Abstract
We aimed to determine the effect of SGI-110 on methylation and expression of the cancer testis antigens (CTAs) NY-ESO-1 and MAGE-A in epithelial ovarian cancer (EOC) cells in vitro and in vivo and to establish the impact of SGI-110 on expression of major histocompatibility (MHC) class I and Intracellular Adhesion Molecule 1 (ICAM-1) on EOC cells, and on recognition of EOC cells by NY-ESO-1-specific CD8+ T-cells. We also tested the impact of combined SGI-110 and NY-ESO-1-specific CD8+ T-cells on tumor growth and/or murine survival in a xenograft setting. EOC cells were treated with SGI-110 in vitro at various concentrations and as tumor xenografts with 3 distinct dose schedules. Effects on global methylation (using LINE-1), NY-ESO-1 and MAGE-A methylation, mRNA, and protein expression were determined and compared to controls. SGI-110 treated EOC cells were evaluated for expression of immune-modulatory genes using flow cytometry, and were co-cultured with NY-ESO-1 specific T-cell clones to determine immune recognition. In vivo administration of SGI-110 and CD8+ T-cells was performed to determine anti-tumor effects on EOC xenografts. SGI-110 treatment induced hypomethylation and CTA gene expression in a dose dependent manner both in vitro and in vivo, at levels generally superior to azacitidine or decitabine. SGI-110 enhanced the expression of MHC I and ICAM-1, and enhanced recognition of EOC cells by NY-ESO-1-specific CD8+ T-cells. Sequential SGI-110 and antigen-specific CD8+ cell treatment restricted EOC tumor growth and enhanced survival in a xenograft setting. SGI-110 is an effective hypomethylating agent and immune modulator and, thus, an attractive candidate for combination with CTA-directed vaccines in EOC.
Abbreviations
| AZA | = | Azacitidine (5-azacytidine) |
| CTA | = | Cancer-testis antigen or cancer-germline antigen |
| CTAG1B | = | Cancer/testis antigen 1B |
| DAC | = | Decitabine (5-aza-2′-deoxycitidine) |
| DNA | = | Deoxyribonucleic acid |
| DNMTi | = | DNA methyltransferase inhibitor |
| EOC | = | Epithelial ovarian cancer |
| HLA | = | Human leukocyte antigen |
| ICAM-1 | = | Intracellular Adhesion Molecule 1 |
| LINE-1 | = | Long interspersed nuclear element-1 |
| MAGE-A | = | Melanoma antigen family A |
| MHC | = | Major histocompatibility complex |
| NY-ESO-1 | = | New york esophageal squamous cell carcinoma 1 |
| RNA | = | Ribonucleic acid |
Introduction
Expression of cancer testis antigen (CTA) genes in epithelial ovarian cancer (EOC) has been extensively characterized.Citation1-3 Patients harboring EOCs that express CTA genes demonstrate humoral and cell mediated immune responses, which can be associated with prolonged survival.Citation1,4-8 A number of vaccine studies targeting the CTA gene product NY-ESO-1 have been undertaken in EOC in order to enhance such anti-tumor immunity.Citation9-12 Variable outcomes in these trials may result from the observation that in CTA-expressing cancers, gene expression is heterogeneous, with some regions of the tumor expressing the antigen robustly, while others do not.Citation1,2 Furthermore, cancers may down-regulate NY-ESO-1 expression during the course of vaccine therapy.Citation13 Tumor-associated expression of CTA genes is often mediated by promoter hypomethylation, which is also influenced by the genome wide DNA methylation status.Citation14-16 Further supporting a role for DNA methylation in CTA gene regulation in EOC cells is the observation that the DNMTi decitabine (DAC, 5-aza-2′-deoxycitidine) induces CTA gene expression.Citation17,18 Importantly, DAC treatment not only leads to CTA gene expression, but also induced protein expression and functional presentation of CTA epitopes on the surface of cancer cells.Citation19,20 Enhancement of CTA-directed immunogenicity by DAC likely also involves the induction of class I Human Leukocyte Antigens (HLA) and co-stimulatory molecules by this agent.Citation20-22
The above results have prompted considerable interest in the potential for clinical combination of CTA vaccine therapy with DNMTi.Citation22 DAC enhances expression of the CTA genes NY-ESO-1 and MAGE-A3/A6 in vitro and in patients; however, the drug is given intravenously and it has a short half-life due to rapid inactivation by cytidine deaminase.Citation23 The other FDA-approved DNMTi, 5-azacytidine (AZA), can have similar effects on gene expression and DNA methylation; however the incorporation of this drug predominantly into RNA (∼85%) complicates its in vivo mechanism of action; like DAC, it has a short half-life.Citation23 Although DAC and AZA have shown significant activity in patients with myeloid cancers, Phase I single agent studies in solid tumors were disappointing, likely due to the relatively slower growth rate of solid tumors and the short half-life of both drugs.Citation24-26 To overcome these pharmacokinetic limitations, a rationally designed novel dinucleotide comprised of deoxy-guanosine complexed through a phosphodiester linker to DAC was synthesized.Citation27 This compound, SGI-110, allows longer half-life and more extended DAC exposure than DAC intravenous infusion, and the agent appears to be at least as active as DAC in inducing CTA genes in vitro.Citation21 Importantly, SGI-110 is administered subcutaneously and has activity in DAC-resistant leukemic patients.Citation28 The effect of SGI-110 on CTA's and its role as an immune modulator has been studied in melanoma and leukemia cell lines, but has not been investigated in ovarian cancer cells.Citation19,21 In the current study, we assessed the activity of SGI-110 for a variety of CTA related measures, using EOC cell lines grown in vitro or in vivo as xenografts. We find that SGI-110 causes CTA promoter and global DNA hypomethylation, CTA mRNA and protein expression, and cell surface expression of key immune-modulatory genes. Furthermore, drug treatment results in CTA-specific CD8+ T-cell cytotoxicity in vitro, and restricts tumor growth in combination with antigen-specific T-cells in vivo. The data thus indicate that SGI-110 is an attractive candidate for combination with CTA-directed vaccines in EOC.
