins. Selenite is reduced via glutathionylation to seleno(/di)glutathione or via glutaredoxin directly to selenide. Metabolism of SelMet takes place via the transsulfuration pathway—catalyzed by the same enzymes that transform methionine to cysteine—to Sec, which is converted by selenocysteine β-lyase (SBL) to selenide and alanine. In a similar reaction, SBL generates methylselenol from Se-methylselenocysteine. The methylseleno-amino acids Se-methylselenocysteine and SelMet can be transaminated by glutamine transaminase K and by L-amino acid oxidase to β-methylselenopyruvate and to α-keto-γ-methylselenobutyrate, respectively (seeCitation24 for references referring to Se metabolism).
Figure 1. Metabolism of dietary selenocompounds. The major organic and inorganic selenocompounds are metabolized by transsulfurations, transaminations and reductions by thioredoxin reductases, glutathione reductases and glutaredoxins. Participating enzymes marked with green background. For details see text. GTK = glutamine transaminase K; AAO = L-amino acid oxidase; GR = glutathione reductase; CGL = cystathionine γ-lyase; CBS = cystathionine β-synthase; SBL = selenocysteine β-lyase.
Influence of Selenium on Epigenetic Modifications of DNA and Histones
Influence of Se on DNA methylation
The methylation of cytosines in genomic DNA is the most common and probably most studied epigenetic modification in higher organisms. The methyl group is transferred in a reaction catalyzed by DNA methyltransferases (DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L) from the donor substrate S-adenosylmethionine (SAM) to the 5-carbon position of cytosine—often when it is bound to guanosine in 3′—, resulting in 5-methylcytosine (5 mC). DNA demethylation, on the other hand, is not catalyzed directly but results from either DNA replication-coupled dilution, wherein 5 mC or 5 mC oxidation products [5-hydroxymethylcytosine (5 hmC), 5-formylcytosine (5 fC) and 5-carboxylcytosine (5 caC)] are not copied to the new DNA strand, or after replacement of 5 mC (or derivatives) together with short stretches of surrounding nucleotides via base excision repair (BER) or nucleotide excision repair (NER).Citation25 Interference of nutrients with DNA methylation can therefore principally occur through modulation of (i) DNMT activity / interaction with accessory factors, (ii) SAM availability, and (iii) demethylation processes. A number of studies have reported effects of Se status or supplementation on global and gene-specific DNA methylation as well as on the expression or activity of DNMTs (listed in ). Arai et al. incubated murine embryonic stem cells with a physiologic, non-toxic Se dose as found in maternal blood serum. Se caused a reversible alteration of the cell heterochromatin status and also changed the DNA methylation status of individual genes with roles in fetal development, including Hnf4α (hepatocyte nuclear factor 4α), Aebp2 (AE binding protein 2), Prickle2 (prickle homolog 2), and Rnd2 (Rho family GTPase 2), without compromising the cell potential to form embryonic bodies.Citation26 These results imply an interesting link between Se and tissue-specific differentiation via effects on gene-specific methylation, as Se is well known to be required for hepatocyte differentiation in vitro, and the transcription factor HNF4α is a key regulator of this process. The change in chromatin structure observed by Arai et al. could be caused by global DNA methylation differences in Se-supplemented versus deficient cells. Studies employing rodentsCitation27-30 and cell linesCitation31 have indeed shown that dietary Se intake levels affect global DNA methylation. The rodent studies gave inconsistent results regarding an increase or decrease of global DNA methylation in response to supplemental Se, although comparable Se diets were used (): Se deficiency resulted in less DNA methylation in rat liverCitation28,29 and colon,Citation29 in contrast to a study by Zeng et al., wherein DNA methylation levels in liver and colon were higher in rats fed supranutritional vs. adequate and deficient Se diets. Zeng et al. pointed to differences in the employed animal strains and contents of the basal diets as possible modifiers of Se effects.Citation27 Additionally, different techniques were applied for the assessment of global DNA methylation: an in vitro methyl group acceptance assay with [3H-methyl]-SAM/SssI methylase and isolated DNA,Citation28,29 a 5 mC ELISA,Citation27 and by HPLC detection of 5 mC monophosphate in enzymatically digested DNA.Citation30 Corresponding data from in vitro studies is limited to one paper, showing that LNCaP prostate tumor cells treated with 1.5 μM selenite for 7 d had a ∼50% decrease in 5 mC immunoreactivity.Citation31 A conclusion that can be drawn is that impacts of selenite and SelMet on global DNA methylation are possibly masked by strain-specific effects and that they are also influenced by the nutritional context (e.g., high fat diet). This topic has been elucidated in a human study (N = 287) that found a significant inverse association of plasma Se and global DNA methylation in leukocytes.Citation32 In addition to effects on global methylation, Se has also been shown to induce differential methylation at regions and specific CpG sites of individual genes. The study by Xiang et al. found that the genes encoding the phase II detoxification enzyme GSTP1 (π-class glutathione S-transferase) and the tumor suppressors APC (adenomatous polyposis coli) and CSR1 (cellular stress response 1), which are frequently silenced in prostate tumors due to hypermethylation of their promoters, were demethylated and re-expressed in LNCaP cells after selenite-treatment. Similarly, a different source of Se (100 μM SelMet) caused promoter demethylation and re-expression of GSTP1.Citation31 Se compounds are commonly applied in in vitro studies at concentrations ranging from 10 nM up to 100 μM. Growth inhibitory and toxic effects of Se are dependent on its chemical form and on the cell type. While Se has low toxicity when it is bound to proteins (as in plasma) or amino acids (e.g., SelMet), many cell lines do not tolerate selenite or methylseleninic acid at doses ≥1 μM.Citation33 In consideration of the physiological concentration range of Se in human plasma (∼0.4–2.5 μM), Se doses of > 5 μM are supraphysiological and not applicable for supplementation trials. The use of Se for cancer prevention is particularly promising for prostate cancer, based on the observation of an inverse association of prostate cancer risk and Se status,Citation34 as well as on supporting findings of accelerated prostate carcinogenesis in selenoprotein-deficient mice.Citation35 In this context, a concept considering Se as an epigenetic drug against cancer progression through targeting of tumor suppressor genes, as implied by Xiang et al., may arise, but certainly in vivo studies need to be performed to strengthen and broaden it to other relevant genes that might also be targeted by Se in different forms and at different stages of carcinogenesis. Methylation of the von-Hippel-Lindau (VHL) gene promoter was found to respond to Se (250 nM Se-methylselenocysteine (SeMSC)) in Caco-2 cells; VHL promoter methylation was decreased by SeMSC in vitro and this was associated with increased VHL expression levels in Caco-2 and in rats fed 2 μg Se as SeMSC.Citation36 VHL is frequently downregulated and mutated in renal cell carcinomas and also found to be deregulated during colorectal carcinogenesis,Citation37 wherein Se has been attributed protective functions based on epidemiological and animal studies.Citation18,38 A study with humans assessed the methylation status of colorectal cancer-related genes in healthy rectal mucosa specimens (84 males, 101 females) in relation