Abstract
The placenta is the principal regulator of the in utero environment, and disruptions to this environment can result in adverse offspring health outcomes. To better characterize the impact of in utero perturbations, we assessed the influence of known environmental pollutants on the expression of microRNA (miRNA) in placental samples collected from the National Children's Study (NCS) Vanguard birth cohort. This study analyzed the expression of 654 miRNAs in 110 term placentas. Environmental pollutants measured in these placentas included dichlorodiphenyldichloroethylene (DDE), bisphenol A (BPA), polybrominated diphenyl ethers (PBDEs), polychlorinated biphenyls (PCBs), arsenic (As), mercury (Hg), lead (Pb), and cadmium (Cd). A moderated t-test was used to identify a panel of differentially expressed miRNAs, which were further analyzed using generalized linear models. We observed 112 miRNAs consistently expressed in >70% of the samples. Consistent with the literature, miRNAs located within the imprinted placenta-specific C19MC cluster, specifically mir-517a, mir-517c, mir-522, and mir-23a, are among the top expressed miRNA in our study. We observed a positive association between PBDE 209 and miR-188–5p and an inverse association between PBDE 99 and let-7c. Both PCBs and Cd were positively associated with miR-1537 expression level. In addition, multiple let-7 family members were downregulated with increasing levels of Hg and Pb. We did not observe DDE or BPA levels to be associated with placental miRNA expression. This is the first birth cohort study linking environmental pollutants and placental expression of miRNAs. Our results suggest that placental miRNA profiles may signal in utero exposures to environmental chemicals.
Introduction
The intrauterine period is defined by gene-environment interactions, making it a critical window of susceptibility in which perturbations have the potential to impact both fetal development and health outcomes later in life. The placenta is the principal regulator of the intrauterine environment, serving as the maternal-fetal interface, playing a vital role in the appropriate growth and development of the fetus, facilitating nutrient exchange and waste removal, providing immune protection, and mediating metabolic and endocrine activity on behalf of the developing fetus. Citation1-5
Exogenous environmental agents, such as metals and organic pollutants, have been shown to cross and accumulate in the placenta with the potential to disrupt processes involved in normal development. Citation6,7 For example, in utero exposures to metals, such as arsenic (As), mercury (Hg), lead (Pb), and cadmium (Cd), as well as organic pollutants, such as polychlorinated biphenyls (PCB), polybrominated diphenyl ethers (PBDE), and dichlorodiphenyldichloroethylene (DDE), have been associated with developmental delays. Citation8-11
MicroRNAs (miRNAs) are labile gene expression regulatory elements that may mediate gene-environment interactions. They are short (21–24 nucleotides), single-stranded RNA molecules that do not code for proteins themselves but are able to control the expression of genes that do.Citation12 They regulate gene expression posttranscriptionally by binding to the 3′-untranslated region of their target mRNA and consequently attenuating protein translation. This repression of gene expression is mediated by either translational repression or mRNA degradation, based on the degree of complementarity to target mRNA sequences.Citation13 As only partial complementarity is needed, a single miRNA can bind multiple mRNA transcripts and each mRNA can be bound be multiple miRNAs. In this way, miRNAs are able to regulate diverse gene networks, including cell proliferation, apoptosis, and differentiation.Citation14
Given the rising concern over the impact of in utero exposures to environmental pollutants, there is an increasing interest to determine whether miRNAs can signal perturbations to the intrauterine environment. In this study, we profiled placental miRNA using samples collected as part of the National Children's Study (NCS) to assess associations between miRNA expression levels and various known environmental pollutants measured in placenta.
Results
Placental miRNA profile
The demographic characteristics of the study population are shown in . As indicated in the table, the study population consists of women with normal term (≥37 weeks) vaginal deliveries with placenta samples collected on average within 1 h.
Table 1. Characteristics of 110 study placentas
The nanoString human miRNA codeset contains 654 miRNA targets, out of which 112 miRNAs were considered to be “expressed” in the placenta using our stringent criteria indicated in the Materials and Methods section. In line with what has been previously reported, placenta-specific miRNAs, particularly those located within the imprinted C19MC cluster, predominate among the expressed miRNAs of our study (Supplementary ). Additionally, similar to previous reports, the most abundantly expressed miRNA is the C19MC derived miR-517a.Citation15 Using LIMMA analysis, we did not detect any differences in miRNA expression by the gender of newborns, maternal age, or gestational age.
Influence of organic pollutants on placental miRNA profile
The organic pollutants investigated were PBDEs, PCBs, DDE, and BPA. These agents were selected based on the known risk of early developmental defects associated with these chemicals and/or their ability to cross the placental barrier and accumulate in the placenta to detectable levels.Citation16–18 The mean placental levels detected in our study population are shown in . While the detected placental levels of these pollutants were relatively low, they fall in line with previous findings in western industrialized nations.Citation19–22
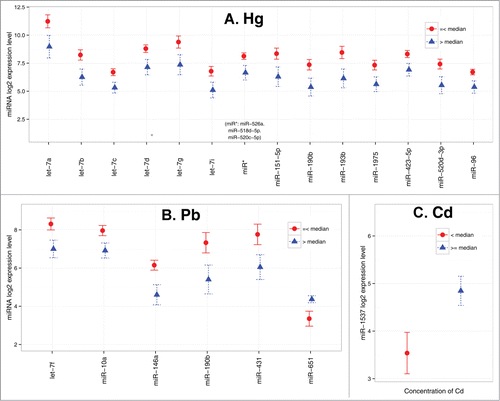
While 10 PBDE congeners were measured in the placenta, we focused our analysis on the low brominated (47, 99) and high brominated (153, 209) congeners previously reported to be the most abundantly detected in the placenta,Citation23,24 as well as total PBDE (summing all the congeners). Using generalized linear models, we observed a positive association between the high-brominated congener 209 and miR-188–5p (Ptrend = 0.01) and an inverse association between the low-brominated congener 99 and let-7c (Ptrend = 0.04) (). No association was observed between total PBDE and any miRNA.