Results
SGI-110 treatment induces DNA hypomethylation and CTA mRNA and protein expression in EOC cell lines in vitro
To evaluate the effect of SGI-110 treatment on CTA methylation, OVCAR3 and A2780 EOC cell lines were treated with 0.1, 1.0 and 5 μM SGI-110 for 6 days as described in Materials and Methods. Cell viability was also assessed after SGI-110 treatment on day 6 to compare with DAC and AZA treatment (Fig. S1). DAC and AZA doses were chosen as they were comparable with the plasma levels in patients.Citation29 LINE-1 was used as a control for effects on global DNA methylation.Citation30 SGI-110 treatment resulted in a significant reduction of both LINE-1 and NY-ESO-1 promoter methylation, as assessed using quantitative bisulfite pyrosequencing (). As pyrosequencing assays were not feasible for the MAGE-A3/6 promoter (data not shown), we used MSP to analyze methylation of this locus. Similar to the other regions, MAGE-A3/6 was hypomethylated following SGI-110 treatment in both EOC cell lines (Fig. S2A). As expected, AZA and DAC treatment also resulted in hypomethylation of these regions (, S2A). To determine if hypomethylation of CTA genes by SGI-110 correlated with gene induction, we determined the mRNA and protein expression of NY-ESO-1 and MAGE-A. Both EOC cell lines demonstrated an increase in NY-ESO-1 and MAGE-A3/6 mRNA following SGI-110 treatment, and gene induction was greater than that observed with AZA or DAC (). Both EOC cell lines also showed marked induction of NY-ESO-1 and MAGE-A protein (). Notably, AZA was less potent at inducing CTA mRNA and protein expression, particularly in A2780 cells, although its effect on DNA methylation was similar. This could be due to the drug's off-target effects related to RNA incorporation.
Figure 1. SGI-110 treatment induces DNA hypomethylation and expression of NY-ESO-1 and MAGE-A3/6 in EOC cell lines in vitro. OVCAR3 and A2780 cells were treated with SGI-110, DAC and AZA for 6 days as described in the Materials and Methods. (A) LINE-1 and NY-ESO-1 promoter methylation were determined by bisulfite pyrosequencing. (B) NY-ESO-1 and MAGE-A3/6 mRNA were quantified by RT-qPCR. (C) NY-ESO-1 and MAGE-A protein expression were determined by western blotting. All experiments were repeated at least 3 times. *P < 0.05 vs. vehicle. S-SGI-110, V-Vehicle, DA-Decitabine, AZ-Azacitidine.
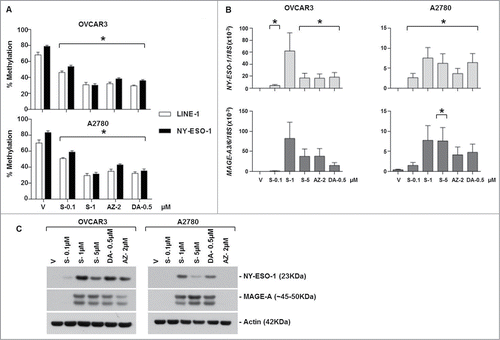
SGI-110 treatment induces DNA hypomethylation and CTA mRNA and protein expression in EOC xenografts using a daily x 5 days treatment schedule
We treated OVCAR3 xenograft-bearing SCID mice with a series of clinically relevant dosing schedules of SGI-110 or DAC (schedules tested in the Phase I/II trial for MDS or AML, see Materials and Methods).Citation31 We did not analyze AZA, as this drug was less potent in vitro in affecting CTA-related endpoints as compared to SGI-110 or DAC (). The effect of SGI-110 treatment on LINE-1, NY-ESO-1, and MAGE-A3/6 methylation was evaluated in excised OVCAR3 tumors as mentioned above. Groups 1 to 5 (see for Group description) were exposed subcutaneously to SGI-110 at doses of 3, 6.1, or 10 mg/kg, or DAC at 2.5 mg/kg, daily for 5 days and tumors were harvested on day 7. Due to differences in molecular weight, the molar equivalent of a 1mg dose of DAC is approximately 2.5 mg of SGI -110, thus the 6.1mg dose of SGI-110, approximates the 2.5 mg/kg DAC dose.Citation27 Mice treated on the 5 day schedule with SGI-110 at the 10 mg/kg/d dose developed significant gastrointestinal toxicity. Both DAC and SGI-110 treatment caused hypomethylation of LINE-1 and NY-ESO-1 at all doses (). MAGE-A3/6 hypomethylation was apparent at the 6.1 mg/kg SGI-110 dose (Fig. S2B). Tumors excised on day 7 demonstrated induction of NY-ESO-1 and MAGE-A3/A6 mRNA with SGI-110 treatment (). Robust induction of CTA protein expression following SGI-110 treatment was also observed, and appeared to be more robust than that seen with DAC treatment ().
