Abstract
With the goal of investigating if epigenetic biomarkers from white blood cells (WBC) are associated with dietary, anthropometric, metabolic, inflammatory and oxidative stress parameters in young and apparently healthy individuals. We evaluated 156 individuals (91 women, 65 men; age: 23.1±3.5 years; body mass index: 22.0±2.9 kg/m2) for anthropometric, biochemical and clinical markers, including some components of the antioxidant defense system and inflammatory response. DNA methylation of LINE-1, TNF-α and IL-6 and the expression of some genes related to the inflammatory process were analyzed in WBC. Adiposity was lower among individuals with higher LINE-1 methylation. On the contrary, body fat-free mass was higher among those with higher LINE-1 methylation. Individuals with higher LINE-1 methylation had higher daily intakes of calories, iron and riboflavin. However, those individuals who presented lower percentages of LINE-1 methylation reported higher intakes of copper, niacin and thiamin. Interestingly, the group with higher LINE-1 methylation had a lower percentage of current smokers and more individuals practicing sports. On the other hand, TNF-α methylation percentage was negatively associated with waist girth, waist-to-hip ratio and waist-to-stature ratio. Plasma TNF-α levels were lower in those individuals with higher TNF-α methylation. This study suggests that higher levels of LINE-1 and TNF-α methylation are associated with better indicators of adiposity status in healthy young individuals. In addition, energy and micronutrient intake, as well as a healthy lifestyle, may have a role in the regulation of DNA methylation in WBC and the subsequent metabolic changes may affect epigenetic biomarkers.
Abbreviations
| BMI | = | Body mass index |
| CI | = | Confidence interval |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| HDL-c | = | High-density lipoprotein |
| IL | = | Interleukin |
| LDL-c | = | Low-density lipoprotein |
| LINE-1 | = | Long interspersed nucleotide element-1 |
| MET | = | Metabolic equivalent |
| MS-HRM | = | Methylation-sensitive high resolution melting |
| OS | = | Oxidative stress |
| OR | = | Odds ratio |
| ox-LDL | = | Oxidized Low-density lipoprotein |
| PBMC | = | Peripheral blood mononuclear cells |
| ROS | = | Oxygen-reactive species |
| TAC | = | Total antioxidant capacity |
| TC/HDL-c | = | Castelli index |
| TC | = | Total cholesterol |
| TNF-α | = | Tumor necrosis factor α |
| UFV | = | Federal University of Viçosa |
| UTR | = | Untranslated region |
| VLDL | = | Very Low-density lipoprotein |
| WBC | = | White blood cell |
Introduction
Lifestyle and nutrients may induce transient or permanent alterations in the epigenetic marks that regulate the expression of genes involved in metabolic processes and networks, which could be one of the factors leading to chronic diseases.Citation1 The most common epigenetic modification is DNA methylation. Approximately 50% of the human genome is composed of repetitive sequences such as Long Interspersed Nuclear Elements (LINE), which are the most common repetitive elements of interspersed DNA repeats.Citation2 These elements are usually methylated, and their transcription and retrotransposition are suppressed by a variety of control mechanisms including methylation and non-coding RNA.Citation3 Because of its high genome dissemination, LINE-1 methylation status has been proposed as a surrogate marker for estimating global DNA methylation level.Citation2
A global decrease in the methylation of peripheral blood DNA was found to be an independent risk factor for many cancersCitation4 and for developmental, autoimmune, and other chronic diseases.Citation5 LINE-1 methylation levels in peripheral blood mononuclear cells (PBMCs) have been reported to predict response to a dietary weight-loss interventionCitation6 and have been associated with serum glucose levels.Citation7 The methylation levels of individual genes, such as those of tumor necrosis factor α (TNF-α), have been also proposed as biomarkers of response to a hypocaloric diet.Citation8 Methylation status may, therefore, serve as a biomarker for early diagnostics, prediction of prognosis, and response to treatments. Moreover, some dietary factors are able to alter the percentage of methylation, both at LINE-1 and in proinflammatory genes. For example, energy restriction has been reported to decrease IL-6 methylation levels in buffy coat DNACitation7 and TNF-α methylation has been associated with n-6 polyunsaturated fatty acids (PUFAs) intake.Citation9 However, the etiopathogenic mechanisms remain poorly understood. Thus, the objective of this study was to explore the relation between DNA methylation levels of LINE-1, TNF-α and IL-6 in white blood cells (WBC) and anthropometric, biochemical, clinical, dietary, inflammatory and oxidative stress parameters in young and apparently healthy adults.