Figure 2. Influence of PBDE congeners on placental miRNA profile (n = 104). (A) Association between let-7c and PBDE 99 (B) Association between miR-188–5p and PBDE 209. Error bars represent standard error interval. P-value is based on generalized linear model.
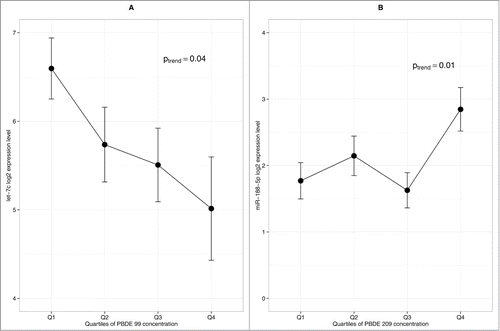
Among the 32 PCB congeners measured in the placenta, we focused on the congeners 52, 101, 105, 118, and 153, previously reported to be the most abundantly detected congeners in the placenta, as well as total PCB (summing all the congeners).Citation25 Generalized linear models indicated a significant positive linear trend between PCB congeners 52, 101, and total PCB and miR-1537 expression (). It is important to point out that PCB congeners 52 and 101 are highly correlated with one another (r = 0.96, P < 0.01) as well as with total PCB, with correlation coefficients of 0.95 and 0.96, respectively (P < 0.01). We did not find any association between miRNA expression and the level of DDE and BPA measured in the placenta.
Figure 3. Influence of PCB congeners on placental miRNA profile (n = 105). (A) Association between miR-1537 and PCB 52 (B) Association between miR-1537 and PCB 101 (C) Association between miR-1537 and total PCB. Error bars represent standard error interval. P-value is based on generalized linear model.
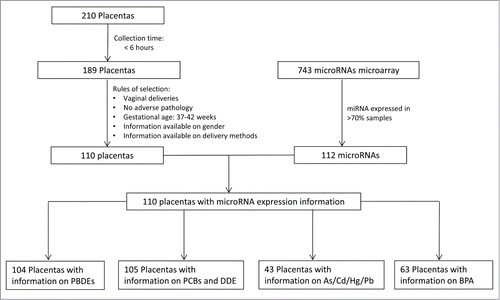
Influence of metals on placental miRNA profile
An array of total metal levels, toxic and essential, were measured in a subset (n = 43) of placenta tissues collected for the NCS. In this investigation, we focused on the known toxic metals, i.e., As, Pb, Hg, and Cd, previously shown to have detrimental effects on development. Due to the reduced sample size and non-normal distribution of metal levels detected, we dichotomized the study samples at the median level detected in the placenta. As shown in Figure 4, we observed that high Hg level was associated with reduced expression of a large number of miRNAs including miR-151–5p, miR-10a, miR-193b, miR-1975, miR-423–5p, miR-520d-3p, miR-96, miR-526a+miR-518d-5p+miR-520c-5p, and most prominently, a large number in the let-7 family (i.e., let-7a, let-7b, let-7c, let-7d, let-7g, and let-7i). The levels of these let-7 isoforms were observed to be highly correlated in the placenta (r = 0.65–0.97). Meanwhile, high placental Pb levels were associated with the reduced expression of 4 miRNAs (let-7f, miR-146a, miR-10a, and miR-431) and an increased expression of miR-651. Interestingly, miR-190b was down regulated by both Pb and Hg. Moreover, high placental Cd levels were associated with increased miR-1537 expression, similar to what was observed with PCBs. We did not find any association between miRNA expression and the level of As measured in the placenta.
Discussion
In the current study, we assessed the global miRNA expression profile of 110 human placentas collected from the NCS. The study focuses on placentas from term, clinically normal pregnancies with vaginal delivery free of pathological complications. The number of miRNA considered to be consistently “expressed” in this study is in line with recent published studies,Citation26 though other studies have reported up to 600 detectable miRNAs in placentas using various different platforms.Citation27-29 Such discrepancies may largely be driven by different criteria of what is considered “expressed." It may also reflect platform-specific differences. As miRNA expression profiles are tissue-specific and the placenta is a known composite of heterogeneous cell types stemming from fetal and maternal origin, the discrepancy may also arise due to differences in tissue sampling.Citation28
In line with previous reports, miRNAs expressed in the placenta from our study largely cluster in distinct chromosomal locations, namely the chromosome 19 miRNA cluster (C19MC).Citation30 MiRNAs located in this cluster have been shown to be abundantly expressed in the placenta while lowly expressed in other tissues.Citation31 Additionally, this cluster overlaps with a paternally expressed imprinted region.Citation32 Imprinted loci belonging to these domains are typically activated at critical developmental stages and involved in controlling cell differentiation and fate in the embryonic growth of placenta tissue.Citation33,34
Our study is one of the few to investigate the potential effects of environmental pollutants on miRNA expression in humans. Our results clearly demonstrate such effects in which both organics and metal- levels are associated with miRNA expression in placenta. Hg shows the most striking effect, with a large number of miRNAs downregulated with increasing level of Hg. Reported postnatal outcomes due to prenatal Hg exposure include birth defects and learning and memory deficits,Citation35,36 with common sources of exposure including maternal fish intakeCitation37,38 and dental amalgams.Citation39 To date, the only epidemiologic study reporting changes in miRNA expression with varying levels of Hg exposure focused on urinary levels in adolescents.Citation40
In our study, Pb was also found to be associated with changes in expression of several miRNAs. While exposure to Pb sharply decreased over the last few decades, residual exposure from lead painting, piping and legacy industrial sources remains of concern. Low levels of prenatal exposure continue to show associations with cognitive deficits.Citation41–43 Our study is the first to report on the impact of prenatal Pb levels on the placental miRNA expression profile. Our finding showing a negative correlation between miR-146a expression and Pb exposure is in agreement with the findings reported in an occupational study of Pb exposure assessed in peripheral blood leukocytes.Citation44
We also observed several instances of individual miRNAs responding to multiple environmental pollutants. For example, members of the let-7 miRNA family were influenced by Hg, Pb, and PBDE 99. Also of note is that all let-7 miRNAs were down regulated with increasing exposure. This family of highly conserved miRNAs has been established to play a critical role in early development, primarily by driving differentiation.Citation45–47 Deregulation of these miRNAs has been linked with several cancers.Citation48 Furthermore, a role in placental development is indicated by a study showing that let-7 family members are up-regulated in 3rd trimester placenta samples compared to 1st trimester placenta samples.Citation49 Hence, the observed downregulation of let-7 isoforms in term placenta with increasing toxicant levels may indicate a state of disrupted placental development in response to chemical exposures.