Table 1. Treatment schedule for xenograft experiments
Figure 2. SGI-110 treatment induces hypomethylation and expression of NY-ESO-1 and MAGE-A3/6 in subcutaneous OVCAR3 xenografts treated using a daily x 5 schedule. SGI-110 at varying doses (3, 6.1,10 mg/kg) or DAC (2.5 mg/kg) was administered for 5 days, and tumors were harvested on day 7. (A) LINE-1 and NY-ESO-1 promoter methylation were determined by bisulfite pyrosequencing. (B) NY-ESO-1 and MAGE-A3/6 mRNA were quantified by RT-qPCR. C) NY-ESO-1 and MAGE-A protein expression were determined by western blotting. Data is representative of 3 animals/group. *P < 0.05 vs. vehicle; **P < 0.05 vs. DAC.
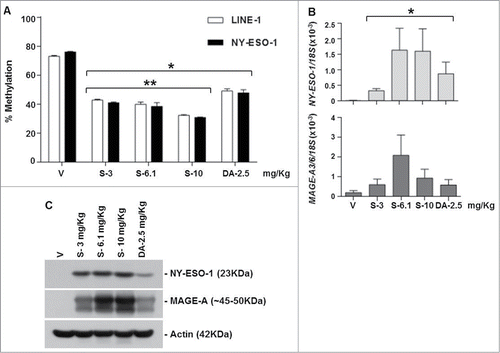
SGI-110 treatment induces DNA hypomethylation and CTA mRNA and protein expression in EOC xenografts using a weekly x 3 treatment schedule
Mice in treatment groups 6 to 10 received SGI-110 at 6.1, 12.2, or 24.4 mg/kg, or DAC at 5 mg/kg, once a week for 3 weeks. SGI-110 treatment resulted in significant LINE-1 and NY-ESO-1 hypomethylation on both days 7 and 16 post-treatment (). SGI-110 at the highest dose (24.4 mg/kg) led to the greatest hypomethylation, without apparent toxicity (data not shown). Using MSP, we evaluated MAGE-A3/6 promoter methylation and observed hypomethylation on both days 7 and 16 (Fig. S2C). This dosing schedule induced mRNA and protein expression of both CTA genes, particularly at the highest dose level ().
Figure 3. SGI-110 treatment induces hypomethylation and expression of NY-ESO-1 and MAGE-A3/6 in subcutaneous OVCAR3 xenografts treated using a weekly x 3 schedule. SGI-110 at varying doses (6.1, 12.2, 24.4 mg/kg) or DAC (5 mg/kg) was administered weekly for 3 weeks; tumors were harvested on days 7 and 16. (A) LINE-1 and NY-ESO-1 promoter methylation were determined by bisulfite pyrosequencing. (B) NY-ESO-1 and MAGE-A3/6 mRNA were determined by RT-qPCR. C) NY-ESO-1 and MAGE-A protein expression were determined by western blotting. Data represent 3 animals/group. *P < 0.05 vs. vehicle; **P < 0.05 vs. DAC.
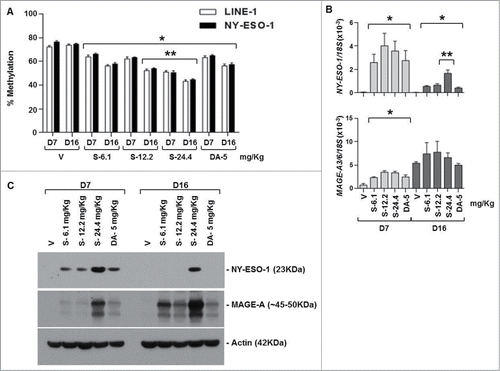
SGI-110 treatment induces DNA hypomethylation and CTA mRNA and protein expression in EOC xenografts using a twice weekly x 3 treatment schedule
Mice in groups 11 to 15 received SGI-110 at doses of 6.1, 12.2, 24.4 mg/kg, or DAC at 2.5 mg/kg, twice per week for 3 weeks, and tumors were harvested on days 7 and 16. Mice treated at the highest SGI-110 dose, 24.4mg/kg demonstrated significant morbidity by day 16 (data not shown). This schedule resulted in significant LINE-1 and NY-ESO-1 promoter hypomethylation at all SGI-110 doses (). Additionally, SGI-110 at the 12.2 (molar equivalent of 4.9 mg DAC) and 24.4 mg/kg (molar equivalent to 9.8 mg/kg DAC) doses resulted in significantly greater hypomethylation than the DAC treatment control (). MAGE-A3/6 hypomethylation was also apparent using this schedule of SGI-110. (Fig. S2D). Again, hypomethylation of CTA promoters resulted in CTA gene induction at both the mRNA and protein levels (). Specific SGI-110 treatment groups showed enhanced expression of CTA genes as compared to DAC. Unexpectedly, there was not a significant change in MAGE-A3/6 mRNA expression at day 7 (); however, MAGE-A protein expression was induced with all DNMTi treatments, and expression was sustained between days 7 and 16. This likely reflects the fact that the western blot assay detects many homologous MAGEA family members, while primers for MAGE A3/6 are relatively specific.Citation32
Figure 4. SGI-110 treatment induces hypomethylation and expression of NY-ESO-1 and MAGE-A3/6 in subcutaneous OVCAR3 xenografts treated using a twice weekly x 3 week schedule. SGI-110 at varying doses (6.1, 12.2, 24.4 mg/kg) or DAC (2.5 mg/kg) was administered twice per week for 3 weeks, and tumors were harvested on days 7 and 16. (A) LINE-1 and NY-ESO-1 promoter methylation were determined by bisulfite pyrosequencing. (B) NY-ESO-1 and MAGE-A3/6 mRNA levels were determined by RT-qPCR. C) NY-ESO-1 and MAGE-A protein expression by western blotting. Data represent 3 animals/group. *P < 0.05 vs. vehicle; **P < 0.05 vs. DAC.