to Se status.Citation39 An association was found for WIF1 (wnt inhibitory factor 1) methylation and plasma Se concentration. Interestingly, Se status was also associated with methylation of other genes and retrotransposons, including LINE1 (long interspersed nucleotide element 1), PCA1 (cation-transporting P-type ATPase), SFRP1/2 (secreted frizzled-related protein 1/2), and APC, and this occurred in a gender-specific manner. It was not analyzed whether differential methylation of these genes was associated with differences in their expression levels. The EPIC (European prospective investigation of cancer and nutrition cohort) study revealed an inverse association of Se status and colorectal cancer risk with the association being stronger in women than men.Citation38 Gender-specific effects of Se are also apparent from different responses of selenoprotein expression levels and biomarkers of Se status to Se supplementation in men and women (summarized inCitation38); the study by Tapp et al. suggests that this gender-specificity of Se-effects extends to epigenetic marks of cancer-related genes, but the relevance of these findings for disease etiology warrants further investigations. A recent study identified the inflammatory-related genes TLR2 (toll-like receptor 2) and ICAM1 (intercellular adhesion molecule 1) as novel targets of Se-dependent epigenetic regulation and proposed a mechanism whereby Se changes the expression of GADD45 (growth arrest and DNA-damage-inducible, α) and DNMT1, leading to epigenetic silencing of TLR2 and ICAM1, and links it to a well-known condition that arises from prolonged periods of severely deficient Se intake: Keshan disease (KD).Citation40 KD is a viral myocarditis with necrotic lesions of the myocardium that was first found in the Keshan County of north-eastern China in 1935 and that is endemic in selenium-deficient areas of China. It was found out that Se deficiency is a causal factor of KD, and the incidence rates lowered dramatically after Se was given as a supplement.Citation41 The mode of action of Se in the etiology of KD is not understood completely, but it is assumed to result from an impaired immune response to viral (Coxsackie virus B) infections in Se-deficiency. Yang et al. compared the DNA methylome in peripheral blood from KD patients and healthy controls using methylated DNA-IP and subsequent analysis of the enriched DNA by a Roche-Nimblegen HG18 CpG Promoter array. The methylome profiles showed differences at several thousand differentially methylated regions (DMRs), which were confirmed for the TLR2 and ICAM1 promoters by methylation-specific PCR. Furthermore, expression levels of both genes were inversely correlated with degrees of promoter methylation and with serum Se concentration. Similar results were obtained with rats fed diets containing 0, 0.1 and 2.0 mg/kg sodium selenite, whereas in contrast to the human subjects methylation/expression of Tlr2 and Icam1 genes were measured in myocardial tissue and in isolated neonatal cardiomyocytes. While a significant reduction of Dnmt1 protein expression level was observed in cardiomyocytes treated with 1.5 μM selenite compared to Se-deficient cells, this was unlikely and against anticipation to be the cause of increased Tlr2 and Icam1 promoter methylation. Instead, the authors suggested that this was due to diminished Gadd45a mRNA and protein expression levels in Se-treated cardiomyocytes. GADD45A has been linked to demethylation of specific genomic loci, e.g., in the genes for RARβ2 (retinoic acid receptor β), via interaction with BER- and NER-executing proteins.Citation25 But a role for GADD45A as mediator of Se-dependent modulation of (site-specific) DNA methylation, as implied by Yang et al.,Citation40 remains speculation until substantiated by additional studies employing, for example, GADD45A gene silencing and chromatin immunoprecipitation. Also, numerous studies have reported that Se enhances DNA damage repair capacity (summarized inCitation42) via enhanced p53 binding activityCitation29 and partly, in contrast to the results obtained with rat cardiomyocytes,Citation40 associated with increased expression of GADD45—in MCF7 cells via GPx-1Citation43—or interaction thereof with DNA repair enzymes, such as AP endonuclease 1.Citation44 Whether these effects of Se on DNA repair enzymes do promote DNA demethylation is however not certain.
Table 1. Studies regarding effects of Se on DNA methylation and DNMT expression or activity
Another possible Se-target to induce differential DNA methylation is the class of DNMT enzymes; the modulation of DNMT expression or activity has been postulated to contribute to the modulation of methylation marks by Se seen in the studies mentioned above. Selenite and 2 synthetic selenocompounds, benzyl selenocyanate (BSC) and 1,4-phenylenebis (methylene) selenocyanate (p-XSC), have been shown to inhibit DNMT activity in nuclear extracts of human colon carcinomas.Citation45 The IC50 values of the 3 compounds were calculated as 3.8 μM (selenite), 8.4 μM (BSC) and 5.2 μM (p-XSC), and the setup of the experiments implies that the compounds acted in their non-metabolized forms. DNMT prepared from rat liver was also inhibited by selenite in an in vitro assay at a Ki of 6.7 μM, and the enzymatic activity was lower when isolated from Se-supplemented compared to control animals.Citation46 In vivo, a non-significant decrease of DNMT activity was seen in the liver and colon of rats fed Se-adequate vs. -deficient diets (0.2 and 0 mg Se/kg diet as sodium selenite).Citation28 Similarly, DNMT1 protein expression was diminished by 1.5 μM sodium selenite in rat cardiomyocytesCitation40 and in LNCaP cellsCitation31 and by 8 μM sodium selenite and 2 μM MSA in MCF-7 cells.Citation47 The in vitro human and animal studies collectively indicate that Se is inversely associated with global DNA methylation and DNMT activity.
Influence of Se on histone acetylation
Histone proteins carry manifold posttranslational modifications (e.g., methylation and acetylation) added by histone methyltransferases, histone acetyltransferases (HATs), etc. at defined amino acid positions. Common histone marks that are studied by the IHEC consortium are methylation and acetylation at lysine residues 4, 9, 27, and 36 of histone 3. The highly diverse code of histone marks controls histone binding to DNA, interaction of DNA with trans-acting factors, and ultimately gene expression. Aberrant histone codes are found at the onset and progression of diseases and have therefore become therapeutic targets. Se and other micronutrients have been shown to induce or to be associated with changes to histone marks, thereby possibly affecting health outcomes. Interference of nutrients with histone marks can principally occur through modulation of histone modifying enzyme activity/expression and via interference with substrate availability. Given the large variety of marks and participating enzymes, the situation is even more complex than for DNA methylation; furthermore, crosstalks exist between DNA methylation and histone marks and together they form a complicated network of epigenetic regulation.Citation48 From a clinical perspective, particular interest has been given to histone deacetylases (HDACs), as their abnormal function and/or expression is linked to cancers and some neurologic and immune disorders. Numerous synthetic HDAC inhibitors have been developed and are currently tested in clinical trials.Citation49 Some naturally occurring dietary factors or metabolites like butyrate, polyphenols and Se have also been shown to act as HDAC inhibitors. Studies reporting the modulation of histone marks and editing enzymes (HDACs and HATs) by dietary and synthetic Se compounds are listed in and discussed below.