Additionally, miR-190b was differentially expressed in the presence of Hg and Pb. The expression of this miRNA was recently reported to regulate Neuregulin 3-mediated inhibitory control processes of the amygdala.Citation50 As dysregulation of inhibitory control is a marker of several mental health disorders, including ADHD, these findings are in line with behavioral defects reported in relation to in utero Hg and Pb exposures.Citation51–54
Similarly, miR-1537 was differentially expressed in the presence of PCB congeners and Cd. While few studies have reported on this miRNA, it has been previously described that miR-1537 is located in a region commonly deleted in a subset of high-risk neuroblastoma cases.Citation55 Few epidemiological studies have reported on the impact of either Cd, a contaminant of growing concern due to its presence in cigarette smoke and electronic-waste, or of PCBs, a persistent class of chemical previously used as a flame retardant and plasticizer, on miRNA expression. While associations between these exposures and alterations in miRNA expression were reported in these studies, the findings were based on the analysis of peripheral blood leukocytes from occupationally-exposed adults.Citation44,56 The current study is the first to report the impact of prenatal Cd and PCB levels on the placental miRNA expression profile.
Neither BPA nor DDE induced changes in miRNA expression profiles. The fact that not all environmental agents elicit alterations in the placental-miRNA profile suggests an inherent specificity in the sensitivity/responsiveness of miRNAs to environmental pollutants.
The findings presented in this study indicate plausible interactions between environmental pollutants and placental epigenetic markers. The observed correlated changes in miRNA expression and environmental pollutant levels could reflect an exposure-induced perturbation on miRNA expression levels. For example, environmental pollutants could interact with the miRNA biosynthesis machinery, thereby directly inducing changes in miRNA expression levels.Citation57,58 Alternatively, altered miRNA expression levels could reflect a host-induced adaptive response, including the upregulation of DNA repair and detoxification genes, to counteract maladaptive effects of environmental pollutants on cellular processes.Citation59 For example, the top Targetscan-derived mRNA targets of miR-146a include TRAF1 and IRAK1, genes involved in the toll-like receptor pathway. Hence, the downregulation of miR-146a we observed with increasing levels of Pb levels could indicate a host cell-mediated immune activation in response to these environmental pollutants.
While suggestive, the observed associations between environmental pollutants and miRNA expression levels warrants further study due to the inherent limitations in the current study design. Outcome variables available in this population were few, and the impact of other exposure-related differences in miRNA expression could not be assessed. Additionally, the metal-related findings are exploratory with limited sample size. Furthermore, similar to the congener-specific effects of organics observed in this study, the impact of metals may also depend upon the specific metal compound, such as methyl mercury vs. total mercury. More refined analyses are warranted that capture these species-specific effects.
As is the case for all biospecimens with a heterogeneous cell-type composition, placental miRNA content can vary based on the sampling method implemented. In the current study, consistency was maintained by removing the maternal decidua prior to sampling and restricting biopsies to villous tissues free from infarcts and calcifications. This sampling strategy minimizes, but does not prevent, the potential for non-random variability in miRNA content across the samples.
Lastly, one important issue that warrants careful consideration is concerning “multiple comparisons.” In this study, we used a step-wise approach to detect gene-environment associations. First, we conducted a moderated t-test across our panel of expressed miRNAs to identify candidate loci that are differentially expressed between the 1st vs. 4th quartiles of environmental exposures, which were then further analyzed with generalized linear models. The candidates selected for further analysis in the first step were chosen based on a cut-off value of P < 0.05. Given that the number of miRNA (n = 112 ) included in our analysis increases the likelihood of false positive findings (Type I error), a more stringent cut-off accounting for the multiple comparisons may be warranted. However, there is continuing debate on whether/when/how multiple comparisons should be taken into account.Citation60 When evaluating results of molecular epidemiology studies, statistical power and the priority of the tested hypothesis also need to be taken into account, in addition to the magnitude of the P value. Conventional multiple comparison adjustment methods, such as the Bonferroni method, may safeguard false positive findings, but at the same time increase Type II error (false negative) and reduce sensitivity.Citation61 Nevertheless, results from our study need to be interpreted with caution and warrant replication in other population studies.
In summary, this study surveys the miRNA profile in human placenta and reported environmental exposure-related differences in expression of miRNA with a moderate sample size. Our findings suggest the potential of miRNAs to serve as markers of prenatal environmental exposures. We observed global changes in miRNA expression levels related to Hg, Pb, and Cd exposures, while PBDE and PCB congeners were associated with specific miRNAs. No changes in miRNA expression were observed in association with BPA or DDE exposure. Furthermore, a single miRNA (such as miR-190b) may be sensitive to multiple exposures. Planned studies will include generating a transcriptome profile of these samples and evaluating the interrelationship between the miRNA and mRNA profile using a systems biology approach to provide further insight on the role of the placental epigenomics in human reproduction and development.