SGI-110 treatment enhances cell surface expression of MHC class I and ICAM-1 in EOC cells
To determine whether SGI-110 treatment enhanced the expression of MHC class I, HLA-A, B, C (HLA-ABC) and/or the costimulatory molecule ICAM-1 in EOC cells, we treated A2780 and OVCAR3 cells in vitro and measured pan HLA-ABC and ICAM-1 using flow cytometry. We observed SGI-110 concentration-dependent upregulation of HLA-ABC and ICAM-1 (). In general, similar effects were observed following AZA and DAC treatment.
Figure 5. SGI-110 enhances HLA-ABC and ICAM-1 expression, and NY-ESO-1-specific CD8+ T cell immune recognition, in EOC cells treated in vitro. OVCAR3 and A2780 cells were treated with SGI-110, DAC, or AZA, and expression of (A) HLA-ABC and (B) ICAM-1 were determined by flow cytometry. Data represent the Log2 transformation of [median fluorescence intensity (MFI) of treatment / MFI of vehicle control]. (C) OVCAR3 cells were treated with SGI-110, DAC, or AZA at the concentrations shown. Following treatment, cells were cultured with NY-ESO-1 specific, HLA compatible CD8+ T cell clones derived from ovarian cancer patients and relative CD107a/b expression was determined. All experiments were repeated 3 independent times. *P < 0.05 vs. vehicle, NP-No peptide control.
![Figure 5. SGI-110 enhances HLA-ABC and ICAM-1 expression, and NY-ESO-1-specific CD8+ T cell immune recognition, in EOC cells treated in vitro. OVCAR3 and A2780 cells were treated with SGI-110, DAC, or AZA, and expression of (A) HLA-ABC and (B) ICAM-1 were determined by flow cytometry. Data represent the Log2 transformation of [median fluorescence intensity (MFI) of treatment / MFI of vehicle control]. (C) OVCAR3 cells were treated with SGI-110, DAC, or AZA at the concentrations shown. Following treatment, cells were cultured with NY-ESO-1 specific, HLA compatible CD8+ T cell clones derived from ovarian cancer patients and relative CD107a/b expression was determined. All experiments were repeated 3 independent times. *P < 0.05 vs. vehicle, NP-No peptide control.](/cms/asset/b875c389-f67f-47fd-b685-33f7f0eba69d/kepi_a_1017198_f0005_b.gif)
SGI-110 treatment promotes NY-ESO-1-specific CD8+ T cell recognition of EOC cells
To determine if SGI-110 mediated induction of NY-ESO-1 and immune-modulatory molecules results in enhanced immune recognition, we co-cultured NY-ESO-1-specific CD8+ T-cells with OVCAR3 cells pretreated with DNMTi, and measured CD107a and CD107b expression, an established measure of antigen-specific CD8+ T cell-mediated cytolytic capacity.Citation33 SGI-110 treatment led to a significant increase in CD107a/b staining in NY-ESO-1 specific CD8+ T-cells as compared to vehicle control (). A similar effect was observed with DAC treatment. These data indicate that SGI-110 treatment enhances NY-ESO-1 specific CD8+ T-cell recognition of HLA compatible tumor cells.
SGI-110 enhances NY-ESO-1-specific CD8+ T cell antitumor response in vivo
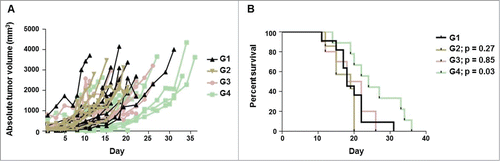
Finally, we sought to determine if the observed increase in CTA expression and other immune molecules translated into enhanced tumor control in vivo. For this task, SCID mice with established OVCAR3 xenografts were treated with SGI-110 and/or NY-ESO-1-specific CD8+ T-cells. As 3.0 mg/kg SGI-110 daily x 5 induced NY-ESO-1 expression in vivo without significant toxicity, we chose this dose for subcutaneous treatment of mice. Three days later, mice were injected intratumorally with NY-ESO-1-specific, HLA compatible, CD8+ T-cells or vehicle control (PBS). The combination of SGI-110 and NY-ESO-1 specific T-cells showed delayed tumor growth in comparison with mice treated with SGI-110 or NY-ESO-1-specific CD8 +T-cells alone (). This delayed tumor growth was associated with a statistically significant increase in survival for mice treated with the combination of SGI-110 and NY-ESO-1-specific CD8+ T-cells (). In these experiments, mice were sacrificed once the tumor diameter reached 2 cm and therefore longer survival reflects slower tumor growth. Together, these data suggest SGI-110 treatment enhances NY-ESO-1-specific antitumor responses in vivo.
Figure 6. SGI-110 treatment enhances NY-ESO-1-specific antitumor response in vivo. SCID mice bearing established OVCAR3 tumors were treated with 3.0 mg/kg/d x 5 days SGI-110 or vehicle control (diluent) subcutaneously and, 3 days later, injected with NY-ESO-1-specific CD8+ T-cells or vehicle control (PBS) intra-tumorally. In total, there were 4 treatment groups: G1, vehicle; G2, NY-ESO-1-specific CD8+ T-cells; G3, SGI-110 and G4, SGI-110 and NY-ESO-1-specific CD8+ T-cells. (A) Absolute tumor volume over time in which drug treatment began on day 1 and NY-ESO-1-specific CD8+ T-cells were injected on day 8 and (B) Kaplan-Meier survival curves. p value as compared to G1.
