 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Polycomblike (Pcl) proteins are important transcriptional regulators and components of the Polycomb Repressive Complex 2 (PRC2). The Tudor domains of human homologs PHF1 and PHF19 have been found to recognize trimethylated lysine 36 of histone H3 (H3K36me3); however, the biological role of Tudor domains of other Pcl proteins remains poorly understood. Here, we characterize the molecular basis underlying histone binding activities of the Tudor domains of the Pcl family. In contrast to a predominant view, we found that the methyl lysine-binding aromatic cage is necessary but not sufficient for recognition of H3K36me3 by these Tudor domains and that a hydrophobic patch, adjacent to the aromatic cage, is also required.
Introduction
Polycomblike (Pcl) proteins are essential components of the Polycomb Repressive Complex 2 (PRC2) that silences genes during development and lineage specification.Citation1 The PRC2 complex methylates lysine 27 of histone H3, producing mono-, di- and tri-methylated H3K27me1/2/3 marks that generally correlate with transcriptionally repressed chromatin.Citation2-5 Impaired PRC2 activity is associated with defects in cell differentiation and growth and can lead to cellular transformation.Citation6
Pcl was originally identified in Drosophila melanogaster as a gene required for the development of body segments in the fly.Citation7 Its product, the Pcl protein, was found to be essential in the generation of H3K27me3 at Polycomb target genes; however, is dispensable for the deposition of H3K27me1/2 marks genome-wide.Citation8 The human genome encodes 3 Pcl orthologs - plant homeodomain (PHD) finger containing protein 1 (PHF1), metal response element binding transcription factor 2 (MTF2), and PHF19. PHF1 modulates PRC2 enzymatic activity and is implicated in DNA damage repair and maintenance of genomic stability.Citation9-12 MTF2 is involved in facilitating PRC2 recruitment to the inactive X-chromosome and, much like PHF19, functions in the transcriptional control of genes implicated in embryonic stem cell renewal and differentiation.Citation13-16 The Pcl family proteins are characterized by similar domain architecture consisting of a Tudor domain followed by 2 PHD fingers. Although the precise function of the PHD fingers remains poorly understood, the region encompassing both domains in Pcl and PHF1 was shown to associate with the catalytic subunit of PRC2, EZH2.Citation17
The Tudor domains of PHF1 and PHF19 have recently been found to recognize trimethylated lysine 36 of histone H3 (H3K36me3).Citation12,14-16,18 The PHF1-H3K36me3 interaction impedes the enzymatic activity of PRC2 and is important for the retention of PHF1 at the sites of DNA damage.Citation12 Binding of PHF19 to H3K36me3 recruits PRC2 and NO66 to embryonic stem cell genes during differentiation and is required for the full enzymatic activity of PRC2 and repression of a subset of PRC2 target genes.Citation14-16 Additionally, it has been demonstrated by a peptide pull-down assay that the Tudor domain of MTF2 associates with H3K36me3, whereas the Tudor domain of Pcl does not.Citation14
Here, we detail the molecular basis underlying histone binding activities of the Tudor domains of the Pcl family and provide explanation for the lack of functional conservation among the members. In contrast to a prevalent view, we found that the methyl lysine-binding aromatic cage is necessary but not sufficient for recognition of H3K36me3 by these Tudor domains and that a hydrophobic patch, adjacent to the aromatic cage, is also required.
Results and Discussion
To characterize the biological function of the Tudor domains of PHF1, Pcl and MTF2, we examined histone binding activities of these proteins using NMR titration experiments. We collected 1H, 15N heteronuclear single quantum coherence (HSQC) spectra of the uniformly 15N-labeled Tudor domains of PHF1, Pcl and MTF2 while gradually adding H3K36me3 peptide (aa 31–40 of H3) to the NMR samples (). Large chemical shift perturbations (CSPs) were observed in the spectra of the PHF1 Tudor domain, revealing direct interaction between the protein and the peptide (, left panel). However titration of the H3K36me3 peptide into the Pcl Tudor domain caused no CSPs even at the protein:peptide ratio of 1:5, indicating that Pcl does not recognize H3K36me3 (, middle panel). Addition of the H3K36me3 peptide to the MTF2 Tudor-PHD1 construct induced substantial CSPs in the protein, although the pattern of CSPs (intermediate-to-fast exchange regime on the NMR time scale) suggested that binding of MTF2 is weaker as compared to the binding of PHF1 (, right panel). Of note, we used the Tudor-PHD1 construct of MTF2 in this study because an isolated Tudor is less soluble; however, as shown in Supplementary Figure S1, the Tudor domain functions independently of the neighboring PHD1 finger and exhibits almost identical CSPs upon addition of H3K36me3 peptide, either without PHD1 or being linked to PHD1.