Materials and Methods
Sample collection
This study is a part of NCS formative research (Project LOI2-BIO-18). Placenta samples were collected from study locations as part of the NCS Vanguard study, a planned large-scale epidemiological cohort study of environmental influences on child health and development. Placenta samples included in the current study were collected from 13 counties across the US. Written informed consent was obtained from all participants. The study protocol was reviewed and approved by the Office of Human Research Protections registered Institutional Review Boards.
All placental specimens were excised from grossly normal areas of the villous parenchyma, excluding the deciduas basalis and chorionic plate. Sampling occurred at the participating collection hospital and a second time at the University of Rochester's Placental Processing Center (URPPC). Dates and times were obtained for each collection. All samples were packaged in a standardized manner and shipped overnight to the URPPC.
Tissue Isolation for Nucleic Acid Studies. Forty to 60 mg of villous tissue was obtained for nucleic acid extraction, by first rinsing twice in sterile RNAse/DNAse free phosphate buffered saline, lightly macerating the tissue with sterile, fine-tipped scissors and then placing the tissue in 5 ml of RNALater (Life Technologies, AM7024) and maintained at 4°C. All samples were labeled with a specific study identification code, which blinded investigators to the collection site and collection time. These sample codes were maintained at the URPPC.
Tissue samples for other environmental analyses. Five to 10 g of villous tissue was collected as above and stored in acid-washed, BPA-free 50-mL tubes using plastic (non-metallic) disposable forceps. Placental sample collection (time 0) occurred within 0–6 h after birth, and samples were stored on dry ice or at −80°C until shipped on dry ice to the URPPC, where they were stored at −80°C.
RNA extraction
Nucleic acids (e.g., genomic DNA and total RNA, microRNAs) were extracted from the samples stored in RNALater at the URPPC using an AllPrep DNA/RNA extraction kit according to manufacturer's instruction (Qiagen, 80204). Genomic DNA and total RNA samples were quantified using a microplate spectrophotometer (Biotek, Winooski, VT). Coded total RNA samples were obtained frozen from the URPPC.
Study Population
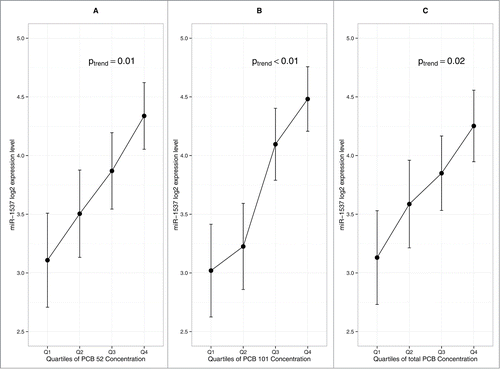
The study design is depicted in . A total of 210 placentas from singleton deliveries were collected. In a pilot study we determined the temporal variability in placental miRNA expression level based on repeat biopsies sampled at varying time-points.Citation62,63 Based on our findings from this prior study, we limited our analysis to placenta samples that were collected within 6 h of delivery to ensure miRNA stability (n = 189). Basic clinical information, including infant gender, gestational age, delivery method, and absence of any pathological abnormalities, was available for 141 subjects. Within this subset, we restricted our analysis to vaginal deliveries with a gestational age of 37 – 42 weeks (n = 110).
miRNA profiling
miRNA expression profiling was conducted using the nCounter Analysis System (Nanostring technologies, Seattle, WA) following the manufacturer's protocol. Briefly, 100 ng total RNA were assayed using the nCounter human miRNA expression assay. Samples were prepared by ligating miRtags onto the 3’ end of mature miRNAs to normalize the melting temperature of the miRNAs and to facilitate the use of the dual probe system. Following the removal of excess tags by restriction digestion at 37°C, the tagged miRNAs were hybridized overnight at 65°C with reporter probes containing color-coded barcodes unique to each target and biotinylated capture probes. Excess reporter and capture probes were sequentially removed using magnetic bead based purification on the nCounter Prep Station II. Following immobilization in the sample cartridge, target molecules were quantified based on the fluorescence of the reporter probes using an nCounter Digital Analyzer.
Chemical analysis
PBDEs, PCBs, DDE
Analytic assessment of the chemicals has been detailed in a previous publication.Citation22 Placenta tissue was freeze-dried, manually ground, and extracted using optimized matrix solid phase dispersion (MSPD) method.Citation64 Sample cleanup was accomplished using a multi-layer silica gel column. PCB congeners and DDE were analyzed using an Agilent 7890A gas chromatograph coupled to an Agilent 7000 tandem mass spectrometer equipped with an electron impact ionization source. PBDE congeners were analyzed using an Agilent 6890 gas chromatograph connected to an Agilent 5973 mass spectrometer (MS) with electron capture negative ionization mode.
BPA
Placental samples were ethyl-acetate extracted 2 times, dried under N2 stream for 1 h and reconstituted with 50% aqueous acetonitrile. Following centrifugation to remove any remaining residual debris, samples were loaded onto a liquid chromatography-tandem mass spectrometry (LC-MS/MS) system for analysis (UltiMate 3000 (Thermo Scientific, Somerset, NJ), micrOTOF II (Bruker Daltonics, Billerica, MA)). Data from the LC-MS/MS run was uploaded to the system's “Quant Analysis” software package (Bruker, Daltonics) to process and quantitate BPA levels. A series of calibration standards was included for quantitation. Quality control samples were run alongside each batch of 10 samples to verify consistent instrument performance.
Metals
All sample preparation for metal analysis was conducted in a class 100 trace metal free clean laboratory facility. Prior to analysis, all samples were thawed at room temperature for approximately 4 h. An aliquot of 1 g from each sample specimen was then individually selected for inductively coupled plasma mass spectrometry (ICP-MS) analysis of metal concentration. Sample aliquots were selected by cutting the placental tissue with a ceramic knife that was cleaned between each sample and blank verified at least once per sample preparation day. Compound specific metals analyses were measured by either high-performance liquid chromatography (HPLC)-ICP-MSCitation65,66 or GC-ICP-MS methodologies.