Table 2. Modulation of histone modifications by dietary and synthetic selenocompounds
Treatment of LNCaP cells with 1.5 μM selenite for 7 d significantly reduced HDAC activity, concomitant with slightly reduced HDAC3 and unchanged HDAC4 and HDAC5 protein levels.Citation31 In line with reduced HDAC activity, the authors found both global and GSTP1 promoter-bound levels of H3K9ac—a repressive mark—to be increased and H3K9me3—an activating mark—to be decreased upon Se treatment. Other studies have confirmed that dietary and synthetic selenocompounds inhibit HDAC activity.Citation50–53 Kassam et al. applied methylseleninic acid (MSA; 5–30 μM) to diffuse large B-cell lymphoma (DLBCL) cell lines and observed decreased HDAC activity and increased total levels of the HDAC targets acetylated histone H3 and acetylated α-tubulin.Citation53 These effects required cellular metabolism of MSA, but the metabolite responsible for HDAC inactivation was not identified. Lee et al. discovered that seleno-α-keto acid metabolites derived from dietary selenocompounds act as inhibitors of HDACs.Citation50 In their study, SeMSC and SelMet reacted in transaminations catalyzed by the enzymes glutamine transaminase K (GTK) and L-amino acid oxidase to β-methylselenopyruvate and to α-keto-γ-methylselenobutyrate, which are structurally similar to the well-known HDAC inhibitor butyrate (). Both seleno-α-keto acids inhibited HDAC activity in cell free assays, in contrast to SeMSC and SelMet, and led to rapid elevation of acetylated histone H3 levels in prostate cancer cells. The anti-cancer effects of high dose Se supplementation observed in studies of experimental carcinogenesis have been partly attributed to the inhibition of HDAC and stimulated the synthesis of more potent Se-containing HDAC inhibitors (HDACi). Based on the structure of suberoylanilide hydroxamic acid (SAHA, commercially available as Vorinostat), a HDACi that is used for the treatment of advanced T-cell lymphoma, 2 Se-based derivatives named SelSA-1 and SelSA-2 () were synthesized.Citation51 SelSA-2 showed higher inhibitory activity against HDAC in HeLa nuclear cell extract (mainly isoforms HDAC1 and HDAC2) than SAHA and trichostatin A.Citation51 Both selenocompounds were also more effective in inhibiting melanoma cell growth and melanocytic lesion development in skin reconstructs,Citation52 but their use as drugs against melanoma or other cancers has not been tested yet. A recent paper has shown that histone H4 acetylation at its lysine residues 5, 8, 12, and 16 was decreased in macrophages treated with selenite (100–500 nM).Citation54 This was concomitant with a decreased abundance of H4K12ac and H4K16ac at the promoters of the pro-inflammatory genes tumor necrosis factor-α (TNF- α) and cyclooxygenase-2 (COX-2), whose expression had been shown before to be diminished in Se-treated RAW264.7 macrophage cells.Citation55 HDAC activity in these cells was unaffected by selenite, and the authors proposed that the selenite-triggered decrease of H4 acetylation levels was rather due to inhibition of p300 HAT activityCitation54 through a Se-dependent and haematopoietic prostaglandin D synthase (H-PGDS)- and COX-mediated production of anti-inflammatory Δ12-PGJ2 and 15d-PGJ2,Citation56 which can covalently bind to p300 and thereby inhibit its activity.Citation57 Importantly, selenite did not change H4 acetylation levels in macrophages that lacked Sec-tRNA[Ser]Sec, showing that selenoprotein biosynthesis was a prerequisite for selenite-induced modulation of H4 acetylation.Citation54 The proposed cascade of events triggered by selenite (involving H-PGDS, COX-2, p300 HAT, and p65 and H4 acetylation) might trigger the switch from the M1 to the M2 phenotype of macrophages. It would thus also add to the understanding of the anti-inflammatory role of Se and selenoproteins in inflammatory bowel diseasesCitation58 and other chronic inflammatory diseases.
Figure 2. Se-containing inhibitors of HDAC activity. Structural formulas of Se metabolites (β-Methylselenopyruvate and α-Keto-γ-methylselenobutyrate) and synthetic selenocompounds (B(PCP)-2Se [Bis(5-phenylcarbamoylpentyl)diselenide], SelSA-1 and PCP-SeCN (5-phenylcarbamoylpentyl selenocyanide), SelSA-2) with HDAC-inhibitory activity.
![Figure 2. Se-containing inhibitors of HDAC activity. Structural formulas of Se metabolites (β-Methylselenopyruvate and α-Keto-γ-methylselenobutyrate) and synthetic selenocompounds (B(PCP)-2Se [Bis(5-phenylcarbamoylpentyl)diselenide], SelSA-1 and PCP-SeCN (5-phenylcarbamoylpentyl selenocyanide), SelSA-2) with HDAC-inhibitory activity.](/cms/asset/85f441ea-184c-47f0-98e0-a94fe7ab36b4/kepi_a_1013792_f0002_b.gif)
Interrelation Between Selenium and One-carbon Metabolism
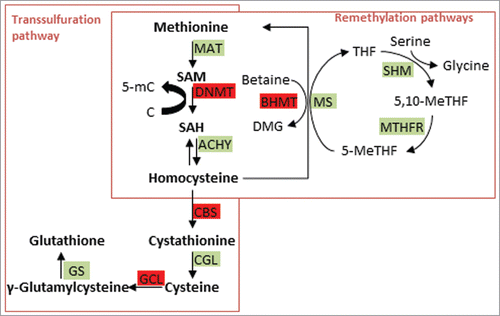
The one-carbon metabolism pathway (depicted in ) provides the methyl donor S-adenosylmethionine (SAM), which is used as substrate by the DNMTs and other methylating enzymes for transfer of the methyl group to cytosines and target proteins. One-carbon metabolism is therefore tightly linked to DNA methylation, and conditions that have an impact on one-carbon metabolism, in particular the availability of the participating enzymes’ cofactors folate, choline/betaine and vitamins B2, B6, and B12, have been shown to result in differential DNA methylation.Citation59 We briefly describe the reactions of the one-carbon metabolism and associated pathways and then assess the current literature regarding findings of Se-dependent modulations thereof. As depicted in , methionine reacts to SAM, which is converted to S-adenosylhomocysteine (SAH) after the DNMT-catalyzed methylation of cytosine or proteins. Removal of the adenosyl group gives homocysteine (HCys). The concentration of HCys in blood or plasma is of clinical importance, as it has been associated with a multitude of complex diseases like cardiovascular and neurodegenerative diseasesCitation60 (and references therein). Clearance of HCys occurs through its remethylation to methionine, in turn serving as a substrate for the formation of SAM, or otherwise via metabolism in the transsulfuration pathway. The first step of this pathway is the condensation reaction of HCys with serine to cystathionine, catalyzed by cystathionine β-synthase (CBS) with pyridoxal phosphate (PLP, the active form of vitamin B6) as cofactor. A variant of the CBS gene (c.844ins68) was found to affect HCys clearance and SAM/SAH ratio following methionine loading.Citation61 The c.844ins68 was also associated with a significantly lower risk of coronary artery disease, baseline HCys levels however appear to be unaffected by the CBS genotype in healthy individuals.Citation61,62 Cystathionine is converted by the PLP-dependent enzyme cystathionine γ-lyase (CGL) to cysteine, which in turn condenses with glutamate to γ-glutamylcysteine, catalyzed by glutamate-cysteine ligase (GCL). Finally, fusion with glycine via glutathione synthetase (GS) results in the formation of glutathione. Remethylation of HCys is facilitated by methionine synthase (MS) in an enzymatic reaction involving vitamin B12 and 5-methyl-THF. THF cycles back to 5-methyl THF via 2 enzymatic steps employing vitamin B6 and flavin adenine dinucleotide (FAD) as cofactors; the methyl group ultimately derives from the essential amino acid serine. A second route to methionine is the methylation of HCys by betaine homocysteine methyltransferase (BHMT), which uses betaine as methyl donor.