Material and methods
Subjects
One hundred 5six healthy subjects were recruited to participate in the study (91 women and 65 men; age: 23.1 ± 3.5 y and BMI: 22.0 ± 2.9 kg/m2). Most of the study population self-reported to be white (n = 131) followed by black (n = 16) and others (n = 9). Initial screening excluded subjects with evidence of any metabolic disease, chronic inflammation, hydric balance disorders, changes in body composition and problems in nutrient absorption or metabolism. Other exclusion criteria were drug or nutritional treatment that affects energy balance, dietary intake, lipid profile, insulin levels or glucose metabolism, contraceptive use up to 2 months before participation in the study and weight loss diet follow-up or unstable weight in the past 6 months. In agreement with the principles of the Helsinki Declaration and following a clear explanation of the study protocol, each participant signed a written informed consent form. The study was approved by the Human Research Ethics Committee of the Federal University of Viçosa, Brazil (protocol number 019/2011).
Anthropometric and body composition assessments
Height was measured with a stadiometer (Seca 206 model, Hamburg, Germany) to the nearest 0.1 cm. Body weight was measured to the nearest 0.1 kg by using an electronic microdigital scale (Tanita TBF-300 A model, Tokyo, Japan). Body mass index (BMI) was calculated by the quotient between body weight and square height (kg/m2). Waist and hip perimeters were measured with an inelastic and flexible tape to the nearest 0.1 m. Triceps, biceps, subscapular and suprailiac skinfold thicknesses were measured to the nearest 1 mm by using a skinfold caliper (Lange caliper, Cambridge Scientific Industries Inc., Maryland, USA). The sum of skinfold thickness was calculated. Total body fat percentage was measured to the nearest 0.1 % using a body composition analyzer (Biodynamics 310 model, Washington, USA). Body fat mass and body fat-free mass were estimated using the same body composition analyzer. Truncal fat percentage was computed as the sum of subscapular and suprailiac skinfold thicknesses divided by the sum of 4 skinfold measurements.Citation10 Finally, truncal adiposity index was calculated by the ratio of subscapular to triceps skinfold thickness.Citation11
Blood pressure assessment
Systolic and diastolic blood pressures were measured with a mercury sphygmomanometer (BIC, SP, Brazil) following World Health Organization criteria.Citation12
Dietary intake and lifestyle assessments
A 72-hour food record was used to collect information about energy and nutrient intake. A booklet was given to the participants to record everything they ate or drank over a period of 3 non-consecutive days, including a weekend day. Dietary intake was computed using specific software (DietPro®, version 5.0, AS Systems).
Covariates about diet and lifestyle, such as vitamin supplementation, smoking status (smokers or non-smokers), number of cigarettes per day, regular physical activity (yes or no) and volume of physical activity, were also collected. To quantify the volume of physical activity, an activity metabolic equivalent (MET) was used.Citation13 This index represents the ratio between energy expenditure during each specific activity and resting metabolic rate. METs were computed by a multiple of resting metabolic rate (MET score) to each activity. The MET scores were provided by Compendium of Physical Activities, a coding scheme that classifies specific physical activity by rate of energy expenditure.Citation14 METs were calculated by multiplying time spent on each activity by a specific MET score to that activity. The scores were then summed over all activities to obtain a mean value of overall week, expressed in hours per day.
Analyses of biological samples
Blood samples were drawn by vein puncture after a 12-hour overnight fast. Ethylenediaminetetraacetic acid (EDTA) plasma, heparin plasma and serum samples were separated from whole blood by centrifugation at 3500 rpm at 5°C for 15 min (Eppendorf AG, 5804 R model, Hamburg, Germany) while erythrocytes were separated from whole blood by centrifugation at 3000 rpm at 5°C for 10 min. All samples were immediately stored at −80°C until assay.