For metal concentrations, each sample was weighed using a calibrated mass balance and the wet tissue weight was recorded. Each sample was then dried for up to 48 h at 60°C in a vacuum oven. After drying, samples were re-weighed to determine the dry weight. Individual samples were then masticated with a trace metal free ceramic mortar and pestle, which was cleaned between each sample. After mastication, sample weights were re-recorded to ± 0.001 mg, and the tissue was digested in Teflon vessels using concentrated (15.9 mol/L) ultra-pure nitric acid (HNO3) at 70°C for approximately 6 h or until digestion was complete as previously described.Citation67 Samples were then digested under temperature- and pressure- controlled conditions using closed SCP Science Class A digestion tubes and a SCP Science 48-sample digestion block with DigiProbe temperature control. After cooling, the remaining solution was baked for approximately 8 h (or until dry) at 50°C to decant excess nitric acid. Residual dried material was then re-digested with ∼0.5 mL of ultra-pure HNO3.
Statistical analysis
The NanoString Norm packageCitation68 was used to normalize nCounter data. Code counts were normalized against the sum of spiked-in positive controls and background corrected based on 2 standard deviations above the mean of the included negative controls. A normalization factor based on the geometric mean of miRNAs with the lowest coefficient of variation in expression across the samples was applied to account for sample content variability. To filter out lowly expressed miRNAs, the background baseline level of 2 standard deviations above the mean of the included negative controls was set as the cut-off value for expression. Using this cut-off, miRNAs with values falling below the background baseline in more than 30% of the samples were considered unexpressed and removed from the analysis. As the levels of environmental pollutants (PCBs, PBDEs, DDE, and BPA) and metals (Hg, Pb, Cd) were not normally distributed, study samples were categorized based on exposure levels; while organics were quartiled, metals were dichotomized at the median due to the reduced size of samples with measured metal content (n = 43). A moderated t-test from the LIMMA packageCitation69 was used to examine the mean miRNA expression differences based on the highest and lowest quartiles for pollutants and above and below the median level for metals. The cut-off value for BPA was set at the detection level (42 samples below the limit of detection and 21 samples above the limit of detection). The relationship between organic pollutants and miRNA was further analyzed by generalized linear model using the quartile-specific geometric mean value of the organics as exposure variables due to the nonlinear nature of the relationship between miRNA expression level and environmental pollutants. All analysis was conducted using R 3.0.2 (http://www.r-project.org/). All statistical tests were 2-sided, and P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1066960_SupplementalTables.docx
Download MS Word (20.9 KB)Acknowledgments
The authors sincerely thank the contributions from other members of the National Children's Study Placenta Consortium (NCS formative research project LOI2-BIO-18) that includes L. Ruffolo, L. Salamone, L. Littman, C. Hobbs, N. Thiex, K. Hao, P. Sheffield, A. Golden, J. Gilbert, S. Teitelbaum, C. Torres, P. Weindenborner, P. Katzman, C. Salafia, S. Wadlinger, B. Specker, C. Holiday, E. Taggert, R. Domalski, A. Penmetsa, J. Tyra, N. Zembiec, H. Gibson, C. Kurlya, L. Baxter, R. Rowley, A. Harris, M. Suter, M. Pacholski, M. A Kent, L. Green, R. Wapner, J. Perou, J. Dudley, A. Cohain, C. Lendor, S. Allen, K. Mantilla, H. Heriveaux, V. Reccoppa, S. Leuthner, S. Szabo, J. Dalton, D. Misra, T. Girardi, P. Getreuer, Y. Y Li, N. Thieux, K. Gutzman, A. Martin, D. MacCloud, C. K Walker, S. E Pontow, B. Fury, C. Holliday, J. Butler, T. Busch, J. Rigdon, E. Campbell, J. Thorp, B. Eucker, C. Bell, E. Taggart, J. Billy, S. Stradling, J. Leavitt, C. Adams, W. Bell, L. Palmer, D. Tharp, M. Durkin, M-N Sandoval, B. O'Brien, M. Layton, D. Todd, K. Wilson, C. Cagnina, L. Merrill, M. Klebanoff, O. Genbacev, and T. Kwong. We also sincerely thank the generosity of participants of the NCS who donated biological samples for this study.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work is part of the National Children's Study Placenta Consortium (NCS formative research project LOI2-BIO-18). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and funded, through its appropriation, by the Office of the Director of the National Institutes of Health, with NICHD Contracts HHSN267200700027C, HHSN275201100002C, and HHSN275200503396C. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institutes of Health or the US Department of Health and Human Services.