Figure 3. Se interferes with one-carbon metabolism. Enzymes are marked with green background; enzymes that have been shown to be affected by Se are marked with red background. SAM = S-adenosylmethionine; MAT = methionine adenosyltransferase; SAH = S-adenosylhomocysteine; DNMT = DNA methyltransferase; ACHY = S-adenosyl-L-homocysteine hydrolase; BHMT = betaine homocysteine methyltransferase; GCL = glutamate-cysteine ligase; MS = methionine synthase; SHM = serine hydroxymethyltransferase; MTHFR = methylenetetrahydrofolate reductase; 5,10-MeTHF = 5,10-methylene-tetrahydrofolate; GTK = glutamine transaminase K; AAO = L-amino acid oxidase; GR = glutathione reductase; CGL = cystathionine γ-lyase; CBS = cystathionine β-synthase; SBL = selenocysteine β-lyase.
Correlations between Se status and plasma homocysteine levels
Several human studies have shown an inverse correlation between plasma or serum Se and HCys levels (). In an Inuit population with very high mean Se status (635.5 μg Se/l blood), plasma HCys was negatively predicted by Se.Citation63 Klapcinska et al. similarly detected an inverse correlation between whole-blood Se and plasma HCys in a population with low mean Se status (62.5 μg Se/l blood).Citation64 While both studies reported similar findings, possible confounders that are known to affect HCys levels, particularly folate and B vitamins, need to be taken into account. In a study where an inverse relationship between Se and HCys was reported for participants of the British National diet and Nutrition Survey aged ≥ 65 years,Citation65 this correlation became insignificant after adjustment for folate, PLP, and vitamin B12.Citation60 In the study by Bekaert et al., however, an association of Se and HCys remained significant after adjustment for the same confounders, but only in males.Citation60 Plasma Se was calculated to account for 1.8% of HCys variance by linear regression analyses. In line with this, serum Se predicted 5.8% variance in HCys in a Spanish population independently of folate and vitamin B12, and individuals within the highest versus lowest Se tertile had a 63% reduced risk of being in the top HCys tertile.Citation66 Ischemic stroke patients aged <55 years that were assessed shortly after stroke onset (third day) were found to have lower Se status than healthy controls, which was inversely correlated with HCys and accounted for 15.4% of HCys variance independent of vitamin B6 levels.Citation67 In light of these baseline associations, trials have been conducted to determine whether Se supplementation affected plasma HCys. Se supplementation in form of daily doses of 100 or 300 μg Se as high-Se yeast for 6 months increased plasma Se but had no effect on HCys levels.Citation60 This lack of effect supported an earlier intervention study where 200 μg Se (as SelMet) given daily for 20 weeks failed to change plasma HCys levels; yet a possible association of Se and HCys at baseline was not assessed in that study.Citation68
Table 3. Human studies of blood Se/homocysteine associations
Animal experiments allow greater flexibility in the design of supplementation studies, a better control for confounding factors, and more options for end point detections. lists studies with rodents that were fed diets with different Se form and content, followed by subsequent assessments of metabolites and enzymes of the one-carbon metabolism. Some general trends, which are independent of the Se form and the species, can be derived from these studies: (I) an inverse correlation between HCys and Se intake/status appears to exist in the liver. This derives from a study with mice fed SelMetCitation69 and a study with rats fed selenate.Citation70 Both studies used diets with comparable Se contents that are alike the range of dietary intakes levels existing in human populations. Strikingly, slightly suboptimal Se intake levels (0.05/0.06 ppm Se) in comparison to adequate intake levels (0.15 ppm Se) caused in both studies a significant increase in liver HCys concentrations, by 30% in ratsCitation70 and 314% in mice.Citation69 The latter study suggested that liver hyperhomocysteinemia was due to Se-dependent regulation of CBS. Other studies have also reported differential expression of enzymes belonging to the transsulfuration (GCL)Citation71 and remethylation pathways (BHMT and glycine N-methyltransferase),Citation28,71,72 but these effects appeared to be species-specific and were only seen under severe Se deficiency. (II) Plasma HCys concentrations were significantly reduced in those animals (mice and rats) that were fed diets considered as Se deficient (≤0.025 ppm).Citation27,28,70-73 (III) A trend toward lower plasma HCys levels was seen in animals fed diets with supranutritional (>0.2 ppm) vs. adequate (0.1–0.2 ppm) Se content.Citation27,29,70,71 Comparison of these 2 dietary groups is particularly interesting in consideration of the human studies showing an inverse correlation of plasma HCys and Se. Increased HCys concentrations result in accumulation of SAH as a consequence of the reversibility of the reaction catalyzed by ACHY (S-adenosyl-L-homocysteine hydrolase; see ). SAH in turn is a competitive inhibitor of methyltransferases and, therefore, increased HCys concentrations would be associated with global DNA hypomethylation. This concept was supported by a study showing that women with normal (mean 7.2 μM; range 5.8–8.7 μM) HCys plasma levels had significantly higher plasma SAM/SAH ratio and lymphocyte DNA methylation levels than women with elevated (mean 12.3 μM; range 9.3–16.5 μM) HCys.Citation74 Importantly, plasma SAH correlated with HCys and lymphocyte DNA methylation but SAM did not, meaning that a measurement of SAM is insufficient for predicting methylation potential. Tissue SAH concentrations were also found to correlate with DNA hypomethylation in testes, brain and liver of mice.Citation75 Data regarding SAH and/or SAM levels in murine Se supplementation studies is limited and inconsistent. Three studies measured lower liver and colon SAM/SAH ratios in groups fed adequate (0.1 or 0.2 ppm) compared to deficient or supranutritional (2 or 4 ppm) Se diets,Citation28,29,71 which was however not seen when SelMet was used instead of selenite.Citation29 Another study found lower SAM/SAH in mice fed 0.5 ppm Se (as selenite) compared to 0.15 and 0.025 ppm Se.Citation73 Two recently conducted genome-wide association studies found associations of SNPs located near genes encoding CBS and BHMT with blood and toenail Se concentrations.Citation76 Exciting data published by the Combs group shows that interference with one-carbon metabolism affects Se metabolism: inhibition of ACHY resulted in reduced SAM/SAH ratios and diminished secretion of the selenium transporter SeP from HepG2 cells as a result of decreased arginine methylation (=less activity) of proteins that are pivotal for transcriptional induction of SeP.Citation77,78 Elimination of Se also requires SAM-dependent methylation (), and, consequently, inhibition of ACHY in rats led to Se retention in liver and kidney with concomitant reduction of excretory Se forms in these organs and in the urine.Citation79 These data together with the findings from animal Se supplementation studies further indicate that whole-body Se and one-carbon metabolism are interconnected.