Figure 1. Binding to histone H3K36me3 is not conserved in the Pcl family. (A) Superimposed 1H, 15N HSQC spectra of PHF1 Tudor, Pcl Tudor, and MTF2 Tudor-PHD1 collected upon titration with H3K36me3 peptide. Spectra are color coded according to the protein:peptide molar ratio. (B) Sequence alignment of the Tudor domains of PHF1, MTF2, Pcl and PHF19: absolutely, moderately and weakly conserved residues are colored pink, wheat, and blue, respectively. The aromatic cage residues are highlighted with red ovals. The aromatic residues of PHF1 are labeled. (C) Structural overlays of the PHF1 Tudor bound to H3K36me3 peptide (modeled as sticks in yellow) (PDB 4HCZ) and either the apo- Pcl Tudor on the left (PDB 2XK0) or the apo- MTF2 Tudor on the right (PDB 2EQJ). The aromatic cage residues of PHF1 Tudor are in brick red, while the corresponding Pcl and MTF2 residues are in purple and gray, respectively. (D) Topology of the aromatic cage in the Pcl family of proteins: the variant residues of Pcl and MTF2 are colored red.
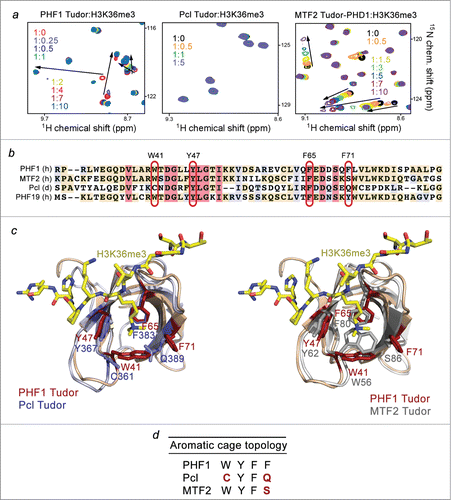
We have previously determined the crystal structure of the PHF1 Tudor domain in complex with H3K36me3 peptide.Citation12 In the complex, the peptide adopts an extended conformation and lays across the open edge of the Tudor's β-barrel. The extended side chain of K36me3 occupies the aromatic cage formed by the Y47, W41, F65 and F71 residues of the protein. The side chains of these aromatic residues are positioned roughly perpendicular to each other and are engaged in cation-π and hydrophobic interactions with the trimethyl ammonium group of K36. The aromatic cage-derived mechanism is utilized by the majority of methyl lysine-binding modules, including chromodomain, MBT, PHD, PWWP and WD40, and thus became a fundamental concept in the epigenetics field (for a review see refs.Citation19,20).
Alignment of the amino acid sequences of the Pcl, PHF1, MTF2, and PHF19 Tudor domains shows that the aromatic cage of Pcl contains 2 aromatic residues, Y367 and F383, but lacks other 2 aromatic residues, corresponding to W41 and F71 of PHF1 (). Structural overlay of the H3K36me3-bound Tudor domain of PHF1 Citation12 and the Pcl Tudor domain in the apo-state Citation21 reveals that C361 and Q389 residues make 2 sides of the aromatic cage in Pcl (). The MTF2 Tudor domain has 3 aromatic residues in the cage, W56, Y62 and F80, with the fourth aromatic residue, corresponding to F71 in PHF1, being replaced by S86 (). All four aromatic residues of the PHF1 Tudor domain are conserved in PHF19, which has been shown to robustly interact with H3K36me3.Citation14-16,18 We reasoned that the incomplete aromatic cages in Pcl and MTF2 prevent and reduce, respectively, binding of these proteins to H3K36me3.