References
- Cross JC. Formation of the placenta and extraembryonic membranes. Ann N Y Acad Sci 1998; 857:23-32; PMID:9917829; http://dx.doi.org/10.1111/j.1749-6632.1998.tb10104.x
- Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H. Molecular cues to implantation. Endocr Rev 2004; 25:341-73; PMID:15180948; http://dx.doi.org/10.1210/er.2003-0020
- Murphy VE, Smith R, Giles WB, Clifton VL. Endocrine regulation of human fetal growth: the role of the mother, placenta, and fetus. Endocr Rev 2006; 27:141-69; PMID:16434511; http://dx.doi.org/10.1210/er.2005-0011
- Pijnenborg R. Implantation and immunology: maternal inflammatory and immune cellular responses to implantation and trophoblast invasion. Reprod Biomed Online 2002; 4 Suppl 3:14-7; PMID:12470559
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet 2001; 2:538-48; PMID:11433360; http://dx.doi.org/10.1038/35080570
- Kozikowska I, Binkowski LJ, Szczepanska K, Slawska H, Miszczuk K, Sliwinska M, Laciak T, Stawarz R. Mercury concentrations in human placenta, umbilical cord, cord blood and amniotic fluid and their relations with body parameters of newborns. Environ Pollut 2013; 182:256-62; PMID:23938449; http://dx.doi.org/10.1016/j.envpol.2013.07.030
- Gascon M, Morales E, Sunyer J, Vrijheid M. Effects of persistent organic pollutants on the developing respiratory and immune systems: a systematic review. Environ Int 2013; 52:51-65; PMID:23291098; http://dx.doi.org/10.1016/j.envint.2012.11.005
- Dallaire R, Dewailly E, Ayotte P, Forget-Dubois N, Jacobson SW, Jacobson JL, Muckle G. Exposure to organochlorines and mercury through fish and marine mammal consumption: associations with growth and duration of gestation among Inuit newborns. Environ Int 2013; 54:85-91; PMID:23422685; http://dx.doi.org/10.1016/j.envint.2013.01.013
- Eskenazi B, Chevrier J, Rauch SA, Kogut K, Harley KG, Johnson C, Trujillo C, Sjodin A, Bradman A. In utero and childhood polybrominated diphenyl ether (PBDE) exposures and neurodevelopment in the CHAMACOS study. Environ Health Perspect 2013; 121:257-62; PMID:23154064; http://dx.doi.org/10.1289/ehp.121-A257
- Wojtyniak BJ, Rabczenko D, Jonsson BA, Zvezday V, Pedersen HS, Rylander L, Toft G, Ludwicki JK, Goralczyk K, Lesovaya A, et al. Association of maternal serum concentrations of 2,2', 4,4'5,5'-hexachlorobiphenyl (CB-153) and 1,1-dichloro-2,2-bis (p-chlorophenyl)-ethylene (p,p'-DDE) levels with birth weight, gestational age and preterm births in Inuit and European populations. Environ Health 2010; 9:56; PMID:20819217; http://dx.doi.org/10.1186/1476-069X-9-56
- Torres-Sanchez L, Rothenberg SJ, Schnaas L, Cebrian ME, Osorio E, Del Carmen Hernandez M, Garcia-Hernandez RM, Del Rio-Garcia C, Wolff MS, Lopez-Carrillo L. In utero p,p'-DDE exposure and infant neurodevelopment: a perinatal cohort in Mexico. Environ Health Perspect 2007; 115:435-9; PMID:17431495; http://dx.doi.org/10.1289/ehp.9566
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001; 294:853-8; PMID:11679670; http://dx.doi.org/10.1126/science.1064921
- Zhang S, Chen L, Jung EJ, Calin GA. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther 2010; 87:754-8; PMID:20428111; http://dx.doi.org/10.1038/clpt.2010.46
- Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci 2008; 65:545-62; PMID:17965831; http://dx.doi.org/10.1007/s00018-007-7355-9
- Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 2012; 18:417-24; PMID:22383544; http://dx.doi.org/10.1093/molehr/gas013
- Corbel T, Gayrard V, Puel S, Lacroix MZ, Berrebi A, Gil S, Viguie C, Toutain PL, Picard-Hagen N. Bidirectional placental transfer of Bisphenol A and its main metabolite, Bisphenol A-Glucuronide, in the isolated perfused human placenta. Reprod Toxicol 2014; 47:51-8; PMID:24933518; http://dx.doi.org/10.1016/j.reprotox.2014.06.001
- Wang SL, Chang YC, Chao HR, Li CM, Li LA, Lin LY, Papke O. Body burdens of polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls and their relations to estrogen metabolism in pregnant women. Environ Health Perspect 2006; 114:740-5; PMID:16675430; http://dx.doi.org/10.1289/ehp.8809
- Zhao Y, Ruan X, Li Y, Yan M, Qin Z. Polybrominated diphenyl ethers (PBDEs) in aborted human fetuses and placental transfer during the first trimester of pregnancy. Environ Sci Technol 2013; 47:5939-46; PMID:23621775; http://dx.doi.org/10.1021/es305349x
- Gundacker C, Frohlich S, Graf-Rohrmeister K, Eibenberger B, Jessenig V, Gicic D, Prinz S, Wittmann KJ, Zeisler H, Vallant B, et al. Perinatal lead and mercury exposure in Austria. Sci Total Environ 2010; 408:5744-9; PMID:20825977; http://dx.doi.org/10.1016/j.scitotenv.2010.07.079
- Osman K, Akesson A, Berglund M, Bremme K, Schutz A, Ask K, Vahter M. Toxic and essential elements in placentas of Swedish women. Clin Biochem 2000; 33:131-8; PMID:10751591; http://dx.doi.org/10.1016/S0009-9120(00)00052-7
- Lafond J, Hamel A, Takser L, Vaillancourt C, Mergler D. Low environmental contamination by lead in pregnant women: effect on calcium transfer in human placental syncytiotrophoblasts. J Toxicol Environ Health A 2004; 67:1069-79; PMID:15205024; http://dx.doi.org/10.1080/15287390490452263
- Nanes JA, Xia Y, Dassanayake RM, Jones RM, Li A, Stodgell CJ, Walker CK, Szabo S, Leuthner S, Durkin MS, et al. Selected persistent organic pollutants in human placental tissue from the United States. Chemosphere 2014; 106:20-7; PMID:24485817; http://dx.doi.org/10.1016/j.chemosphere.2013.12.080
