Abstract
The proto-oncogene c-Jun plays crucial roles in tumorigenesis, and its aberrant expression has been implicated in many cancers. Previous studies have shown that the c-Jun gene is positively autoregulated by its product. Notably, it has also been reported that c-Jun proteins are enriched in its gene body region. However, the role of c-Jun proteins in its gene body region has yet to be uncovered. HP1a is an evolutionarily conserved heterochromatin-associated protein, which plays an essential role in heterochromatin-mediated gene silencing. Interestingly, accumulating evidence shows that HP1a is also localized to euchromatic regions to positively regulate gene transcription. However, the underlying mechanism has not been defined. In this study, we demonstrate that HP1a is involved in the positive autoregulatory loop of the Jra gene, the c-Jun homolog in Drosophila. Jra recruits the HP1a/KDM4A complex to its gene body region upon osmotic stress to reduce H3K36 methylation levels and disrupt H3K36 methylation-dependent histone deacetylation, resulting in high levels of histone acetylation in the Jra gene body region, thus promoting gene transcription. These results not only expand our knowledge toward the mechanism of c-Jun regulation, but also reveal the mechanism by which HP1a exerts its positive regulatory function in gene expression.
Abbreviations
| AP-1 | = | Activating Protein 1; HP1a, Heterochromatin protein 1a; Jra, Jun-related antigen; KDM4A, lysine-specific demethylase 4A |
Introduction
c-Jun belongs to the Activating Protein 1 (AP-1) transcription factor family, which includes Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra1, and Fra2) subfamilies, as well as Activating Transcription Factor (ATF) proteins ATF2, LRF1/ATF3, B-ATF, and JDP1. The Maf subfamily members, including c-Maf, MafB, MafA, MafG/F/K, and Nrl, have also been shown to be members of the AP-1 transcription factor family.Citation1 c-Jun plays essential roles in inflammation, differentiation, apoptosis, cellular migration, and wound healing.Citation2-4 As an oncogene, c-Jun has been shown to be amplified in a fraction of undifferentiated, aggressive sarcomas.Citation5 Similarly, c-Jun has been found to be overexpressed in tumor cells of patients with classical Hodgkin's disease and in anaplastic large cell lymphoma (ALCL).Citation6 Moreover, c-Jun has also been implicated in the progression of skin cancer.Citation7,8 Because of its crucial role in tumorigenesis, c-Jun continues to be a topic of intense investigation.
Heterochromatin Protein 1 (HP1) was first characterized in Drosophila as a heterochromatin-binding protein,Citation9 and its homologues have since been identified in most eukaryotic species. In fact, there are multiple HP1 paralogs in many organisms.Citation10-12 In Drosophila, 3 ubiquitously expressed HP1 proteins have been identified so far: HP1a, HP1b, and HP1c.Citation9,13,14 Accordingly, there are 3 corresponding HP1 paralogs in mammals, HP1α, HP1β, and HP1γ.Citation15,16 Although these HP1 paralogs share high similarity in amino acid sequence and all contain 2 conserved domains, the chromodomain (CD) and chromoshadow domain (CSD), their functions are quite different. HP1b is localized to both heterochromatin and euchromatin and its aberrant expression impairs both heterochromatin formation and gene expression from euchromatic regions.Citation13 HP1c seems solely localized to euchromatin and positively regulates gene expression.Citation14,17 Although HP1a was initially identified as a heterochromatin-binding protein and plays an essential role in heterochromatin-mediated gene silencing, many studies showed that HP1a is also localized to euchromatin and is involved in euchromatic gene expression. For instance, HP1a was shown to bind to heat shock gene hsp70 as well as developmentally regulated genes, and is positively involved in the activation of these genes.Citation18 Despite much evidence demonstrating that HP1a may also play a positive role in gene expression, the mechanism by which HP1a facilitates gene expression has not yet been uncovered.
In this study, we demonstrate that Jra, the Drosophila homolog of c-Jun, interacts and recruits HP1a to the gene body region of Jra. HP1a in turn recruits histone demethylase KDM4A to reduce H3K36 methylation levels and disrupt H3K36 methylation-dependent histone deacetylation, resulting in high levels of histone acetylation in the Jra gene body region, and thus promotes gene transcription. Our data not only expand our knowledge toward the autoregulatory loop of c-Jun gene, but also reveal the mechanism by which HP1a is positively involved in gene expression.