Discussion
We demonstrate herein that SGI-110 efficiently induces CTA promoter hypomethylation and gene expression in EOC cell lines in vitro. Furthermore we show that subcutaneous exposure of EOC tumor bearing mice to clinically relevant doses and schedules of SGI-110 leads to CTA promoter hypomethylation, mRNA, and protein expression. SGI-110 treatment also increases expression of HLA-ABC and ICAM-1, likely further promoting in vitro recognition of EOC cells by antigen specific CD8+ T-cell clones. Tumor-bearing SCID mice treated with SGI-110 and subsequently intra-tumorally injected with HLA-compatible NY-ESO-1 specific T-cell clones showed delayed tumor growth compared with SGI-110 treatment or CD8+ T-cells alone. Taken together, these data suggest that SGI-110 enhances immune recognition of cancer cells both by inducing the expression of tumor antigens and by upregulating MHC class I and immunomodulatory molecules. Interestingly, SGI-110 appeared to be more potent than DAC at inducing expression of the CTAs MAGE-A and NY-ESO-1, particularly at the protein level. All 3 of the tested in vivo administration schedules of SGI-110 (daily x 5; weekly x 3; twice weekly x 3) were efficient at causing CTA promoter hypomethylation and inducing CTA gene expression. All 3 schedules have been investigated in the recently completed Phase I/II clinical trial for AML and MDS, and the daily x 5 schedule has been taken forward; in contrast the weekly x 3 schedule produced minimal methylation changes and was discontinued.Citation32 In contrast, our current data support either of these schedules for potential combination with NY-ESO-1 vaccination in EOC patients. Prior reports supporting the notion that tumor specific T-cells and/or in combination with DAC can suppress tumor growth are consistent with our observations using SGI-110.Citation34,35 Additionally, Fang et al. recently reported that SGI-110 reduces ovarian tumors growth both alone and in combination with cisplatin, and can act as a chemosensitizer, demonstrating activity in other contexts for this agent in EOC.Citation36 Here we found that SGI-110 enhances NY-ESO-1 specific CD8+ T cell responses, and delayed tumor growth in a manor analogous to that reported in other contexts with DAC. Overall, these data have 2 major implications: i) induction of CTAs and related immune-modulatory activities should be considered as a potential mechanism of action of SGI-110 in ongoing trials of EOC and other solid tumors,Citation37 and ii) SGI-110 should be considered as a potential agent to augment the efficacy of CTA-directed vaccines in trials of EOC and other relevant tumor types, including melanoma and lung cancer.Citation21,38 The immunological effects of DNMTi are also supported by previous clinical studies of DAC.Citation39,40 In this context, we have recently shown in a Phase I trial that DAC combination therapy with NY-ESO-1 vaccine and liposomal doxorubicin is safe, promotes immunological responses, and may provide clinical benefit in EOC patients.Citation38 In addition to DNA methylation, CTA genes are regulated by other epigenetic mechanisms including histone acetylation, histone methylation, and nucleosome occupancy.Citation41-43 Thus, agents that impinge on these processes may also be useful for combination with CTA-directed vaccination strategies. In summary, the current data suggest that SGI-110 is a promising agent to incorporate into the spectrum of novel epigenetic/immunological approaches for EOC.
Materials and Methods
Cell lines and in vitro drug treatments
OVCAR3 and A2780 EOC cell lines were propagated in DMEM supplemented with 10% Fetal Bovine Serum (FBS), 2 mM L, and 1% Penicillin-Streptomycin. SGI-110 was provided by Astex Pharmaceuticals, Inc.. (Dublin, CA). AZA and DAC were obtained from Sigma. Cell lines were treated with SGI-110 at concentration 0.1, 1.0 and 5 μmol/L (in PBS) on day 2 and day 4, and harvested on day 6. As controls, cells were treated with vehicle alone, 0.5 μmol/L DAC, and 2.0 μmol/L AZA using the same treatment schedule as SGI-110. Trypan blue exclusion was used to evaluate cell viability (Fig. S1).
Xenograft experiments
A total of 1 × 106 OVCAR3 cells suspended in Matrigel (BD Biosciences) were implanted into the hindquarters of 6 week old female SCID mice using an IACUC-approved protocol. After ∼2–3 weeks, when macroscopic tumors were formed (∼50 mm3), mice were assigned into the different groups (). All treatment groups were exposed to vehicle, SGI-110 or DAC subcutaneously. For groups 1–5 a single tumor was implanted in one hindquarter and for groups 6–15 2 tumors were implanted, one in each hindquarter. Tumors were excised for molecular analysis on day 7 for Groups 1–5, and on days 7 and 16 for Groups 6–15.
DNA Methylation Analysis
Genomic DNA was isolated using the Puregene kit (Qiagen) and sodium bisulfite conversion was performed using the EZ DNA Methylation Kit (Zymo Research). NY-ESO-1 promoter and LINE-1 repetitive element DNA methylation were determined by sodium bisulfite pyrosequencing as described previously.Citation2 To evaluate MAGE-A3/6 promoter methylation, MSP was performed as previously described.Citation44 Primers were designed using MethPrimer.Citation45 Primer sequences were used as previously reported.Citation19 Unmethylated and methylated PCR products were amplified with initial denaturation at 95˚C for 5 min, then 35 cycles of denaturation at 95˚C for 30 s, annealing at 60˚C for 30 s and extension at 72˚C for 60 s, followed by a final 5 min extension at 72˚C. PCR products were analyzed on 2% agarose gel by ethidium bromide staining.