Table 4. Murine studies on interference of Se with one-carbon metabolism
Regulation of microRNA expression by selenium
The regulation of gene expression by targeting of mRNA through non-coding RNA molecules such as microRNA (miRNA) is sometimes considered as an additional epigenetic mechanism. To date only one study has examined whether Se has an effect on microRNA (miRNA) expression. Microarray analysis (737 miRNAs in total) of the miRNA profiles of Caco-2 cells grown in Se-deficient or Se-supplemented medium revealed that the expression of 12 miRNAs was affected by Se supply.Citation80 Expression levels of 50 mRNAs were also Se-responsive in the same study, and numerous of these mRNAs were predicted to be targeted by the Se-responsive miRNAs. One of these, miRNA-185, whose expression decreased under Se deficiency, was confirmed to regulate expression of glutathione peroxidase 2 (GPx-2) and selenophosphate synthetase 2 (SPS-2). As the enzymatic product of SPS-2 is part of the selenoprotein biosynthesis machinery, these findings indicate that Se availability affects the selenoproteome in part through epigenetic mechanisms involving miRNA-185 and possibly other miRNAs. miRNA-185 is a particularly interesting target of Se, as it has recently emerged as a tumor suppressor that is frequently downregulated in ovarian, breast, renal,Citation81 prostate,Citation82 and gastric cancers,Citation83 and targets oncogenes, e.g., Six1,Citation81 androgen receptor,Citation82 and apoptosis repressor with caspase recruitment domain (ARC).Citation83 Se has been shown to act anticarcinogenic in experimental settings and also in some human studies (Citation18 for an overview); it will therefore be an interesting area of future work to uncover putative roles of miRNAs as mediators of Se-dependent tumor protection against malignant transformation.
Conclusions and Future Directions
Investigation of epigenetic effects elicited by Se species remains a relatively new field that has not been comprehensively studied. The current data from mainly murine and cell-based, but also some human studies show that Se supplementation and status modify DNA methylation globally and at specific gene regions or loci. DNMT inhibition by Se and its interaction with one-carbon metabolism are possible routes through which this occurs. Additionally, histone modifications are altered by Se, and this has been shown to occur -at least in vitro- via inhibition of HDAC activity by the Se metabolism products seleno-α-keto acids. There is a need to systematically assess Se-related effects on epigenetic marks -DNA methylation and histone modifications- at a genome-wide level in murine and human intervention studies. These studies will ideally use different dietary forms of Se, subjects with low initial Se status, so that dose-response relationships can be observed, and include transcriptome assessments. The combined readouts will give important information about the role of Se in epigenetic and transcriptional regulation, and exploit interference of Se with epigenetic marks that are associated with or predict the risk of diseases. This is particularly relevant to better understand the role of Se in the prevention and progression of certain diseases (e.g., prostate cancer and type 2 diabetes), which has become unclear after publication of diverging results from large-scale Se intervention trials.Citation18,84-86 Another research priority is to gain more insight into the inhibition of DNMT and HDAC enzymes by Se, to identify the active Se species, their possible DNMT/HDAC isoform specificity, mechanism of inhibition and the required doses for inhibition in vitro and in vivo. Se-responsive signaling pathways that influence nuclear proteins linked to epigenetic mechanisms, e.g., through nucleosomal remodeling, transcription or DNA repair, as exemplified for GADD45A, also need to be examined closer. We propose that a detailed genome-wide and mechanistic understanding of epigenetic processes elicited by Se is required to complete the picture of selenium's systems biology and to clarify and predict its impact on health outcomes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012; 150:12-27; PMID:22770212; http://dx.doi.org/10.1016/j.cell.2012.06.013
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 2012; 8:e1002629; PMID:22532803; http://dx.doi.org/10.1371/journal.pgen.1002629
- Tsai PC, Spector TD, Bell JT. Using epigenome-wide association scans of DNA methylation in age-related complex human traits. Epigenomics 2012; 4:511-26; PMID:23130833; http://dx.doi.org/10.2217/epi.12.45
- Arai E, Kanai Y. DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics 2010; 2:467-81; PMID:22121905; http://dx.doi.org/10.2217/epi.10.16
- Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 2013; 13:497-510; PMID:23760024; http://dx.doi.org/10.1038/nrc3486
- Dozmorov MG, Wren JD, Alarcon-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics 2014; 9:276-85; PMID:24213554; http://dx.doi.org/10.4161/epi.27021
- Hasler R, Feng Z, Backdahl L, Spehlmann ME, Franke A, Teschendorff A, Rakyan VK, Down TA, Wilson GA, Feber A, et al. A functional methylome map of ulcerative colitis. Genome Res 2012; 22:2130-7; PMID:22826509; http://dx.doi.org/10.1101/gr.138347.112
- Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, Poulsen P, Ribel-Madsen R, Pedersen NL, Almgren P, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 2014; 63:2962-76; PMID:24812430; http://dx.doi.org/10.2337/db13-1459
- Whayne TF. Epigenetics in the development, modification, and prevention of cardiovascular disease. Mol Biol Rep 2014; PMID:25205125; http://dx.doi.org/10.1007/s11033-014-3727-z
- McKay JA, Mathers JC. Diet induced epigenetic changes and their implications for health. Acta Physiol 2011; 202:103-18; PMID:21401888; http://dx.doi.org/10.1111/j.1748-1716.2011.02278.x