Materials and Methods
Cell culture and RNAi
Drosophila melanogaster S2 cells were maintained at 25°C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal calf serum. dsRNAs were generated using MEGAscript Kit (Ambion, AM1334) according to the manufacturer's protocol. For RNAi assay, S2 cells were seeded into 6-well plate in serum-free medium followed by addition of dsRNA to a final concentration of 10 μg/ml. Cells were then incubated at 25°C for 1 h followed by addition of 2 ml of complete medium and fetal calf serum (final concentration, 10%). After 5 days, cells were collected for further analyses.
Immunopurification of Jra complex for Mass Spectrometry
S2 cells expressing Jra-FLAG were first treated with or without 500 mM sorbitol for 30 minutes. The resultant cells were collected and washed with cold PBS, and then lysed with the lysis buffer [10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 1% NP-40, 1 mM DTT, 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, and complete protease inhibitor cocktail tablet (Roche)]. Nuclei were collected by centrifugation at 5,000 rpm for 5 min at 4°C and extracted with extraction buffer [20 mM HEPES-NaOH (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% (v/v) glycerol, 1 mM DTT, 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, and complete protease inhibitor cocktail tablet (Roche)]. Nuclear extracts were subjected to centrifugation at 14,000 rpm for 15 min at 4°C and then at 45,000 rpm for 1.5 hours at 4°C. The NaCl concentration of the nuclear extract was then adjusted to 300 mM. Nuclear extracts were incubated with anti-FLAG (M2) agarose beads (Sigma) overnight at 4°C. The beads were washed 4 times for 10 min in washing buffer [10 mM HEPES-NaOH (pH 7.9), 300 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.2% Triton X-100, 1 mM PMSF). The proteins were eluted in elution buffer (0.5 mg/ml FLAG peptide in 10 mM HEPES-NaOH (pH 7.9), 100 mM NaCl, 1.5 mM MgCl2, 0.05% Triton X-100 and complete protease inhibitor cocktail tablet (Roche)]. Proteins were TCA precipitated and subjected to mass spectrometry analysis.
Co-immunoprecipitation
S2 cells were treated with 500 mM sorbitol for 30 minutes to induce osmotic stress, before harvested and subjected to nuclear extract preparation according to the protocol previously described.Citation19 The following antibodies were used in co-immunoprecipitation assays and Western blot assays: anti-FLAG (Sigma, F3165), c-Jun (Santa Cruz, sc-74543), anti-HP1a (Covance, PRB291C), anti-β-tubulin (DSHB, E7), and anti-JNK (Santa Cruz, sc-571).
Chromatin immunoprecipitation
ChIP assays were performed as described previously.Citation13 The following antibodies were used in ChIP assays: anti-HP1a (Covance, PRB291C), anti-Histone H3 (di methyl K9) (Abcam, ab1220), anti-Histone H3 (tri methyl K36) (Abcam, ab9050), H3 pan-acetyl antibody (Abcam, ab47915), H4 pan-acetyl antibody (Abcam, ab177790), anti-histone H3 (Abcam, ab1791), and anti-histone H4 (Abcam, ab10807), anti-FLAG (Sigma, F3165). The specificity of the histone modification antibodies was confirmed by Western blot assay (Fig. S1). For all the ChIP-qPCR assays, at least 3 independent experiments were performed on 3 independent samples.
Quantitative real-time PCR
Total RNA was isolated using Trizol reagent following the manufacturer's protocol (Invitrogen). Total RNA samples were treated with DNase I before reverse transcription. Real-time PCR analyses were performed using RealMasterMix (SYBR Green; Tiangen FP202). All primers used in this study are provided in Supplementary Materials. For qRT-PCR assays, at least 3 independent experiments were performed on 3 independent samples. Relative levels of mRNA expression were measured by using the cycle threshold (CT) method. Housekeeping gene Actin5c was used as internal control for normalization.
Results and Discussion
Jra interacts with HP1a under osmotic stress
In order to identify Jra interacting partners in Drosophila, we purified the Jra complex from S2 cells expressing FLAG-tagged Jra. Cells were first treated with or without 500 mM sorbitol for 30 minutes, which induces osmotic stress and activates the MAPK signaling pathway and Jra (Fig. S2). The nuclear extracts prepared from these cells were subjected to complex purification using anti-FLAG antibody. The mass spectrometry data showed that, among other Jra interacting partners, heterochromatin protein HP1a co-purifies with Jra-FLAG under osmotic stress (). The interaction between Jra-FLAG and HP1a was further confirmed by Western blot analysis (). Interestingly, Jra only co-immunoprecipitates with HP1a under osmotic stress, but not under unstressed conditions.