Reverse Transcriptase quantitative PCR (RT-qPCR)
Total RNA was purified using Trizol (Invitrogen) and cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad). Absolute quantification was performed using PCR Master Mix for SYBR Green assays (Eurogentech) on 7300 Real-Time PCR System (Applied Biosciences). All samples were run in triplicate, and NY-ESO-1 and MAGE-A-3/6 gene expression data were normalized to 18SrRNA. Primer sequences used were reported previously.Citation19
Western blot analysis
Total cellular protein was extracted from cell lines and/ or tissue samples using NP-40 lysis buffer and quantitated, and protein expression for NY-ESO-1 and MAGE-A was evaluated as described previously.Citation19 Figure S3 shows representative blots with molecular weight of the proteins studied along with molecular weight marker.
Flow cytometry
Cells were harvested and stained with anti-human HLA-ABC (clone W6/32) or anti-human CD54 (alternatively known as ICAM-1, clone HA58) antibodies (eBioscience) and changes in the expression of HLA-ABC and ICAM-1 following DNMTi treatment was determined as described previously.Citation19 Representative gating is shown in Figure S4.
NY-ESO-1-specific CD8+ T cell recognition assay
NY-ESO-1-specific HLA-A2-restricted CD8+ T-cells were prepared as described previouslyCitation11, and HLA-A2 positive OVCAR3 cells were treated with AZA, DAC, or varying doses of SGI-110 as described above. Following treatment, cells were co-cultured for 6 hours at 37°C in the presence of anti-CD107a and CD107b antibodies (BD Biosciences), monensin, and brefeldin A (Sigma). Negative control stimulations with no peptide and positive control stimulations with peptide (NY-ESO-1157-165) or phorbol myristate acetate and ionomycin were analyzed in parallel. Using NY-ESO-1-specific CD8+ T-cells (identified as tetramer+ CD8+ cells), CD107a/b+ was determined and normalized to positive control which served as an internal control from each independent experiment.
NY-ESO-1-specific CD8 +T cell antitumor response
OVCAR3 cells (1 × 106) suspended in PBS were subcutaneously implanted into the flanks of 5 week old female SCID mice using an IACUC-approved protocol. After ∼2–3 weeks, when macroscopic tumors formed, mice were treated daily with either 3.0 mg/kg/d SGI-110 or vehicle control (diluent) and, 3 days later, intra-tumorally injected with 2 × 106 NY-ESO-1-specific CD8 +T-cells or vehicle control (PBS). In total, there were 4 treatment groups (which included at least 7 mice/group): G1, vehicle (diluent and PBS); G2, NY-ESO-1-specific CD8+ T-cells (diluent and NY-ESO-1-specific CD8+ T-cells); G3, SGI-110 (SGI-110 and PBS) and G4, SGI-110 and NY-ESO-1-specific CD8 +T-cells. Tumors were measured according to the shortest (A) and longest diameter (B) to determine tumor volume (V) in which V = (A2B)/2. Survival was determined on the basis of tumor diameter. If a tumor reached 2cm in diameter, the mouse was euthanized.
Statistical analysis
Statistical analysis was performed by analysis of variance using GraphPad Prism. Differences between the 2 groups were analyzed using the t test. All data are presented as mean ± standard error. Difference between HLA-ABC and ICAM1 expression fold change was analyzed using one sample t test. The statistical significance of Kaplan-Meier survival curves was determined using log-rank (Mantel-Cox) test.
Disclosure of Conflicts of Interest
EAG and ARK received research funding from Astex Pharmaceuticals. EAG is a principal investigator on the Phase I/II clinical trial of SGI-110 sponsored by Astex Pharmaceuticals and has received honoraria from Celgene, Inc.. P.T. is an employee of Astex Pharmaceuticals. Other authors have no conflicts to disclose.
Supplemental_Material.zip
Download Zip (490.7 KB)Acknowledgments
The authors would like to thank Drs. Michael Moser from the animal core facility at Roswell Park Cancer Institute and Dr. Michael Nemeth in the Department of Immunology at the Roswell Park Cancer Institute for helpful assistance.
Funding
BEP is supported by the Ruth L. Kirschstein National Research Service Award Institutional Training Grant (T32 CA009072). A.R.K. is supported by the Betty J. and Charles D. McKinsey Ovarian Cancer Research Fund and the Otis Glebe Medical Research Foundation.
Supplemental Material
Supplemental data for this article is available on the publisher's website.