We tested whether the restoration of the aromatic cage would enable binding of Pcl Tudor to H3K36me3 through mutating C361 to a tryptophan and Q389 to a phenylalanine. As shown in , amide resonances of neither C361W mutant nor Q389F mutant of Pcl Tudor were perturbed upon addition of H3K36me3, implying that the individual substitutions are insufficient. Furthermore, the double C361W/Q389F mutant of Pcl, in which the aromatic cage fully mimics the aromatic cage of the PHF1 Tudor domain, failed to interact with H3K36me3 (, right panel). Together these results demonstrate that while required, the intact aromatic cage is not sufficient for the recognition of H3K36me3.
Figure 2. The aromatic cage is necessary but not sufficient for Pcl Tudor to bind H3K36me3. (A) Superimposed 1H, 15N HSQC spectra of indicated Pcl Tudor mutants color coded according to the protein:H3K36me3 peptide molar ratio. (B) Structure of the PHF1 Tudor domain bound to H3K36me3 peptide (yellow sticks) (PDB 4HCZ) with the aromatic cage residues in red and the hydrophobic patch residues in pale green. Electrostatic surface potential of the PHF1 Tudor domain is shown on the right with acidic and basic surfaces colored red and blue, respectively. (C) Structure of apo- Pcl Tudor (PDB 2XK0), with the aromatic cage and hydrophobic patch residues colored blue and mustard yellow, respectively. Electrostatic surface potential of the Pcl Tudor domain is shown on the right. (D) Overlaid 1H, 15N HSQC spectra of the indicated mutants of Pcl Tudor recorded in the presence of increasing concentrations of H3K36me3. (E) Representative binding isotherms used to determine binding affinity of C361W/R365L/Q389F Pcl Tudor for H3K36me3 via normalized CSPs.
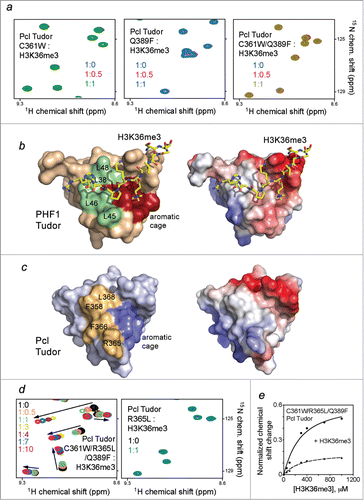
To identify additional elements that are necessary for the interaction with H3K36me3, we examined the histone-binding site of PHF1 beyond the aromatic cage. Notably, the PHF1 Tudor domain has an extensive hydrophobic patch, located next to the aromatic cage and composed of 4 solvent exposed leucine residues (colored pale green in ). This patch forms a binding site for the hydrophobic side chain of P38 and the neutral side chain of H39 of the peptide in the PHF1 Tudor-H3K36me3 complex. In contrast, the Pcl Tudor domain contains a positively charged R365 and 2 bulky albeit hydrophobic F358 and F366 residues in place of 3 leucine residues of PHF1 (colored mustard yellow in ). We generated a triple mutant of Pcl, C361W/R365L/Q389F, in which R365 is substituted with a leucine, and assayed its binding by NMR. Titration of the H3K36me3 peptide into the 15N-labeled C361W/R365L/Q389F Pcl Tudor domain induced substantial CSPs, indicative of a robust interaction of this gain-of-function mutant (, left panel). Plotting normalized CSPs vs. peptide concentration for each protein amide yielded binding curves and a Kd of 230 µM ( and ). The fact that the single R365L mutant of Pcl was incapable of binding to H3K36me3 reinforced the idea that the aromatic cage is absolutely required (, right panel).