Figure 1. Jra interacts with HP1a under osmotic stress. (A) S2 cells stably expressing Jra-FLAG were treated with or without sorbitol. Nuclear extracts prepared from these cells were then subjected to co-immunoprecipitation assay using anti-FLAG antibody. The eluate was analyzed by Western blot assay using antibodies as indicated. (B) Nuclear extracts prepared from wild type S2 cells treated with or without sorbitol were subjected co-immunoprecipitation assay using anti-Jra antibody. The eluate was analyzed by Western blot assay using antibodies as indicated.
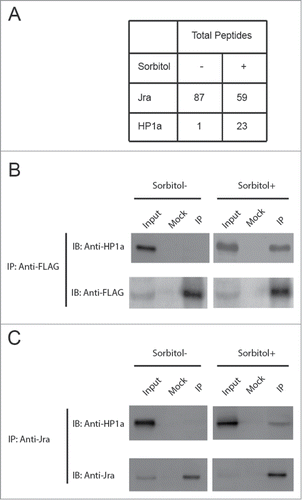
To further determine whether HP1a interacts with endogenous Jra, nuclear extracts from S2 cells treated with or without osmotic stress were subjected to co-immunoprecipitation assay using anti-Jra antibody. The results confirmed that endogenous Jra co-immunoprecipitates with HP1a under osmotic stress (). Taken together, our data demonstrate that HP1a interacts with Jra under osmotic stress.
Jra recruits HP1a to its gene body, independently of H3K9 methylation, under osmotic stress
Jra has been previously shown to bind to its gene body region.Citation20 Having found that Jra interacts with HP1a, we reasoned that Jra might recruit HP1a to its gene body region. To answer this question, we performed a chromatin immunoprecipitation (ChIP) assay to determine whether HP1a binds to the Jra gene locus. As shown in , HP1a is not localized to either the promoter or the gene body region of Jra under unstressed conditions. However, under osmotic stress, HP1a is enriched in the gene body region of Jra, but not in the promoter region (). Western blot assay confirmed that Jra is phosphorylated under osmotic stress (Fig. S2). Phosphorylation is an important post-translational modification for c-Jun regulation, which has previously been shown to regulate its transcriptional activity and its protein stability.Citation21,22 Our ChIP result combined with co-immunoprecipitation data indicate that phosphorylated Jra, but not unphosphorylated Jra, interacts with HP1a.
Figure 2. Jra recruits HP1a to its gene body region under osmotic stress. (A) HP1a is not localized to Jra gene under unstressed conditions. S2 cells treated with JNK dsRNA were directly subjected to ChIP analysis using anti-HP1a antibody followed by quantitative Real-Time PCR analysis. S2 cells treated with GFP dsRNA or without any dsRNA are used as negative control. 1360 element and Tub56D gene body region served as a positive control and a negative control for HP1a binding. Three independent experiments were represented as mean ± s.d. (B) HP1a is enriched in Jra gene body region under osmotic stress. S2 cells were treated with JNK dsRNA to deplete JNK, and then treated with sorbitol before ChIP-qRT-PCR analysis. As a control, S2 cells were treated with GFP dsRNA or without any dsRNA, and then treated with sorbitol before ChIP-qRT-PCR analysis. Three independent experiments were represented as mean ± s.d. *P < 0.001 (unpaired Student's t-test). (C) Western blot assay was performed to verify JNK knockdown efficiency. (D) The recruitment of HP1a to the Jra gene body region is independent of H3K9 methylation. S2 cells treated with or without sorbitol were subjected to ChIP assay using anti-H3K9me2 antibody followed by qRT-PCR. The enrichment of H3K9m2 was normalized to that of histone H3. Three independent experiments were represented as mean ± s.d.