References
- Odunsi K, Jungbluth AA, Stockert E, Qian F, Gnjatic S, Tammela J, Intengan M, Beck A, Keitz B, Santiago D, et al. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res 2003; 63:6076-83; PMID:14522938
- Woloszynska-Read A, Mhawech-Fauceglia P, Yu J, Odunsi K, Karpf AR. Intertumor and intratumor NY-ESO-1 expression heterogeneity is associated with promoter-specific and global DNA methylation status in ovarian cancer. Clin Cancer Res 2008; 14:3283-90; PMID:18519754; http://dx.doi.org/10.1158/1078-0432.CCR-07-5279
- Imura M, Yamashita S, Cai LY, Furuta J, Wakabayashi M, Yasugi T, Ushijima T. Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 ovarian cancer cell lines. Cancer Lett 2006; 241:213-20; PMID:16303245; http://dx.doi.org/10.1016/j.canlet.2005.10.010
- Gnjatic S, Atanackovic D, Jager E, Matsuo M, Selvakumar A, Altorki NK, Maki RG, Dupont B, Ritter G, Chen YT, et al. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: Correlation with antibody responses. Proc Natl Acad Sci U S A 2003; 100:8862-7; PMID:12853579; http://dx.doi.org/10.1073/pnas.1133324100
- Stone B, Schummer M, Paley PJ, Thompson L, Stewart J, Ford M, Crawford M, Urban N, O'Briant K, Nelson BH. Serologic analysis of ovarian tumor antigens reveals a bias toward antigens encoded on 17q. Int J Cancer 2003; 104:73-84; PMID:12532422; http://dx.doi.org/10.1002/ijc.10900
- Huarte E, Karbach J, Gnjatic S, Bender A, Jager D, Arand M, Atanackovic D, Skipper J, Ritter G, Chen YT, et al. HLA-DP4 expression and immunity to NY-ESO-1: Correlation and characterization of cytotoxic CD4+ CD25- CD8- T cell clones. Cancer Immun 2004; 4:15; PMID:15600300
- Qian F, Gnjatic S, Jager E, Santiago D, Jungbluth A, Grande C, Schneider S, Keitz B, Driscoll D, Ritter G, et al. Th1/Th2 CD4+ T cell responses against NY-ESO-1 in HLA-DPB1*0401/0402 patients with epithelial ovarian cancer. Cancer Immun 2004; 4:12; PMID:15521719
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A 2005; 102:18538-43; PMID:16344461; http://dx.doi.org/10.1073/pnas.0509182102
- Odunsi K, Qian F, Matsuzaki J, Mhawech-Fauceglia P, Andrews C, Hoffman EW, Pan L, Ritter G, Villella J, Thomas B, et al. Vaccination with an NY-ESO-1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc Natl Acad Sci U S A 2007; 104:12837-42; PMID:17652518; http://dx.doi.org/10.1073/pnas.0703342104
- Karbach J, Gnjatic S, Bender A, Neumann A, Weidmann E, Yuan J, Ferrara CA, Hoffmann E, Old LJ, Altorki NK, et al. Tumor-reactive CD8+ T-cell responses after vaccination with NY-ESO-1 peptide, CpG 7909 and montanide ISA-51: Association with survival. Int J Cancer 2010; 126:909-18; PMID:19728336; http://dx.doi.org/10.1002/ijc.24850
- Odunsi K, Matsuzaki J, Karbach J, Neumann A, Mhawech-Fauceglia P, Miller A, Beck A, Morrison CD, Ritter G, Godoy H, et al. Efficacy of vaccination with recombinant vaccinia and fowlpox vectors expressing NY-ESO-1 antigen in ovarian cancer and melanoma patients. Proc Natl Acad Sci U S A 2012; 109:5797-802; PMID:22454499; http://dx.doi.org/10.1073/pnas.1117208109
- Sabbatini P, Tsuji T, Ferran L, Ritter E, Sedrak C, Tuballes K, Jungbluth AA, Ritter G, Aghajanian C, Bell-McGuinn K, et al. Phase I trial of overlapping long peptides from a tumor self-antigen and poly-ICLC shows rapid induction of integrated immune response in ovarian cancer patients. Clin Cancer Res 2012; 18:6497-508; PMID:23032745; http://dx.doi.org/10.1158/1078-0432.CCR-12-2189
- Nicholaou T, Chen W, Davis ID, Jackson HM, Dimopoulos N, Barrow C, Browning J, Macgregor D, Williams D, Hopkins W, et al. Immunoediting and persistence of antigen-specific immunity in patients who have previously been vaccinated with NY-ESO-1 protein formulated in ISCOMATRIX. Cancer Immunol Immunother 2011; 60:1625-37; PMID:21698545; http://dx.doi.org/10.1007/s00262-011-1041-3
- De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol 2004; 24:4781-90; PMID:15143172; http://dx.doi.org/10.1128/MCB.24.11.4781-4790.2004
- Akers SN, Odunsi K, Karpf AR. Regulation of cancer germline antigen gene expression: Implications for cancer immunotherapy. Future Oncol 2010; 6:717-32; PMID:20465387; http://dx.doi.org/10.2217/fon.10.36
- Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem 2006; 281:10118-26; PMID:16497664; http://dx.doi.org/ M510469200 [pii]
- Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, Treisman J, Rosenberg SA. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res 1994; 54:1766-71; PMID:7511051
- Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol 2004; 65:18-27; PMID:14722233; http://dx.doi.org/10.1124/mol.65.1.18
- Srivastava P, Paluch BE, Matsuzaki J, James SR, Collamat-Lai G, Karbach J, Nemeth MJ, Taverna P, Karpf AR, Griffiths EA. Immunomodulatory action of SGI-110, a hypomethylating agent, in acute myeloid leukemia cells and xenografts. Leuk Res 2014; 38:1332-41; PMID:25260825; http://dx.doi.org/10.1016/j.leukres.2014.09.001
- Adair SJ, Hogan KT. Treatment of ovarian cancer cell lines with 5-aza-2′-deoxycytidine upregulates the expression of cancer-testis antigens and class I major histocompatibility complex-encoded molecules. Cancer Immunol Immunother 2009; 58:589-601; PMID:18791715; http://dx.doi.org/10.1007/s00262-008-0582-6