Lipid and glucose profile
Serum glucose, total cholesterol, high-density lipoprotein cholesterol (HDL-c), and triacylglycerol concentrations were assessed in an automated biochemical analyzer (BS-200, Shenzhen Mindray Bio-medical Electronics Co., Nanshan, China) using specific colorimetric kits (Bioclin, Quibasa, Minas Gerais, Brazil). Low-density lipoprotein cholesterol (LDL-c) data were calculated by the Friedewald equation as previously validated.Citation15 The total cholesterol-to-HDL-c ratio was also assessed.Citation16 Plasma insulin concentrations (sensitivity 2 µU/mL) were measured by an enzyme-linked immunosorbent assay (ELISA) kit as described by the supplier (Linco Research, St. Charles, USA). The homeostasis model assessment of insulin resistance (HOMA-IR), calculated as fasting glucose (nmol/L) x fasting insulin (µU/mL)/22.5,Citation17 was used to estimate insulin resistance.
Antioxidant markers
Plasma total antioxidant capacity (TAC) was assessed by a colorimetric assay, which relies on the ability of antioxidants in the sample to inhibit the oxidation of ABTS (2,2′-azino-di-[3-ethylbenzthiazoline sulphonate]) to ABTS•+ by metmyoglobin (Cayman Chemical, Ann Arbor, MI, USA). Plasma oxidized-LDL (ox-LDL) concentrations were determined by ELISA (Mercodia, Uppsala, Sweden). Glutathione peroxidase (GPx) activity [nmol/(mL/min)] was measured in erythrocytes using a commercially available kit as described by the supplier (Cayman Chemical, Cat. 703102). Uric acid and ceruloplasmin concentrations were assessed with an automated biochemical analyzer (BS-200, Shenzhen Mindray Bio-medical Electronics Co., China) using specific commercially available kits (Bioclin).
Inflammatory markers
Plasma IL-6, TNF-α and C-reactive protein (CRP) levels were determined by using commercial ELISA kits from Cayman Chemical. Adiponectin levels were also assessed by ELISA (SPIBIO, Montigny le Bretonneux, France). Serum complement factor-3 (C3) was quantified with an automated biochemical analyzer (model BS-200) using a specific colorimetric kit (Bioclin).
Trace elements in nails
Nail samples were collected at the time of interview and stored at room temperature in clean polypropylene bags. Fingernail and toenail samples were treated with sub boiling nitric acid in a high-pressure Teflon digestion vessel using a microwave digestion system (Ethos Plus, Millestone, Sorisole, Italy). A Perkin Elmer Analyst 800 atomic absorption spectrometer (Norwalk, CT, USA), equipped with transverse-heated graphite atomizer, Zeeman background corrector and AS-800 autosampler, was used for measuring selenium at 196.0 nm with a spectral band width of 2.0 nm.Citation18 An electrodeless discharge lamp (Perkin Elmer) was used as a light source operated at 280 mA. Pyrolytic coated graphite tubes with end caps supplied by Perkin Elmer were used. Zinc and copper concentrations in digested acid solutions were analyzed by flame atomic absorption spectrophotometry. Zinc and copper hollow cathode lamps provided resonance lines of 213.9 and 324.8 and were operated both at 15 mA with a slit width seat at 0.7 nm.
RNA extraction and real time quantitative PCR
Total RNA was extracted from WBC using Trizol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). The concentration and purity of RNA were determined at 260/280 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Total RNA (2 μg) was reverse-transcribed using the SYBR® Green RT-PCR Reagents Kit (Cat. 4306736, Life Technologies, Waltham, MA, USA) according to the manufacturer's protocol. cDNA was amplified in triplicate with SYBR® Green PCR Master Mix (Cat. No. 4309155) and the respective specifics primers (see Supplementary Table 1). The analyzed genes were selected on the basis of previous studies supporting their possible involvement in inflammatory pathways related to metabolic syndrome. mRNA levels were normalized to the endogenous control glycerol-3-phosphate dehydrogenase (GAPDH). The ΔΔCt (crossing threshold) method was used for quantification (ABI) and the fold changes reported as 2−ΔΔCt.Citation9 All quantitative real-time RT-PCR measurements were performed using a 7900 HT Fast Real-Time PCR system (Life Technologies).