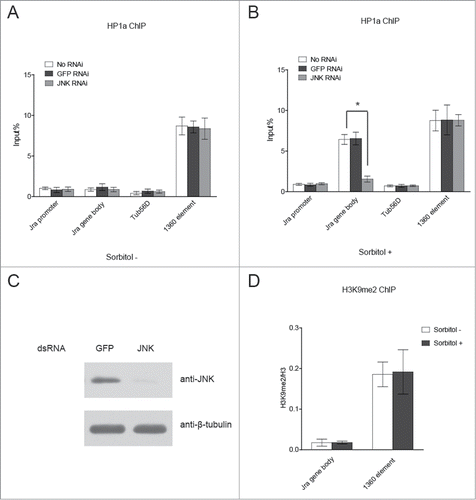
Next, we tried to determine if HP1a binding to the Jra gene body region is dependent on Jra phosphorylation. To address this concern, we treated S2 cells with JNK dsRNA to knock down JNK expression. JNK is the protein kinase in MAPK signaling pathway, which is responsible for c-Jun phosphorylation. S2 cells were also treated with GFP dsRNA as a control. Western blot result confirmed that the expression levels of JNK were dramatically reduced in in JNK dsRNA treated cells (), and that the phosphorylation of Jra was greatly reduced in JNK-depleted cells under osmotic stress (Fig. S2). These cells were then analyzed by ChIP assay using anti-HP1a antibody. The data showed that upon the depletion of JNK, HP1a lost its binding to the Jra gene body under osmotic stress, suggesting that phosphorylated Jra recruits HP1a to its gene body region ().
In the heterochromatin region, HP1a binding is dependent on H3K9 methylation. Therefore, we examined if H3K9 methylation also plays a role in HP1a recruitment to the Jra gene body. Cells treated with or without osmotic stress were subjected to ChIP assay using anti-H3K9me2 antibody. The data showed that there was no significant change in H3K9me2 levels in the Jra gene body region, eliminating the regulatory role of H3K9 methylation in the recruitment of HP1a to the Jra gene body region (). Taken together, our results showed that Jra recruits HP1a to its gene body region under osmotic stress, which is independent of H3K9 methylation.
Jra recruits the HP1a/KDM4a complex to its gene body region to reduce H3K36 methylation levels and disrupts H3K36 methylation-dependent histone deacetylation
Since we observed an increased enrichment of HP1a in the Jra gene body region under osmotic stress, we next tried to examine the effect of HP1a on Jra gene expression. S2 cells were treated with HP1a dsRNA to knock down HP1a expression. S2 cells treated with GFP dsRNA were used as a control. The depletion of HP1a was confirmed by Western blot assay (). The total RNA extracted from these cells was then analyzed by qRT-PCR. The results showed that HP1a knockdown significantly reduced Jra mRNA levels, indicating HP1a is positively involved in Jra transcription ().
Figure 3 (See previous page). Jra recruits the HP1a/KDM4a complex to its gene body region to reduce H3K36 methylation levels and disrupts H3K36 methylation-dependent histone deacetylation. (A) Western blot assay was performed to verify HP1a knockdown efficiency. (B) HP1a knockdown impairs Jra expression under osmotic stress. S2 cells were first treated with HP1a dsRNA or GFP dsRNA, then with sorbitol to induce osmotic stress. Total RNA prepared from these cells was subjected to qRT-PCR analysis. Three independent experiments were represented as mean ± s.d. *P = 0.0105 (unpaired Student's t-test). (C) HP1a knockdown elevates H3K36me3 levels in Jra gene body region. S2 cells were first treated with HP1a dsRNA or GFP dsRNA, then with sorbitol to induce osmotic stress. These cells were analyzed by ChIP assay using anti-H3K36me3 antibody followed by qRT-PCR. The enrichment of H3K36m3 was normalized to that of histone H3. Three independent experiments were represented as mean ± s.d. **P < 0.01 (unpaired Student's t-test). (D) The recruitment of KDM4A in Jra gene body region is dependent on HP1a. S2 cells stably expressing KDM4A-FLAG were first treated with HP1a dsRNA, then with sorbitol to induce osmotic stress. These cells were analyzed by ChIP assay using anti-FLAG antibody followed by qRT-PCR. Three independent experiments were represented as mean ± s.d. **P < 0.01 (unpaired Student's t-test). (E, F) HP1a knockdown reduces histone acetylation levels in Jra gene body region. S2 cells were first treated with HP1a dsRNA or GFP dsRNA, and then with sorbitol to induce osmotic stress. These cells were analyzed by ChIP assay using anti-H3 acetylation antibody or anti-H4 acetylation antibody, followed by qRT-PCR analysis. The enrichment of H3 acetylation and H4 acetylation was normalized to that of histone H3 and H4 respectively. Three independent experiments were represented as mean ± s.d. **P < 0.01 (unpaired Student's t-test).