- Schwenk RW, Vogel H, Schurmann A. Genetic and epigenetic control of metabolic health. Mol Metab 2013; 2:337-47; PMID:24327950; http://dx.doi.org/10.1016/j.molmet.2013.09.002
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science 2003; 300:1439-43; PMID:12775843; http://dx.doi.org/10.1126/science.1083516
- Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal 2007; 9:775-806; PMID:17508906; http://dx.doi.org/10.1089/ars.2007.1528
- Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med 2003; 34:496-502; PMID:12566075; http://dx.doi.org/10.1016/S0891-5849(02)01360-6
- Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, et al. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol 2004; 24:9414-23; PMID:15485910; http://dx.doi.org/10.1128/MCB.24.21.9414-9423.2004
- Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc Natl Acad Sci U S A 1997; 94:5531-4; PMID:9159106; http://dx.doi.org/10.1073/pnas.94.11.5531
- Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, Rajanayagam O, Padidela R, Ceron-Gutierrez L, Doffinger R, Prevosto C, et al. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Invest 2010; 120:4220-35; PMID:21084748; http://dx.doi.org/10.1172/JCI43653
- Steinbrenner H, Speckmann B, Sies H. Toward understanding success and failures in the use of selenium for cancer prevention. Antioxid Redox Signal 2013; 19:181-91; PMID:23421468; http://dx.doi.org/10.1089/ars.2013.5246
- Fairweather-Tait SJ, Bao Y, Broadley MR, Collings R, Ford D, Hesketh JE, Hurst R. Selenium in human health and disease. Antioxid Redox Signal 2011; 14:1337-83; PMID:20812787; http://dx.doi.org/10.1089/ars.2010.3275
- Rayman MP. The use of high-selenium yeast to raise selenium status: how does it measure up? Br J Nutr 2004; 92:557-73; PMID:15522125; http://dx.doi.org/10.1079/BJN20041251
- Xia Y, Hill KE, Byrne DW, Xu J, Burk RF. Effectiveness of selenium supplements in a low-selenium area of China. Ame J Clin Nutr 2005; 81:829-34; PMID:15817859
- Hurst R, Armah CN, Dainty JR, Hart DJ, Teucher B, Goldson AJ, Broadley MR, Motley AK, Fairweather-Tait SJ. Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr 2010; 91:923-31; PMID:20181815; http://dx.doi.org/10.3945/ajcn.2009.28169
- Rayman MP, Stranges S. Epidemiology of selenium and type 2 diabetes: can we make sense of it? Free Radic Biol Med 2013; 65:1557-64; PMID:23597503; http://dx.doi.org/10.1016/j.freeradbiomed.2013.04.003
- Weekley CM, Harris HH. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem Soc Rev 2013; 42:8870-94; PMID:24030774; http://dx.doi.org/10.1039/c3cs60272a
- Schomacher L. Mammalian DNA demethylation: multiple faces and upstream regulation. Epigenetics 2013; 8:679-84; PMID:23803967; http://dx.doi.org/10.4161/epi.24977
- Arai Y, Ohgane J, Yagi S, Ito R, Iwasaki Y, Saito K, Akutsu K, Takatori S, Ishii R, Hayashi R, et al. Epigenetic assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev 2011; 57:507-17; PMID:21606628; http://dx.doi.org/10.1262/jrd.11-034A
- Zeng H, Yan L, Cheng WH, Uthus EO. Dietary selenomethionine increases exon-specific DNA methylation of the p53 gene in rat liver and colon mucosa. J Nutr 2011; 141:1464-8; PMID:21653573; http://dx.doi.org/10.3945/jn.111.140715
- Uthus EO, Ross SA, Davis CD. Differential effects of dietary selenium (se) and folate on methyl metabolism in liver and colon of rats. Biol Trace Elem Res 2006; 109:201-14; PMID:16632891; http://dx.doi.org/10.1385/BTER:109:3:201
- Davis CD, Uthus EO, Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr 2000; 130:2903-9; PMID:11110844
- Armstrong KM, Bermingham EN, Bassett SA, Treloar BP, Roy NC, Barnett MP. Global DNA methylation measurement by HPLC using low amounts of DNA. Biotechnol J 2011; 6:113-7; PMID:21053336; http://dx.doi.org/10.1002/biot.201000267
- Xiang N, Zhao R, Song G, Zhong W. Selenite reactivates silenced genes by modifying DNA methylation and histones in prostate cancer cells. Carcinogenesis 2008; 29:2175-81; PMID:18676679; http://dx.doi.org/10.1093/carcin/bgn179
- Pilsner JR, Hall MN, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Associations of plasma selenium with arsenic and genomic methylation of leukocyte DNA in Bangladesh. Environ Health Perspect 2011; 119:113-8; PMID:21205583; http://dx.doi.org/10.1289/ehp.1001937
- Pinto A, Speckmann B, Heisler M, Sies H, Steinbrenner H. Delaying of insulin signal transduction in skeletal muscle cells by selenium compounds. J Inorg Biochem 2011; 105:812-20; PMID:21497580; http://dx.doi.org/10.1016/j.jinorgbio.2011.03.010
- Hurst R, Hooper L, Norat T, Lau R, Aune D, Greenwood DC, Vieira R, Collings R, Harvey LJ, Sterne JA, et al. Selenium and prostate cancer: systematic review and meta-analysis. Am J Clin Nutr 2012; 96:111-22; PMID:22648711; http://dx.doi.org/10.3945/ajcn.111.033373
- Diwadkar-Navsariwala V, Prins GS, Swanson SM, Birch LA, Ray VH, Hedayat S, Lantvit DL, Diamond AM. Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model. Proc Natl Acad Sci U S A 2006; 103:8179-84; PMID:16690748; http://dx.doi.org/10.1073/pnas.0508218103
- Uthus E, Begaye A, Ross S, Zeng H. The von Hippel-Lindau (VHL) tumor-suppressor gene is down-regulated by selenium deficiency in Caco-2 cells and rat colon mucosa. Biol Trace Elem Res 2011; 142:223-31; PMID:20632125; http://dx.doi.org/10.1007/s12011-010-8764-4
- Giles RH, Lolkema MP, Snijckers CM, Belderbos M, van der Groep P, Mans DA, van Beest M, van Noort M, Goldschmeding R, van Diest PJ, et al. Interplay between VHL/HIF1alpha and Wnt/beta-catenin pathways during colorectal tumorigenesis. Oncogene 2006; 25:3065-70; PMID:16407833; http://dx.doi.org/10.1038/sj.onc.1209330