Figure 3. The histone H3K36me3 binding activity is fully restored in the F61L/S86F mutant of MTF2 Tudor-PHD1. (A) Binding affinities of the Tudor domains as measured by tryptophan fluorescence (a) or NMR (b). (B) Representative binding curves used to determine the Kd values by fluorescence. (C) Superimposed 1H, 15N HSQC spectra of the wild type and mutated MTF2 Tudor-PHD1 color coded according to the protein:H3K36me3 peptide molar ratio. (D) Overlays of the aromatic cage residues of PHF1 Tudor (brick red) (PDB 4HCZ), MTF2 Tudor (gray) (PDB 2EQJ), and ING2 PHD (green) (2G6Q). Trimethylated lysine 36 from the PHF1 Tudor-H3K36me3 complex and trimethylated lysine 4 from the ING2 PHD-H3K4me3 complex are shown in yellow and green, respectively. (E) Superimposed 1H, 15N HSQC spectra of the S86Q mutant of MTF2 Tudor-PHD1 color coded according to the protein:H3K36me3 peptide molar ratio. (F) Structure of apo- MTF2 Tudor (PDB 2EQJ), with the aromatic cage residues colored dark gray. The hydrophobic patch residues, leucines and a phenylalanine, are colored wheat and pale green, respectively.
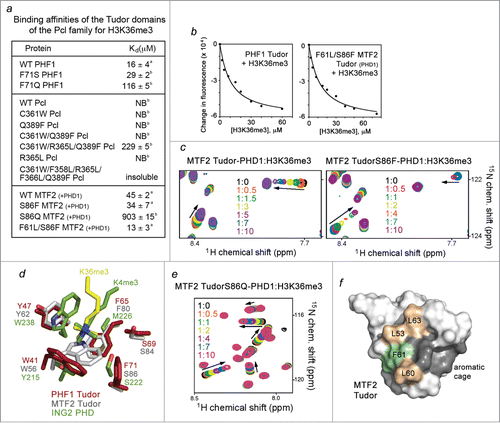
Despite the high sequence similarity of the Tudor domains of PHF1 and MTF2, association of MTF2 with H3K36me3 was found to be ∼ 3-fold weaker. We measured binding affinities of MTF2 Tudor-PHD1 for H3K36me3 (Kd = 45 µM) and of the PHF1 Tudor domain for H3K36me3 (Kd = 16 µM) using fluorescence spectroscopy (). Interestingly, the aromatic cage and the hydrophobic patch of MTF2 and PHF1 each differ by one amino acid. The aromatic cage of MTF2 contains S86 in place of the F71 residue in PHF1. We assessed the histone binding ability of the S86F mutant of MTF2 Tudor-PHD1 by NMR titration experiments. An intermediate exchange regime observed for the interaction of the S86F mutant pointed to an enhanced binding as compared to binding of the wild type protein (), and this was supported by a Kd of 34 µM measured using tryptophan fluorescence (). Together these data demonstrate a critical role of the intact 4-aromatic residue cage of the Tudor domain in the interaction with H3K36me3.
We note that a serine residue is also present in the aromatic cage of the ING2 PHD finger that recognizes H3K4me3 with high specificity and affinity.Citation22 The 2 most important for the binding aromatic residues of ING2 superimpose well with W41 and Y47 of MTF2; however, the methyl lysine-binding cage in MTF2 is wider, with S86 positioned further from the trimethylammonium group than the serine residue is positioned in ING2 (). The greater distance may account for the less pronounced effect of S86 on the interaction, still mutation of this residue to a Pcl−like glutamine further diminished binding of S86Q MTF2 Tudor-PHD1 (). In the PHF1 Tudor-H3K36me3 complex the aromatic side chain of F71 is slightly rotated, most likely contributing more to the hydrophobic contacts and less to the cation-π contacts. Nevertheless, the presence of an aromatic residue at this position is important, because binding affinities of the F71S (MTF2-like) and F71Q (Pcl−like) mutants of PHF1 decreased ∼ 2- and 7-fold, respectively ( and Fig. S2).