DNA isolation and methylated DNA standards
DNA was isolated from the WBC using the Master Pure DNA Purification Kit for Blood Version II (Epicenter, Madison, WI, USA) according to the instructions provided by the manufacturer. Purified DNA was stored at −20°C until use. Purified DNA was quantified by PicoGreen dsDNA Quantitation Reagent (Invitrogen). Cells-to-CpG™ Methylated gDNA Control Kit (Life Technologies) and DNA from placenta cells (D3160, Sigma Aldrich, St. Louis, MO, USA) were used as methylated and non-methylated DNA standards, respectively. To generate a range of methylated and unmethylated DNA standards, the 2 standard DNA controls were mixed in 0, 20, 40, 60, 80, and 100% methylated to unmethylated template ratios. One microgram of each standard and sample DNA was bisulfite-converted (BSC) by using the Epitect Fast Bisulfite Conversion Kit (Qiagen, Venlo, Limburg, The Netherlands) according to the manufacturer's instructions, thus converting non-methylated cytosines into uracil. All bisulfite-converted DNAs were diluted to 5 ng/μL for use in PCR.
Methylation-sensitive high resolution melting (MS-HRM) analysis
Specific primers against the completely methylated sense strand sequence were designed according to the recommendations of Wojdacz and Dobrovic (2007)Citation19 in order to minimize PCR bias. The promoter region of the consensus LINE-1 sequence (GenBank: X58075) was used to design primer sets with the Primer3 website (http://frodo.wi.mit.edu/primer3/). Thus, the primers for analyzing LINE-1 methylation were: forward, 5′-GCGAGGTATTGTTTTATTTGGGA-3′; reverse, 5′-CGCCGTTTCTTAAACC-3′. They screened 8 CpGs in an amplicon length of 141 bp and were first employed by Tse et al.Citation20 The primers for analyzing IL-6 methylation were: forward, 5′-TTATGTAGGAAAGAGAATTTGGTTTAG-3′ and reverse, 5′-AAAAAATAA AATCATCCATTCTTCAC-3′. They covered 5 CpGs in an amplicon length of 181 bp as mentioned elsewhere.Citation7 The primers for TNF-α were: forward, 5′-TTTTGGAAAGGATATTATGAGTATTGA-3′ and reverse, 5′-CTAAAACCCTA AAACCCCCCTAT-3′. They covered 4 CpGs in an amplicon length of 99 bp as mentioned elsewhere.Citation9 Genomic sequence and CpGs sites covered by the MS-HRM primers studying TNF-α and IL-6 methylation are shown in Figs. S1 and S2. PCR amplification of the DNA was carried out using a 7900 HT Fast Real-Time PCR System (Life Technologies) equipped with the SDS Software (Version 2.4.1, Life Technologies). PCR was performed in a 10-μL reaction volume, and 5 ng of bisulfite-converted DNA templates for LINE-1 assay were added to each well, which contained 1×MeltDoctor™ HRM Master Mix (HRM) (Life Technologies) and 0.2 μM each primer. The cycling protocol conditions included a single enzyme activation step of 10 minutes at 95°C followed by 40 cycles of the following steps: denaturation 95°C, 15 seconds, and annealing 60°C, 1 minute. The MS-HRM step was performed after 40 cycles of amplification and the MS-HRM analysis was initiated by denaturing all products at 95°C for 1 minute, followed by annealing at 55°C for 1 minute. Samples were slowly warmed to 95°C at 0.1°C/second. The High ReSolution Melt Software v2.0 (Life Technologies) was employed for end-product analysis. This algorithm allowed the raw melt curves to be normalized for fluorescence intensity, and a temperature shift was applied to align the normalized melt curves, which facilitated the analysis of samples with varying Ct values. A difference curve was then derived from the first derivative of the melt curves. Data for the difference melt curves were exported to Excel (Office 2007; Microsoft Corp., Redmond, WA) for further analyses. Graphs were plotted and inverted vertically. Both peak-height and area-under-the-curve from the normalized, temperature-shifted, difference curves were used to generate a standard curve and determine the degree of methylation of each DNA sample. All participant DNA samples were analyzed on a 384-well plate, which included a no-template control (NTC) and a set of reference methylation standards. Reference methylation standard curves and experimental samples were tested in triplicate.