- Hughes DJ, Fedirko V, Jenab M, Schomburg L, Meplan C, Freisling H, Bueno-de-Mesquita HB, Hybsier S, Becker NP, Czuban M, et al. Selenium status is associated with colorectal cancer risk in the European prospective investigation of cancer and nutrition cohort. Int J Cancer 2014; 136:1149-61; PMID: 25042282; http://dx.doi.org/10.1002/ijc.29071
- Tapp HS, Commane DM, Bradburn DM, Arasaradnam R, Mathers JC, Johnson IT, Belshaw NJ. Nutritional factors and gender influence age-related DNA methylation in the human rectal mucosa. Aging Cell 2013; 12:148-55; PMID:23157586; http://dx.doi.org/10.1111/acel.12030
- Yang G, Zhu Y, Dong X, Duan Z, Niu X, Wei J. TLR2-ICAM1-Gadd45alpha axis mediates the epigenetic effect of selenium on DNA methylation and gene expression in Keshan disease. Biol Trace Elem Res 2014; 159:69-80; PMID:24811888; http://dx.doi.org/10.1007/s12011-014-9985-8
- Chen J. An original discovery: selenium deficiency and Keshan disease (an endemic heart disease). Asia Pac J Clin Nut 2012; 21:320-6; PMID:22705420
- Bera S, De Rosa V, Rachidi W, Diamond AM. Does a role for selenium in DNA damage repair explain apparent controversies in its use in chemoprevention? Mutagenesis 2013; 28:127-34; PMID:23204505; http://dx.doi.org/10.1093/mutage/ges064
- Nasr MA, Fedele MJ, Esser K, Diamond AM. GPx-1 modulates Akt and P70S6K phosphorylation and Gadd45 levels in MCF-7 cells. Free Radic Biol Med 2004; 37:187-95; PMID:15203190; http://dx.doi.org/10.1016/j.freeradbiomed.2004.04.038
- Jung HJ, Kim HL, Kim YJ, Weon JI, Seo YR. A novel chemopreventive mechanism of selenomethionine: enhancement of APE1 enzyme activity via a Gadd45a, PCNA and APE1 protein complex that regulates p53-mediated base excision repair. Oncol Rep 2013; 30:1581-6; PMID:23846616
- Fiala ES, Staretz ME, Pandya GA, El-Bayoumy K, Hamilton SR. Inhibition of DNA cytosine methyltransferase by chemopreventive selenium compounds, determined by an improved assay for DNA cytosine methyltransferase and DNA cytosine methylation. Carcinogenesis 1998; 19:597-604; PMID:9600343; http://dx.doi.org/10.1093/carcin/19.4.597
- Cox R. Selenite: a good inhibitor of rat-liver DNA methylase. Biochem Int 1985; 10:63-9; PMID:3985987
- de Miranda JX, Andrade FD, Conti AD, Dagli ML, Moreno FS, Ong TP. Effects of selenium compounds on proliferation and epigenetic marks of breast cancer cells. J Trace Elem Med Biol 2014; 28:486-91; http://dx.doi.org/10.1016/j.jtemb.2014.06.017
- Du J, Patel DJ. Structural biology-based insights into combinatorial readout and crosstalk among epigenetic marks. Biochim Biophys Acta 2014; 1839:719-27; PMID:24747177; http://dx.doi.org/10.1016/j.bbagrm.2014.04.011
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014; 13:673-91; PMID:25131830; http://dx.doi.org/10.1038/nrd4360
- Lee JI, Nian H, Cooper AJ, Sinha R, Dai J, Bisson WH, Dashwood RH, Pinto JT. Alpha-keto acid metabolites of naturally occurring organoselenium compounds as inhibitors of histone deacetylase in human prostate cancer cells. Cancer Prev Res 2009; 2:683-93; PMID:19584079; http://dx.doi.org/10.1158/1940-6207.CAPR-09-0047
- Desai D, Salli U, Vrana KE, Amin S. SelSA, selenium analogs of SAHA as potent histone deacetylase inhibitors. Bioorg Med Chem Lett 2010; 20:2044-7; PMID:20167479; http://dx.doi.org/10.1016/j.bmcl.2009.07.068
- Gowda R, Madhunapantula SV, Desai D, Amin S, Robertson GP. Selenium-containing histone deacetylase inhibitors for melanoma management. Cancer Biol Ther 2012; 13:756-65; PMID:22669577; http://dx.doi.org/10.4161/cbt.20558
- Kassam S, Goenaga-Infante H, Maharaj L, Hiley CT, Juliger S, Joel SP. Methylseleninic acid inhibits HDAC activity in diffuse large B-cell lymphoma cell lines. Cancer Chemother Pharmacol 2011; 68:815-21; PMID:21519842; http://dx.doi.org/10.1007/s00280-011-1649-1
- Narayan V, Ravindra KC, Liao C, Kaushal N, Carlson BA, Prabhu KS. Epigenetic regulation of inflammatory gene expression in macrophages by selenium. J Nutr Biochem 2014; 26:138-45; PMID:25458528; http://dx.doi.org/10.1016/j.jnutbio.2014.09.009
- Zamamiri-Davis F, Lu Y, Thompson JT, Prabhu KS, Reddy PV, Sordillo LM, Reddy CC. Nuclear factor-kappaB mediates over-expression of cyclooxygenase-2 during activation of RAW 264.7 macrophages in selenium deficiency. Free Radic Biol Med 2002; 32:890-7; PMID:11978490; http://dx.doi.org/10.1016/S0891-5849(02)00775-X
- Gandhi UH, Kaushal N, Ravindra KC, Hegde S, Nelson SM, Narayan V, Vunta H, Paulson RF, Prabhu KS. Selenoprotein-dependent up-regulation of hematopoietic prostaglandin D2 synthase in macrophages is mediated through the activation of peroxisome proliferator-activated receptor (PPAR) gamma. J Biol Chem 2011; 286:27471-82; PMID:21669866; http://dx.doi.org/10.1074/jbc.M111.260547
- Ravindra KC, Narayan V, Lushington GH, Peterson BR, Prabhu KS. Targeting of histone acetyltransferase p300 by cyclopentenone prostaglandin Delta(12)-PGJ(2) through covalent binding to Cys(1438). Chem Res Toxicol 2012; 25:337-47; PMID:22141352; http://dx.doi.org/10.1021/tx200383c
- Speckmann B, Steinbrenner H. Selenium and selenoproteins in inflammatory bowel diseases and experimental colitis. Inflamm Bowel Dis 2014; 20:1110-9; PMID:24694793
- Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 2012; 23:853-9; PMID:22749138; http://dx.doi.org/10.1016/j.jnutbio.2012.03.003
- Bekaert B, Cooper ML, Green FR, McNulty H, Pentieva K, Scott JM, Molloy AM, Rayman MP. Effect of selenium status and supplementation with high-selenium yeast on plasma homocysteine and B vitamin concentrations in the UK elderly. Mol Nutr Food Res 2008; 52:1324-33; PMID:18683819; http://dx.doi.org/10.1002/mnfr.200700353
- Janosikova B, Pavlikova M, Kocmanova D, Vitova A, Vesela K, Krupkova L, Kahleova R, Krijt J, Kraml P, Hyanek J, et al. Genetic variants of homocysteine metabolizing enzymes and the risk of coronary artery disease. Mol Genet Metab 2003; 79:167-75; PMID:12855221; http://dx.doi.org/10.1016/S1096-7192(03)00079-9