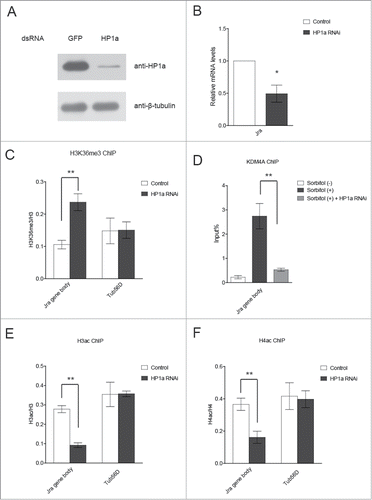
Having defined a role for HP1a in positively regulating Jra transcription, we sought to explore the mechanism by which this occurs. A previous study reported that HP1a interacts with Drosophila KDM4A demethylase and stimulates its H3K36 demethylation activity.Citation23 Since we observed the enrichment of HP1a in the Jra gene body region, and H3K36me3 modification has been reported to be enriched in the gene body region, we wondered whether HP1a collaborates with KDM4A to reduce H3K36me3 levels in the Jra gene body region. To test this idea, we measured the H3K36me3 levels of the Jra gene body region upon HP1a depletion (). The results showed that HP1a depletion significantly elevated H3K36me3 levels in the Jra gene body region, indicating a potential involvement of KDM4A in Jra transcription. To verify the recruitment of KDM4A to the Jra gene body, we established an S2 cell line stably expressing KDM4A-FLAG. These cells were treated with or without osmotic stress and subjected to ChIP analysis. The results demonstrate that KDM4A is enriched in Jra Jra gene body region upon osmotic stress, and the depletion of HP1a abolishes its binding to the Jra gene body region ().
H3K36me3 has been shown to be essential in preventing cryptic transcription from the gene body region.Citation24,25 Following RNAP II elongation, histone acetylation marks are removed by the Rpd3S complex, which is directed by H3K36me3 in the gene body region.Citation26 Because we observed elevated levels of H3K36me3 upon HP1a depletion, we then examined the histone acetylation levels in this region. As expected, the overall histone acetylation levels were significantly reduced upon HP1a depletion ().
It should be noted that a previous study has demonstrated that HP1a positively regulates euchromatic gene expression through its involvement in RNA packaging and stability.Citation27 To determine if the reduced mRNA levels of Jra was due the decreased mRNA stability in HP1a-depleted cells, we first treated cells with 500mM sorbitol and then with actinomycin D for 30 minutes, 60 minutes, and 90 minutes. Total RNA was subsequently isolated for quantitative real-time PCR analysis. The result showed that in both wild-type S2 cells and HP1a-depleted S2 cells, Jra mRNA levels were slightly reduced. However, HP1a depletion did not significantly accelerate Jra mRNA turnover after actinomycin D treatment (Supplemental Figure S3). Therefore, we proposed that the reduced Jra expression we observed is primarily due to the disruption of the positive role of HP1a in Jra transcription.
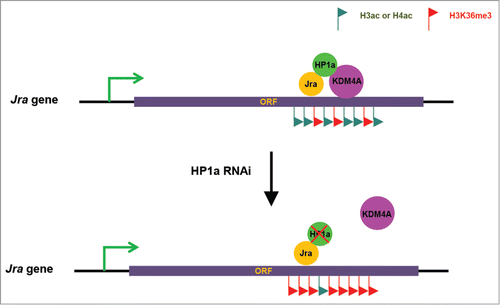
Although many studies reported that HP1a is not only localized to heterochromatin, but also localized to euchromatic regions to regulate gene transcription, the mechanism by which HP1a facilitates gene transcription is still unclear. It has also been established that c-Jun gene is under the control of a positive autoregulatory loop.Citation27 In addition, c-Jun proteins have also been shown to bind to its own gene body region.Citation20 However, what c-Jun does in its own gene body region has yet to be uncovered. In this study, we presented evidence showing that HP1a is involved in the positive autoregulatory loop of the Jra gene. Based on the data we obtained, we proposed a model in which Jra recruits the HP1a/KDM4A complex to its gene body region upon osmotic stress to reduce H3K36 methylation levels and disrupt H3K36 methylation-dependent histone deacetylation, resulting in high levels of histone acetylation in the Jra gene body region, and thus promotes gene transcription (). These results not only expand our knowledge of the mechanism by which the c-Jun oncogene is regulated, but also reveal the mechanism by which HP1a exerts its positive regulatory function in gene expression.