Statistical analysis
The Kolmogorov-Smirnov normality test was used to determine variable distribution. Accordingly, the parametric Student t test or nonparametric Mann-Whitney U test was performed to detect differences between subjects with higher and lower DNA methylation percentage than the median value. Dichotomous variables were analyzed by X2 test. Contrasts and Tukey's post-hoc tests for one-way ANOVA were performed to analyze differences among tertiles and quartiles. P for trend was calculated. The Spearman correlation coefficients were used to screen the statistical associations between DNA methylation and interest variables. Linear regression model, used to identify the predictors of DNA methylation, was adjusted for covariates such as calories, sex, age, smoking status and regular physical activity, when they showed significant effect. Nutrients from the diet were adjusted by total energy intake by using the residual method.Citation21 Results are presented as mean ± SD (standard deviation). Confidence intervals (95% CIs) were used to describe linear regression coefficients (β). P < 0.05 was considered statistically significant. Statistical analyses were performed by using SAS software system version 8.0 for Windows (SAS Institute Inc., Cary, NC 27513, USA). GraphPad Prism® version 6.0 C (La Jolla, CA, USA) was used to show graphically the results.
Results
Baseline anthropometric, clinical and biochemical measurements from participants are presented in Supplementary Table 2. The study population was considered healthy and some expected differences in anthropometric variables (i.e., body weight, BMI, waist circumference, truncal fat percentage, etc.) were found between genders.
LINE-1 methylation levels were significantly lower in women (about 7.3% less) (P < 0.01). In addition, LINE-1 methylation were positively associated with body weight (r = 0.296; P = 0.032; n = 120). These results were confirmed when the subjects were divided according to the median of body weight (61.0 kg), even when analyses were made separately by gender (P < 0.05 for both). More detailed analyses showed that indicators of adiposity, such as skinfolds and total body fat, were lower among individuals with higher LINE-1 methylation (P < 0.05 for all) (). On the contrary, body fat-free mass was higher (P for trend = 0.016) among those with higher LINE-1 methylation. These individuals also exhibited lower plasma levels of ceruloplasmin (P for trend = 0.002) and higher IL-6 in plasma (P for trend = 0.006), as shown in .
Table 1. Anthropometric, clinical and biochemical data (mean ± SD) of all individuals (n=120) categorized by quartiles of LINE-1 methylation (%).
Calorie (kcal) intake was higher (P for trend = 0.04) among individuals with higher LINE-1 methylation even after adjusting for body weight (). A similar result was observed for daily iron intake (mg) even after adjusting by calories and body weight (P < 0.05). It is worth emphasizing that iron intake explained, through the r values, about 8.4% of LINE-1 methylation, even after adjusting by energy, gender and smoking (). In the same way, higher daily riboflavin (B2 vitamin) intake was found in those individuals with bigger values to LINE-1 methylation (P < 0.05 inter quartiles). However, those individuals who presented lowers percentages of LINE-1 methylation reported eating higher amounts of copper, niacin (B3 vitamin) and thiamin (B1 vitamin) (P for trend <0.05 for all). Interestingly, the group with higher LINE-1 methylation (<83.02%) had a lower percentage of current smokers (P = 0.012) and more individuals practicing sports (P < 0.05) (). These results are confirmed with the lower number of cigarettes smoked per day (P = 0.041) and higher physical activity per day (P = 0.047) in those subjects with bigger values to LINE-1 methylation. These results suggest that high LINE-1 methylation could be associated to a healthier lifestyle.
Figure 1. Linear regression model showing association between daily iron intake (refined by calories - residue method)Citation21 and LINE-1 methylation (%) after adjustment for smoking and gender. The dotted lines represent the confidence interval limit (95% CI).
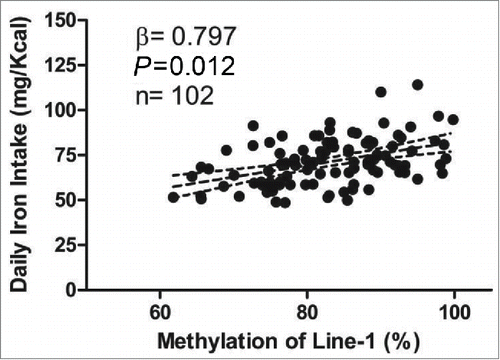
Table 2. Daily nutrient intake (mean ± SD) for all individuals categorized according to the quartiles of LINE-1 methylation (%) (n=115).
Table 3. Lifestyle features (mean ± SD) for all individuals categorized according to the median of LINE-1 methylation (%).