- Frederiksen M, Thomsen M, Vorkamp K, Knudsen LE. Patterns and concentration levels of polybrominated diphenyl ethers (PBDEs) in placental tissue of women in Denmark. Chemosphere 2009; 76:1464-9; PMID:19682725; http://dx.doi.org/10.1016/j.chemosphere.2009.07.017
- Gomara B, Herrero L, Ramos JJ, Mateo JR, Fernandez MA, Garcia JF, Gonzalez MJ. Distribution of polybrominated diphenyl ethers in human umbilical cord serum, paternal serum, maternal serum, placentas, and breast milk from Madrid population, Spain. Environ Sci Technol 2007; 41:6961-8; PMID:17993135; http://dx.doi.org/10.1021/es0714484
- Gomara B, Athanasiadou M, Quintanilla-Lopez JE, Gonzalez MJ, Bergman A. Polychlorinated biphenyls and their hydroxylated metabolites in placenta from Madrid mothers. Environ Sci Pollut Res Int 2012; 19:139-47; PMID:21698361; http://dx.doi.org/10.1007/s11356-011-0545-x
- Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li YX, Zhu X, Yao Y, Wang H, Qiao J, et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 2014; 63:1276-84; PMID:24664294; http://dx.doi.org/10.1161/HYPERTENSIONAHA.113.02647
- Wang W, Feng L, Zhang H, Hachy S, Satohisa S, Laurent LC, Parast M, Zheng J, Chen DB. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a, and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab 2012; 97:E1051-9; PMID:22438230; http://dx.doi.org/10.1210/jc.2011-3131
- Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol 2011; 204:178 e12-21; PMID:21093846; http://dx.doi.org/10.1016/j.ajog.2010.09.004
- Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci 2011; 18:46-56; PMID:21079238; http://dx.doi.org/10.1177/1933719110374115
- Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol 2013; 97:51-61; PMID:23432872; http://dx.doi.org/10.1016/j.jri.2012.11.001
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007; 8:166; PMID:17565689; http://dx.doi.org/10.1186/1471-2164-8-166
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 2010; 19:3566-82; PMID:20610438; http://dx.doi.org/10.1093/hmg/ddq272
- Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 2009; 4:587-92; PMID:19923923; http://dx.doi.org/10.4161/epi.4.8.10230
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 2004; 36:1291-5; PMID:15516931; http://dx.doi.org/10.1038/ng1468
- Debes F, Budtz-Jorgensen E, Weihe P, White RF, Grandjean P. Impact of prenatal methylmercury exposure on neurobehavioral function at age 14 years. Neurotoxicol Teratol 2006; 28:536-47; PMID:17067778; http://dx.doi.org/10.1016/j.ntt.2006.02.005
- Oken E, Radesky JS, Wright RO, Bellinger DC, Amarasiriwardena CJ, Kleinman KP, Hu H, Gillman MW. Maternal fish intake during pregnancy, blood mercury levels, and child cognition at age 3 years in a US cohort. Am J Epidemiol 2008; 167:1171-81; PMID:18353804; http://dx.doi.org/10.1093/aje/kwn034
- Needham LL, Grandjean P, Heinzow B, Jorgensen PJ, Nielsen F, Patterson DG, Jr., Sjodin A, Turner WE, Weihe P. Partition of environmental chemicals between maternal and fetal blood and tissues. Environ Sci Technol 2011; 45:1121-6; PMID:21166449; http://dx.doi.org/10.1021/es1019614
- Hsu CS, Liu PL, Chien LC, Chou SY, Han BC. Mercury concentration and fish consumption in Taiwanese pregnant women. BJOG 2007; 114:81-5; PMID:17081179; http://dx.doi.org/10.1111/j.1471-0528.2006.01142.x
- Watson GE, van Wijngaarden E, Love TM, McSorley EM, Bonham MP, Mulhern MS, Yeates AJ, Davidson PW, Shamlaye CF, Strain JJ, et al. Neurodevelopmental outcomes at 5 years in children exposed prenatally to maternal dental amalgam: the Seychelles Child Development Nutrition Study. Neurotoxicol Teratol 2013; 39:57-62; PMID:23856391; http://dx.doi.org/10.1016/j.ntt.2013.07.003
- Kong AP, Xiao K, Choi KC, Wang G, Chan MH, Ho CS, Chan I, Wong CK, Chan JC, Szeto CC. Associations between microRNA (miR-21, 126, 155 and 221), albuminuria and heavy metals in Hong Kong Chinese adolescents. Clin Chim Acta 2012; 413:1053-7; PMID:22405870; http://dx.doi.org/10.1016/j.cca.2012.02.014
- Mazumdar M, Bellinger DC, Gregas M, Abanilla K, Bacic J, Needleman HL. Low-level environmental lead exposure in childhood and adult intellectual function: a follow-up study. Environ Health 2011; 10:24; PMID:21450073; http://dx.doi.org/10.1186/1476-069X-10-24
- Zhang N, Baker HW, Tufts M, Raymond RE, Salihu H, Elliott MR. Early childhood lead exposure and academic achievement: evidence from Detroit public schools, 2008-2010. Am J Pub Health 2013; 103:e72-7; PMID:23327265; http://dx.doi.org/10.2105/AJPH.2012.301164
- Lanphear BP, Hornung R, Khoury J, Yolton K, Baghurst P, Bellinger DC, Canfield RL, Dietrich KN, Bornschein R, Greene T, et al. Low-level environmental lead exposure and children's intellectual function: an international pooled analysis. Environ Health Perspect 2005; 113:894-9; PMID:16002379; http://dx.doi.org/10.1289/ehp.7688
- Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 2010; 118:763-8; PMID:20061215; http://dx.doi.org/10.1289/ehp.0901300
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 2007; 21:415-26; PMID:17167072; http://dx.doi.org/10.1096/fj.06-6130com
- Ventayol M, Vinas JL, Sola A, Jung M, Brune B, Pi F, Mastora C, Hotter G. miRNA let-7e targeting MMP9 is involved in adipose-derived stem cell differentiation toward epithelia. Cell Death Dis 2014; 5:e1048; PMID:24503540; http://dx.doi.org/10.1038/cddis.2014.2