One of the leucine residues in the 4-leucine hydrophobic patch of PHF1 is replaced by a phenylalanine, F61, in MTF2. To examine the effect of this residue, we produced the F61L/S86F mutant of Tudor-PHD1, substituting F61 with a leucine and fully recreating the PHF1-like binding site. As we predicted, the F61L/S86F mutant of MTF2 Tudor-PHD1 was able to bind to H3K36me3 as strong as the PHF1 Tudor bound to this PTM (Kd = 13 µM, ). We concluded that the hydrophobic patch plays an essential role in binding of these Tudors to methylated chromatin. Furthermore, the physiological relevance of this patch is underscored by the fact that L60 of MTF2 (which corresponds to R365 in Pcl) is found mutated to a phenylalanine in head and neck cancer (cBioportal).
The mechanism for the H3K36me-P38-H39 readout by the Tudor domain of the Pcl family differs from the binding mechanisms employed by other H3K36me-recognizing modules, including PWWP of BRPF1 and the EAF6 chromobarrel domain (Fig. S3).Citation23,24 However the important contribution of the hydrophobic or negatively-charged pockets adjacent to the aromatic cage to the association with methyl lysine-containing histone peptides has been reported for WD40 domain of EED, Tandem Tudor Domain of JMJD2A, and a number of PHD fingers.Citation25-28 In conclusion, our data reveal a critical role of the hydrophobic patch in the interaction of the Pcl family Tudors with H3K36me3. These findings underscore that while the aromatic cage is essential, it is not the only determinant in fine-tuned recognition of epigenetic marks.
Materials and Methods
Cloning and protein purification
The MTF2 (aa 40–101 and aa 40–155) constructs were cloned from full-length cDNA (Open Biosystems) into pDEST15 using Gateway® cloning technology. The PHF1 (aa 14–87) construct in pGex6P1 was described previously.Citation12 dPcl (aa 339–405) construct in pGex4T3 was a kind gift from Luciano Di Croce. Point mutants were generated using the Stratagene QuickChange XL Site Directed Mutagenesis kit. Wild type and mutant proteins were expressed in Rosetta2 (DE3) pLysS or BL21 (DE3) RIL in either Luria Broth or M19 minimal media supplemented with 50 μM ZnCl2 (for MTF2 Tudor-PHD1) and 15NH4Cl. Cells were induced with 0.5 mM IPTG, grown for 16 h at 18°C, and harvested by centrifugation at 6k rpm. Cells were lysed by sonication in lysis buffer containing 25 mM Tris pH 7.5 at 4°C, 150 mM NaCl, 3 mM dithiothreitol, 0.5 % Triton X-100, 1 mM PMSF, 5 mM MgCl2, DNaseI and clarified by centrifugation at 15k rpm for 30 min. Proteins were purified on glutathione agarose beads (Pierce®cat# 16101) and the GST-tag cleaved using either PreScission or Thrombin protease for at least overnight at 4°C. Cleaved proteins were concentrated into 25 mM Tris pH 7.5, 150 mM NaCl, and 3 mM dithiothreitol. Unlabeled proteins were purified by size exclusion chromatography using a HiPrep 16/60 Sephacryl S-100 column.
Nuclear Magnetic Resonance
Experiments were collected on a Varian INOVA 600 MHz spectrometer at the University of Colorado School of Medicine NMR Core facility. Chemical shift perturbation experiments were carried out at 298K using uniformly 15N-labeled wild type or mutant proteins. 1H, 15N HSQC spectra were recorded in the presence of increasing concentrations of H3K36me3 peptide (synthesized by the University of Colorado Denver Biophysics Core Facility.) Kd values were calculated by a nonlinear least-squares analysis in Kaleidagraph using the following equation:where [L] is the concentration of the peptide, [P] is the concentration of the protein, Δδ is the observed normalized chemical shift change and Δδmax is the normalized chemical shift change at saturation, calculated as
where δ is the chemical shift in p.p.m.
Fluorescence binding assay
Tryptophan fluorescence measurements were performed on a Fluoromax-3 spectrofluorometer at room temperature. The samples containing 1 or 2 µM wild type or mutated protein in 25 mM Tris pH 7.5, 150 mM NaCl, 3 mM DTT and increasing concentrations of histone H3K36me3 peptide (31–40) were excited at 295 nm. Emission spectra were recorded from 320 to 380 nm with a 0.5 nm step size and a 0.5 sec integration time, averaged over 3 scans. The Kd values were determined using a nonlinear least-squares analysis and the equation:where [L] is the concentration of the peptide, [P] is the concentration of the protein, ΔI is the observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states. The Kd values were averaged over 3 separate experiments, with error calculated as the standard deviation between runs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Supplemental_Material.zip
Download Zip (6.1 MB)Acknowledgments
We thank Catherine Musselman for help with experiments.