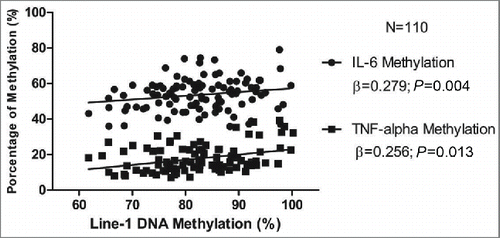
LINE-1 methylation positively associated with TNF-α and IL-6 methylation percentage not only when linear regression was adjusted by gender, smoking and age (r = 0.204; P = 0.048 and r = 0.332; P = 0.003, respectively) (), but also when the subjects were divided according to the median of LINE-1 methylation (83.02%; P<0.05 for both). Results from logistic regression analysis (odds ratio-OR) confirmed that individuals with higher LINE-1 methylation levels (third tertile, >83.1 %) were more likely to have higher percentage of IL-6 methylation (3.08 times) than those with lower methylation levels (first tertile, <77.10 %; IC: 1.13−8.36; P = 0.013).
Figure 2. Linear regression model (r) showing association among LINE-1 methylation (in %) and IL-6 (r=0.204; p=0.048) and TNF-α (r=0.332; p=0.003) methylation percentage after adjusting by gender and smoking.
Individuals whose TNF-α methylation percentage was above the tertile 3 (T3) showed lowers values of waist girth, waist-to-hip ratio, waist-to-stature ratio and selenium in nails (P < 0.05) (). Moreover, they presented lower TNF-α expression in WBC (P for trend = 0.041), suggesting that the hypermethylation of this gene was associated with inhibition of its transcription ( and ). It should be noted that the plasma levels of TNF-α were also lower in those individuals with greater methylation for this gene (P for trend = 0.048). On the other hand, these individuals presented higher expression of IL-18 and plasma values of IL-6 (P < 0.05 for all) (). As expected, the individuals with higher TNF-α methylation showed also higher methylation of IL-6 and LINE-1 ().
Figure 3. Associations between relative expression and methylation levels of TNF-α and IL-6. A) Pearson's correlation (r2) between relative expression in arbitrary units (AU) and methylation levels (%) of TNF-α. B) Pearson's correlation between relative expression (AU) and methylation levels (%) of IL-6 (only in men). The dotted lines represent the confidence rating limit (IC-95%).
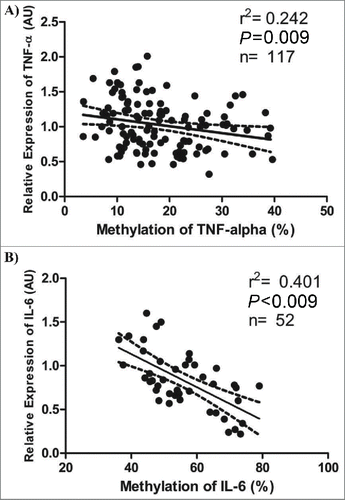
Table 4. Anthropometric, clinical and biochemical data (mean ± SD) of all individuals (n=107) categorized by tertiles of TNF-α methylation (%).
Table 5. Relative expression and percentage of methylation data (mean ± SD) for all participants (n=105) categorized by median of TNF-α methylation (%).
No differences were found between the groups whose IL-6 methylation levels were above and below the median (≤ or >56.57 %). However, when only men were taken into account, the levels of selenium in nails and mRNA levels of IL-6 and TNF-α were lower in those individuals with higher IL-6 methylation values (P < 0.05 for all). On the other hand, a negative association was found between IL-6 methylation in white blood cells and relative expression (mRNA) of IL-6 in the same cells ().
Discussion
There is growing evidence of the involvement of epigenetic mechanisms in disease onset, including obesity and type 2 diabetes.Citation1 A decrease in global DNA methylation is associated with increased genomic instability and chromosomal rearrangements,Citation22 a common biological mechanism in several diseases including cancer. Reduced DNA methylation in WBCs may be an indicator of systemic hypomethylation and of cumulative environmental impacts. Conversely, an increase in DNA methylation levels of repetitive elements may have a protective effect against genomic instability and unwanted chromosomal rearrangements.Citation23 However, there are many gaps about the determinants of DNA methylation levels in healthy people prior to disease onset, and there is great interest in finding new, early biomarkers for disease risk reduction and health promotion.