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell stem cell 2014; 14:40-52; PMID:24239284; http://dx.doi.org/10.1016/j.stem.2013.11.001
- Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell cycle 2008; 7:1529-38; PMID:18469525; http://dx.doi.org/10.4161/cc.7.11.5977
- Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol Endocrinol Metab 2013; 304:E836-43; PMID:23443922; http://dx.doi.org/10.1152/ajpendo.00660.2012
- Pietrzykowski AZ, Spijker S. Impulsivity and comorbid traits: a multi-step approach for finding putative responsible microRNAs in the amygdala. Front Neurosci 2014; 8:389; PMID:25561905; http://dx.doi.org/10.3389/fnins.2014.00389
- Roy A, Bellinger D, Hu H, Schwartz J, Ettinger AS, Wright RO, Bouchard M, Palaniappan K, Balakrishnan K. Lead exposure and behavior among young children in Chennai, India. Environ Health Perspect 2009; 117:1607-11; PMID:20019913; http://dx.doi.org/10.1289/ehp.0900625
- Wu J, Ying T, Shen Z, Wang H. Effect of low-level prenatal mercury exposure on neonate neurobehavioral development in China. Pediatr Neurol 2014; 51:93-9; PMID:24938141; http://dx.doi.org/10.1016/j.pediatrneurol.2014.03.018
- Suzuki K, Nakai K, Sugawara T, Nakamura T, Ohba T, Shimada M, Hosokawa T, Okamura K, Sakai T, Kurokawa N, et al. Neurobehavioral effects of prenatal exposure to methylmercury and PCBs, and seafood intake: neonatal behavioral assessment scale results of Tohoku study of child development. Environ Res 2010; 110:699-704; PMID:20673887; http://dx.doi.org/10.1016/j.envres.2010.07.001
- Wasserman GA, Staghezza-Jaramillo B, Shrout P, Popovac D, Graziano J. The effect of lead exposure on behavior problems in preschool children. Am J Pub Health 1998; 88:481-6; PMID:9518990; http://dx.doi.org/10.2105/AJPH.88.3.481
- Fieuw A, Kumps C, Schramm A, Pattyn F, Menten B, Antonacci F, Sudmant P, Schulte JH, Van Roy N, Vergult S, et al. Identification of a novel recurrent 1q42.2-1qter deletion in high risk MYCN single copy 11q deleted neuroblastomas. Int J Cancer J Int du Cancer 2012; 130:2599-606; PMID:21796619; http://dx.doi.org/10.1002/ijc.26317
- Guida M, Marra ML, Zullo F, Guida M, Trifuoggi M, Biffali E, Borra M, De Mieri G, D'Alessandro R, De Felice B. Association between exposure to dioxin-like polychlorinated biphenyls and miR-191 expression in human peripheral blood mononuclear cells. Mutat Res 2013; 753:36-41; PMID:23500661; http://dx.doi.org/10.1016/j.mrgentox.2012.12.018
- Ligorio M, Izzotti A, Pulliero A, Arrigo P. Mutagens interfere with microRNA maturation by inhibiting DICER. An in silico biology analysis. Mutat Res 2011; 717:116-28; PMID:21889945; http://dx.doi.org/10.1016/j.mrfmmm.2011.07.020
- Izzotti A, Pulliero A. The effects of environmental chemical carcinogens on the microRNA machinery. Int J Hygiene Environ Health 2014; 217:601-27; PMID:24560354; http://dx.doi.org/10.1016/j.ijheh.2014.01.001
- Suzuki HI, Miyazono K. Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway. J Mol Med 2010; 88:1085-94; PMID:20614100; http://dx.doi.org/10.1007/s00109-010-0650-1
- Michels KB, Rosner BA. Data trawling: to fish or not to fish. Lancet 1996; 348:1152-3; PMID:8888172; http://dx.doi.org/10.1016/S0140-6736(96)05418-9
- Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology 1990; 1:43-6; PMID:2081237; http://dx.doi.org/10.1097/00001648-199001000-00010
- Stodgell CJ, Miller RK, Salamone L, Murray J, Chen J, Lambertini L, Schadt E, Littman L, Landrigan P, Aagaard K, et al. LACK OF CORRELATION BETWEEN PLACENTAL GENE EXPRESSION AND RNA INTEGRITY NUMBER (RIN) OR TIME TO COLLECTION. Placenta 2014; 35:A46; http://dx.doi.org/10.1016/j.placenta.2014.06.152
- Miller RK, Stodgell CJ, Katzman P, Friedman A, Friedman M, Ruffolo L, Penmetsa A, Salamone L, Salafia C, Chen J, et al. Human placental study of genetics/genomic, environmental contaminant and morphology assessments from 12 U.S. counties e methods and results from the U.S. national children's study (ncs). Placenta 2014; 35:A2; http://dx.doi.org/10.1016/j.placenta.2014.06.007
- Dassanayake RM, Wei H, Chen RC, Li A. Optimization of the matrix solid phase dispersion extraction procedure for the analysis of polybrominated diphenyl ethers in human placenta. Anal Chem 2009; 81:9795-801; PMID:19863067; http://dx.doi.org/10.1021/ac901805d
- Kahakachchi CL, Moore DA. Identification and characterization of gadolinium(III) complexes in biological tissue extracts. Metallomics 2010; 2:490-7; PMID:21072349; http://dx.doi.org/10.1039/b915806e
- Kohlmeyer U, Jantzen E, Kuballa J, Jakubik S. Benefits of high resolution IC-ICP-MS for the routine analysis of inorganic and organic arsenic species in food products of marine and terrestrial origin. Anal Bioanal Chem 2003; 377:6-13; PMID:12830352; http://dx.doi.org/10.1007/s00216-003-2064-1
- Darrah TH, Prutsman-Pfeiffer JJ, Poreda RJ, Ellen Campbell M, Hauschka PV, Hannigan RE. Incorporation of excess gadolinium into human bone from medical contrast agents. Metallomics 2009; 1:479-88; PMID:21305156; http://dx.doi.org/10.1039/b905145g
- Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012; 28:1546-8; PMID:22513995; http://dx.doi.org/10.1093/bioinformatics/bts188
- Smyth GK. Limma: linear models for microarray data. New York: Springer, 2005:397-420.