- Coral S, Parisi G, Nicolay HJ, Colizzi F, Danielli R, Fratta E, Covre A, Taverna P, Sigalotti L, Maio M. Immunomodulatory activity of SGI-110, a 5-aza-2′-deoxycytidine-containing demethylating dinucleotide. Cancer Immunol Immunother 2013; 62:605-14; PMID:23138873; http://dx.doi.org/10.1007/s00262-012-1365-7
- Karpf AR. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics 2006; 1:116-20; PMID:17786175
- Griffiths EA, Gore SD. Epigenetic therapies in MDS and AML. Adv Exp Med Biol 2013; 754:253-83; PMID:22956506; http://dx.doi.org/10.1007/978-1-4419-9967-2_13
- Samlowski WE, Leachman SA, Wade M, Cassidy P, Porter-Gill P, Busby L, Wheeler R, Boucher K, Fitzpatrick F, Jones DA, et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: Inhibition of promoter-specific and global genomic DNA methylation. J Clin Oncol 2005; 23:3897-905; PMID:15753459; http://dx.doi.org/ JCO.2005.06.118 [pii]
- Sessa C, Suinink W, Stoter G, Renard J, Cavalli F. Phase II study of 5-aza-2′-deoxycytidine in advanced ovarian carcinoma. the EORTC early clinical trials group. Eur J Cancer 1990; 26:137-8; PMID:1691012
- Bellet RE, Catalano RB, Mastrangelo MJ, Berd D. Phase II study of subcutaneously administered 5-azacytidine (NSC-102816) in patients with metastatic malignant melanoma. Med Pediatr Oncol 1978; 4:11-5; PMID:75498
- Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res 2007; 67:6400-8; PMID:17616700; http://dx.doi.org/ 67/13/6400 [pii]
- Griffiths EA, Choy G, Redkar S, Taverna P, Azab M, Karpf AR. SGI-110. DNA methyltransferase inhibitor, oncolytic. Drugs Fut 2013; 38:535.
- Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer 2008; 123:8-13; PMID:18425818; http://dx.doi.org/10.1002/ijc.23607
- Woloszynska-Read A, Zhang W, Yu J, Link PA, Mhawech-Fauceglia P, Collamat G, Akers SN, Ostler KR, Godley LA, Odunsi K, et al. Coordinated cancer germline antigen promoter and global DNA hypomethylation in ovarian cancer: Association with the BORIS/CTCF expression ratio and advanced stage. Clin Cancer Res 2011; 17:2170-80; PMID:21296871; http://dx.doi.org/10.1158/1078-0432.CCR-10-2315
- Kantarjian H, Roboz G, Rizzieri D, Stock W, O’Connell C, Griffiths EA, Yee K, Tibes R, Garcia-Manero G, Ravandi F, et al. Results from the dose escalation phase of a randomized phase 1–2 first-in-human (FIH) study of SGI-110, a novel low volume stable subcutaneous (SQ) second generation hypomethylating agent (HMA) in patients with relapsed/refractory MDS and AML. Blood 2012; 120:414
- De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics 1994; 40:360-9; PMID:7927540
- Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods 2003; 281:65-78; PMID:14580882
- Oflazoglu E, Elliott M, Takita H, Ferrone S, Henderson RA, Repasky EA. Adoptively transferred human lung tumor specific cytotoxic T cells can control autologous tumor growth and shape tumor phenotype in a SCID mouse xenograft model. J Transl Med 2007; 5:29; PMID:17592641; http://dx.doi.org/ 1479-5876-5-29 [pii]
- Natsume A, Wakabayashi T, Tsujimura K, Shimato S, Ito M, Kuzushima K, Kondo Y, Sekido Y, Kawatsura H, Narita Y, et al. The DNA demethylating agent 5-aza-2′-deoxycytidine activates NY-ESO-1 antigenicity in orthotopic human glioma. Int J Cancer 2008; 122:2542-53; PMID:18240144; http://dx.doi.org/10.1002/ijc.23407
- Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, Pilrose J, Choy G, Azab M, Pawelczak KS, et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin Cancer Res 2014; 20:6504-16; PMID:25316809; http://dx.doi.org/10.1158/1078-0432.CCR-14-1553
- Clinicaltrials.gov. A randomized, controlled, open-label, phase 2 trial of SGI-110 and carboplatin in subjects with platinum-resistant recurrent ovarian cancer: NCT01696032. 2014; 2014
- Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, Zhang W, Akers SN, Griffiths EA, Miliotto A, et al. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer Immunol Res 2014; 2:37-49
- Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res 2012; 72:2197-205; PMID:22549947; http://dx.doi.org/10.1158/0008-5472.CAN-11-3909
- Schrump DS, Fischette MR, Nguyen DM, Zhao M, Li X, Kunst TF, Hancox A, Hong JA, Chen GA, Pishchik V, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res 2006; 12:5777-85; PMID:17020984; http://dx.doi.org/10.1158/1078-0432.CCR-06-0669
- Link PA, Gangisetty O, James SR, Woloszynska-Read A, Tachibana M, Shinkai Y, Karpf AR. Distinct roles for histone methyltransferases G9a and GLP in cancer germ-line antigen gene regulation in human cancer cells and murine embryonic stem cells. Mol Cancer Res 2009; 7:851-62; PMID:19531572; http://dx.doi.org/10.1158/1541-7786.MCR-08-0497
- Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res 2006; 4:339-49; PMID:16687489; http://dx.doi.org/ 4/5/339 [pii]
- James SR, Cedeno CD, Sharma A, Zhang W, Mohler JL, Odunsi K, Wilson EM, Karpf AR. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epigenetics 2013; 8:849-63; PMID:23839233; http://dx.doi.org/10.4161/epi.25500
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821-6; PMID:8790415
- Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002; 18:1427-31; PMID:12424112