Our findings support previous results showing that women have significantly lower levels of LINE-1 methylation.Citation24-26 Lower levels of global methylation in women may be due to different levels of dietary folate or other one-carbon nutrients in men and women.Citation27 Women may also have a higher folate requirement than men because of regular loss of red blood cells through menstruation, but it can not explain the gender-specific difference in global methylation in postmenopausal women.Citation27,28 There is still discussion about hormonal factors and the importance of DNA methylation for X-chromosome inactivation in women.Citation24,29 Further studies are needed to decipher the relationship between gender and LINE-1 methylation.
The results from this study are also in line with other studies that reported association between LINE-1 methylation and smoking status.Citation25 Cigarette smoke is considered one of the most powerful environmental modifiers of DNA methylation.Citation30,31 For example, an experimental study showed that cigarette smoke could induce DNA demethylation in repeat elements such as LINE-1.Citation32 The specific mechanisms of how cigarette smoke may alter DNA methylation are becoming better understood and may be reviewed elsewhere.Citation33 Our findings suggest that the toxic effects of tobacco could be at least partly mediated by modulation of the epigenetic landscape. On the other hand, higher LINE-1 methylation has been previously described in individuals with higher exercise levels,Citation34,35 suggesting that an increase in LINE-1 methylation might be associated with healthy lifestyle habits. As epigenetic marks are potentially reversible, this result may have public health implications and merits further investigation.
Methylation of DNA is a biochemical process in which a methyl group is added to DNA nucleotides. Several nutrients act as key enzyme cofactors and play essential roles in methyl group metabolism and DNA methylation in particular, being riboflavin, vitamin B12 and folate the major determinants of one-carbon metabolism.Citation36 In this sense, our results confirm the importance of riboflavin in DNA methylation process.
Iron intake was positively associated with LINE-1 methylation level even after adjustment for gender, calories and body weight. A study with 892 individuals also showed a similar association between iron intake and LINE-1 methylation in leukocytes.Citation25 Iron, together with 2-oxoglutarate and oxygen, is an essential cofactor for the 10–11 translocation (TET) family of proteins that hydroxylates 5-methylcytosine to 5-hydroxymethylcytosine and further oxidizes to 5-carboxylcytosine and 5-formylcytosine, which have all been suggested to be precursors for both active and passive DNA demethylation.Citation37 The mechanisms are not well understood, but these data reinforce the role of dietary iron on the methylation status.
A recent trial explored the associations between changes in lifestyle modifications, such as diet, and global epigenetic biomarkers in blood of overweight female breast cancer survivors. After a weight loss intervention consisting on dietary and physical activity changes, LINE-1 methylation levels were significantly elevated as compared to baseline.Citation38 Particularly, a 10 % increase in the frequency of fruit consumption was associated with an increase in LINE-1 methylation levels of 0.42 %. In a study with 177 young healthy women, those whose consumption of fruit was below the median value (<201 g/day) were 3.7 times more likely to display LINE-1 hypomethylation than women whose consumption was above the median (OR 3.7; 95% CI 1.4–9.5).Citation39
Several studies have examined the association between BMI and LINE-1 methylation levels with conflicting results.Citation40 Elevated BMI has been shown to be associated with lower LINE-1 methylation in some investigations,Citation41,42 but, in other study, participants with BMI ≥40 kg/m2 had higher LINE-1 methylation levels than those with BMI ≤25.Citation43 On the other hand, some studies found no associations between BMI and LINE-1 methylation.Citation44,45 Although the current study did not find an association between BMI and LINE-1 status, body weight and LINE-1 were positively associated. A recent report from our group has shown that LINE-1 methylation was a biomarker of weight loss in obese subjects.Citation6 In a recent randomized, crossover study of a 6-month weight loss intervention, over a 12-month period, changes in body fat percentage were positively associated with LINE-1 methylation (β=0.19, p=0.001).Citation38 In that study, LINE-1 methylation was statistically significantly elevated at 6 and 12 months compared to baseline. Moreover, analysis of rectal biopsies free of colorectal disease obtained from 185 individuals showed that higher waist and hip perimeters were significantly associated with lower methylation of LINE-1.Citation26 Interestingly the present study found that central obesity and fat mass percent were significant predictors for low LINE-1 methylation levels, although no associations remained after adjusting for gender and smoking.