Funding
This work was supported by NIH grants R01 GM106416 and GM100907 to TGK. JG is an NIH NRSA predoctoral fellow (F31 CA189487).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343-9; PMID:21248841; http://dx.doi.org/10.1038/nature09784
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039-43; PMID:12351676; http://dx.doi.org/10.1126/science.1076997
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 2002; 16:2893-905; PMID:12435631; http://dx.doi.org/10.1101/gad.1035902
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111:185-96; PMID:12408863; http://dx.doi.org/10.1016/S0092-8674(02)00975-3
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002; 111:197-208; PMID:12408864; http://dx.doi.org/10.1016/S0092-8674(02)00976-5
- Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev 2012; 26:751-5; PMID:22508723; http://dx.doi.org/10.1101/gad.191163.112
- Duncan IM. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 1982; 102:49-70; PMID:6813190
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J 2007; 26:4078-88; PMID:17762866; http://dx.doi.org/10.1038/sj.emboj.7601837
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 2008; 28:2718-31; PMID:18285464; http://dx.doi.org/10.1128/MCB.02017-07
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 2008; 28:1862-72; PMID:18086877; http://dx.doi.org/10.1128/MCB.01589-07
- Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, Yasui A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res 2008; 36:2939-47; PMID:18385154; http://dx.doi.org/10.1093/nar/gkn146
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 2012; 19:1266-72; PMID:23142980; http://dx.doi.org/10.1038/nsmb.2435
- Li X, Isono K, Yamada D, Endo TA, Endoh M, Shinga J, Mizutani-Koseki Y, Otte AP, Casanova M, Kitamura H, et al. Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes. Mol Cell Biol 2011; 31:351-64; PMID:21059868; http://dx.doi.org/10.1128/MCB.00259-10
- Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 2012; 19:1257-65; PMID:23104054; http://dx.doi.org/10.1038/nsmb.2434
- Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 2012; 19:1273-81; PMID:23160351; http://dx.doi.org/10.1038/nsmb.2449
- Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 2013; 49:571-82; PMID:23273982; http://dx.doi.org/10.1016/j.molcel.2012.11.026
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem 2001; 276:43065-73; PMID:11571280; http://dx.doi.org/10.1074/jbc.M104294200
- Qin S, Guo Y, Xu C, Bian C, Fu M, Gong S, Min J. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun 2013; 430:547-53; PMID:23228662; http://dx.doi.org/10.1016/j.bbrc.2012.11.116
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012; 19:1218-27; PMID:23211769; http://dx.doi.org/10.1038/nsmb.2436
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 2007; 14:1025-40; PMID:17984965; http://dx.doi.org/10.1038/nsmb1338
- Friberg A, Oddone A, Klymenko T, Muller J, Sattler M. Structure of an atypical Tudor domain in the Drosophila Polycomblike protein. Protein Sci: Pub Protein Soc 2010; 19:1906-16; PMID:20669242; http://dx.doi.org/10.1002/pro.476
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006; 442:100-3; PMID:16728977
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol 2010; 17:617-9; PMID:20400950; http://dx.doi.org/10.1038/nsmb.1797
- Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 2008; 16:1740-50; PMID:18818090; http://dx.doi.org/10.1016/j.str.2008.08.008
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009; 461:762-7; PMID:19767730; http://dx.doi.org/10.1038/nature08398
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, Mackenzie F, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci USA 2010; 107:19266-71; PMID:20974918; http://dx.doi.org/10.1073/pnas.1008937107
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 2006; 312:748-51; PMID:16601153; http://dx.doi.org/10.1126/science.1125162
- Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 2011; 39:9061-71; PMID:21813457; http://dx.doi.org/10.1093/nar/gkr613