Figure 4. Working model. Jra recruits the HP1a/KDM4A complex to its gene body region upon osmotic stress to reduce H3K36 methylation levels and disrupt H3K36 methylation-dependent histone deacetylation, resulting in high levels of histone acetylation in the Jra gene body region, and thus promotes gene transcription. Upon HP1a depletion, Jra fails to recruit KDM4A to its gene body region, resulting in high levels of H3K36me3 and low levels of histone acetylation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.docx
Download MS Word (551.5 KB)Funding
This work was supported by the National Natural Science Foundation of China (No. 81302324) and the Instruction Plan of the Science and Technology Research and Development Project of Tangshan (No. 13130282z, No. 13130261a, No. 13130262a; Science and Technology Project of Tangshan Science and Technology Bureau).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer 2008; 8:683-93; PMID:19143053; http://dx.doi.org/10.1038/nrc2460
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 2004; 117:5965-73; PMID:15564374; http://dx.doi.org/10.1242/jcs.01589
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol 2002; 4:E131-6; PMID:11988758; http://dx.doi.org/10.1038/ncb0502-e131
- Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev 2005; 208:126-40; PMID:16313345; http://dx.doi.org/10.1111/j.0105-2896.2005.00332.x
- Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, Stern MH, Aurias A. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 2007; 11:361-74; PMID:17418412; http://dx.doi.org/10.1016/j.ccr.2007.02.007
- Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, Bommert K, Mechta-Grigoriou F, Stein H, Dorken B, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J 2002; 21:4104-13; PMID:12145210; http://dx.doi.org/10.1093/emboj/cdf389
- Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, Sibilia M, Wagner EF. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell 2003; 4:879-89; PMID:12791272; http://dx.doi.org/10.1016/S1534-5807(03)00161-8
- Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, Horvath CM, Ronai Z. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell 2001; 7:517-28; PMID:11463377; http://dx.doi.org/10.1016/S1097-2765(01)00199-X
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 1986; 6:3862-72; PMID:3099166
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 2004; 5:296-304; PMID:15071554; http://dx.doi.org/10.1038/nrm1355
- Lomberk G, Wallrath L, Urrutia R. The Heterochromatin protein 1 family. Genome Biol 2006; 7:228; PMID:17224041; http://dx.doi.org/10.1186/gb-2006-7-7-228
- Zeng W, Ball AR, Jr., Yokomori K. HP1: heterochromatin binding proteins working the genome. Epigenetics 2010; 5:287-92; PMID:20421743; http://dx.doi.org/10.4161/epi.5.4.11683
- Zhang D, Wang D, Sun F. Drosophila melanogaster heterochromatin protein HP1b plays important roles in transcriptional activation and development. Chromosoma 2011; 120:97-108; PMID:20857302; http://dx.doi.org/10.1007/s00412-010-0294-5
- Font-Burgada J, Rossell D, Auer H, Azorin F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev 2008; 22:3007-23; PMID:18981478; http://dx.doi.org/10.1101/gad.481408
- Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 2014; 24:377-86; PMID:24618358; http://dx.doi.org/10.1016/j.tcb.2014.01.002
- Hediger F, Gasser SM. Heterochromatin protein 1: don't judge the book by its cover! Curr Opin Genet Dev 2006; 16:143-50; PMID:16503133; http://dx.doi.org/10.1016/j.gde.2006.02.013
- Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev 2010; 24:2133-45; PMID:20889714; http://dx.doi.org/10.1101/gad.1959110
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 2003; 161:707-14; PMID:12756231; http://dx.doi.org/10.1083/jcb.200303012
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR 3rd, Washburn MP, Abmayr SM, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol 2006; 26:871-82; PMID:16428443; http://dx.doi.org/10.1128/MCB.26.3.871-882.2006
- Suganuma T, Mushegian A, Swanson SK, Abmayr SM, Florens L, Washburn MP, Workman JL. The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell 2010; 142:726-36; PMID:20813260; http://dx.doi.org/10.1016/j.cell.2010.07.045
- Davis RJ. Signal Transduction by the JNK Group of MAP Kinases. Cell 2000; 103: 239-52; PMID:11057897; http://dx.doi.org/10.1016/S0092-8674(00)00116-1
- Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 1997; 275: 400-2; PMID:8994040; http://dx.doi.org/10.1126/science.275.5298.400
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 2008; 32:696-706; PMID:19061644; http://dx.doi.org/10.1016/j.molcel.2008.11.008
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 2012; 489:452-5; PMID:22914091; http://dx.doi.org/10.1038/nature11326
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 2007; 21:1422-30; PMID:17545470; http://dx.doi.org/10.1101/gad.1539307
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005; 123:581-92; PMID:16286007; http://dx.doi.org/10.1016/j.cell.2005.10.023
- Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 1988; 55:875-85; PMID:3142689; http://dx.doi.org/10.1016/0092-8674(88)90143-2