- Tsai MY, Loria CM, Cao J, Kim Y, Siscovick DS, Schreiner PJ, Hanson NQ. Polygenic association with total homocysteine in the post-folic acid fortification era: the CARDIA study. Mol Genet Metab 2009; 98:181-6; PMID:19577940; http://dx.doi.org/10.1016/j.ymgme.2009.05.012
- Belanger MC, Dewailly E, Berthiaume L, Noel M, Bergeron J, Mirault ME, Julien P. Dietary contaminants and oxidative stress in Inuit of Nunavik. Metabolism 2006; 55:989-95; PMID:16839831; http://dx.doi.org/10.1016/j.metabol.2006.03.007
- Klapcinska B, Poprzecki S, Danch A, Sobczak A, Kempa K. Selenium levels in blood of upper Silesian population: evidence of suboptimal selenium status in a significant percentage of the population. Biol Trace Elem Res 2005; 108:1-15; PMID:16327055; http://dx.doi.org/10.1385/BTER:108:1-3:001
- Bates CJ, Thane CW, Prentice A, Delves HT. Selenium status and its correlates in a British national diet and nutrition survey: people aged 65 years and over. J Trace Elem Med Biol 2002; 16:1-8; http://dx.doi.org/10.1016/S0946-672X(02)80002-5
- Gonzalez S, Huerta JM, Alvarez-Uria J, Fernandez S, Patterson AM, Lasheras C. Serum selenium is associated with plasma homocysteine concentrations in elderly humans. J Nutr 2004; 134:1736-40; PMID:15226462
- Angelova EA, Atanassova PA, Chalakova NT, Dimitrov BD. Associations between serum selenium and total plasma homocysteine during the acute phase of ischaemic stroke. Eur Neurol 2008; 60:298-303; PMID:18824858; http://dx.doi.org/10.1159/000157884
- Venn BJ, Grant AM, Thomson CD, Green TJ. Selenium supplements do not increase plasma total homocysteine concentrations in men and women. J Nutr 2003; 133:418-20; PMID:12566477
- Geillinger KE, Rathmann D, Kohrle J, Fiamoncini J, Daniel H, Kipp AP. Hepatic metabolite profiles in mice with a suboptimal selenium status. J Nutr Biochem 2014; 25:914-22; PMID:24917045; http://dx.doi.org/10.1016/j.jnutbio.2014.04.003
- Wolf NM, Mueller K, Hirche F, Most E, Pallauf J, Mueller AS. Study of molecular targets influencing homocysteine and cholesterol metabolism in growing rats by manipulation of dietary selenium and methionine concentrations. Br J Nutr 2010; 104:520-32; PMID:20350341; http://dx.doi.org/10.1017/S0007114510000899
- Uthus EO, Ross SA. Dietary selenium affects homocysteine metabolism differently in Fisher-344 rats and CD-1 mice. J Nutr 2007; 137:1132-6; PMID: 17449570
- Uthus EO, Yokoi K, Davis CD. Selenium deficiency in Fisher-344 rats decreases plasma and tissue homocysteine concentrations and alters plasma homocysteine and cysteine redox status. J Nutr 2002; 132:1122-8; PMID:12042420
- Metes-Kosik N, Luptak I, Dibello PM, Handy DE, Tang SS, Zhi H, Qin F, Jacobsen DW, Loscalzo J, Joseph J. Both selenium deficiency and modest selenium supplementation lead to myocardial fibrosis in mice via effects on redox-methylation balance. Mol Nutr Food Res 2012; 56:1812-24; PMID:23097236; http://dx.doi.org/10.1002/mnfr.201200386
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000; 275:29318-23; PMID:10884384; http://dx.doi.org/10.1074/jbc.M002725200
- James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr 2002; 132:2361S-6S; PMID:12163693
- Cornelis MC, Fornage M, Foy M, Xun P, Gladyshev VN, Morris S, Chasman DI, Hu FB, Rimm EB, Kraft P, et al. Genome-wide association study of selenium concentrations. Hum Mol Genet 2014; 24:1469-77; PMID:25343990; http://dx.doi.org/10.1093/hmg/ddu546
- Jackson MI, Cao J, Zeng H, Uthus E, Combs GF, Jr. S-adenosylmethionine-dependent protein methylation is required for expression of selenoprotein P and gluconeogenic enzymes in HepG2 human hepatocytes. J Biol Chem 2012; 287:36455-64; PMID:22932905; http://dx.doi.org/10.1074/jbc.M112.412932
- Speckmann B, Walter PL, Alili L, Reinehr R, Sies H, Klotz LO, Steinbrenner H. Selenoprotein P expression is controlled through interaction of the coactivator PGC-1alpha with FoxO1a and hepatocyte nuclear factor 4alpha transcription factors. Hepatology 2008; 48:1998-2006; PMID:18972406; http://dx.doi.org/10.1002/hep.22526
- Jackson MI, Lunoe K, Gabel-Jensen C, Gammelgaard B, Combs GF, Jr. Metabolism of selenite to selenosugar and trimethylselenonium in vivo: tissue dependency and requirement for S-adenosylmethionine-dependent methylation. J Nutr Biochem 2013; 24:2023-30; PMID:24139672; http://dx.doi.org/10.1016/j.jnutbio.2013.04.007
- Maciel-Dominguez A, Swan D, Ford D, Hesketh J. Selenium alters miRNA profile in an intestinal cell line: evidence that miR-185 regulates expression of GPX2 and SEPSH2. Mol Nutr Food Res 2013; 57:2195-205; PMID:23934683; http://dx.doi.org/10.1002/mnfr.201300168
- Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, Rao MK. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 2010; 29:4971-9; PMID:20603620; http://dx.doi.org/10.1038/onc.2010.233
- Qu F, Cui X, Hong Y, Wang J, Li Y, Chen L, Liu Y, Gao Y, Xu D, Wang Q. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol Cell Biochem 2013; 377:121-30; PMID:23417242; http://dx.doi.org/10.1007/s11010-013-1576-z
- Li Q, Wang JX, He YQ, Feng C, Zhang XJ, Sheng JQ, Li PF. MicroRNA-185 regulates chemotherapeutic sensitivity in gastric cancer by targeting apoptosis repressor with caspase recruitment domain. Cell Death Dis 2014; 5:e1197; PMID:24763054; http://dx.doi.org/10.1038/cddis.2014.148
- Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Jama 2009; 301:39-51; PMID:19066370; http://dx.doi.org/10.1001/jama.2008.864
- Clark LC, Combs GF, Jr., Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. Jama 1996; 276:1957-63; PMID:8971064; http://dx.doi.org/10.1001/jama.1996.03540240035027
- Steinbrenner H, Speckmann B, Pinto A, Sies H. High selenium intake and increased diabetes risk: experimental evidence for interplay between selenium and carbohydrate metabolism. J Clin Biochem Nutr 2011; 48:40-5; PMID:21297910; http://dx.doi.org/10.3164/jcbn.11-002FR