In the present research, TNF-α methylation analysis demonstrates a negative association with central adiposity. In this sense, a cross-sectional study with 40 normal-weight young women showed that those with higher truncal fat (≥52.3%) presented lower methylation of the TNF-α gene promoter than those with lower truncal adiposity.Citation9 In two other studies, authors concluded that TNF-α methylation levels could be used as epigenetic biomarker concerning the response to a low-calorie diet.Citation8,46 Indeed, methylation profile could help predict susceptibility to weight loss as well as some obesity-related comorbidities, such as hypertension or type 2 diabetes.Citation47 These results are in line with the findings from the present work, suggesting an important relationship between epigenetic mechanisms and body composition.
Recent studies also reported a good correlation between leukocyte DNA methylation level and degree of inflammation, suggesting that cytosine methylation may represent a novel mechanism underlying the association between obesity, inflammation and disease risk.Citation48,49 In this context, our study observed that the methylation levels of both, TNF-α and IL-6, negatively correlated with the mRNA levels of both cytokines in white blood cells. However, it is important to note that a relationship between IL-6 mRNA and plasma levels was not found, which can be explained by this cytokine not only being released to plasma by white blood cells, but also by muscle, adipose tissue and other organs. It is important to highlight that the present study was conducted in young, apparently healthy individuals, with no physiopathological conditions. Thus, it is possible to consider this negative association as a compensatory mechanism, with the purpose of restoring homeostatic balance.
This study presents some limitations that should be considered. Firstly, the dietary assessment software did not provide important information in order to discuss the data deeply. We only examined consumption of specific nutrients, which does not account for the combinations and interactions of multiple nutrients in the human diet. Moreover, random measurement error in ascertaining dietary intake with the food frequency questionnaire could have resulted in some misclassification of intake. Secondly, some individuals were not included in the present analyses due to lack of methylation data. However, there was no relevant difference in selected characteristics (except age and gender) between those with and without those data. To minimize the potential bias, all the models were adjusted for age, gender and smoking status when necessary. Thirdly, we did not account for the proportion of white blood cell subtypes from the buffy coat in the analyses. DNA methylation is tissue- and cell-specific, and it could depend on the cell type distribution. In this sense, there is some evidence that DNA methylation is inversely related to the proportion of lymphocytes.Citation44 Finally, the cross-sectional design of this research does not allow determination of causality and the associations observed should be checked carefully. Therefore, type I/II errors cannot be ruled out, nor can the involvement of other mechanisms in the regulation of methylation status. Future studies are required to replicate and extend these findings to different populations.
As a conclusion, our findings contribute to the growing evidence that the health impacts of changes in dietary habits and other lifestyle factors may be mediated through epigenetic modifications. Moreover, our work provides preliminary evidence for the use of WBC DNA methylation biomarkers to monitor lifestyle interventions trials and identify early biomarkers of metabolic complications.
Conclusions
This study suggests that higher LINE-1 and TNF-α methylation could be associated with some indicators of adiposity, especially body fat, waist girth and waist-to-hip ratio. In addition, high LINE-1 methylation levels are directly associated with some dietary factors (calories, iron) but inversely associated with total fat mass. Moreover, high LINE-1 methylation is associated with a healthier lifestyle (physical activity and not smoking) and might be an early indicator of resistance to win adiposity even when eating more food.
Authors' Contributions
Contributors JLM-R: field work, data collection, analysis, and writing of the manuscript; FIM and MLM: design, analysis, and editing of the manuscript; DMM: design, field work, and data collection; JB: project leader, scientific interpretation, financial management, and editing of the manuscript; JAM: design, field work, scientific interpretation, financial management, and editing of the manuscript. All authors read and approved the final manuscript.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank the volunteers of this study and the excellent technical assistance of Ana Lorente and Caroline Müller.
KEPI_A_1135286_s02.docx
Download MS Word (268.7 KB)Funding
This study was supported by the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG-APQ-01609-10), CAPES Foundation (Ministry of Education of Brazil, process n° 6409-13-0), CIBERobn, MINECO (ref. AGL2013-45554-R) and the Health Department of the Government of Navarra (482009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Martinez JA, Milagro FI, Claycombe KJ, Schalinske KL. Epigenetics in adipose tissue, obesity, weight loss, and diabetes. Adv Nutr 2014; 5:71-81; PMID:24425725; http://dx.doi.org/10.3945/an.113.004705
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33:6823-36; PMID:16326863; http://dx.doi.org/10.1093/nar/gki987
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006; 6:107-16; PMID:16491070; http://dx.doi.org/10.1038/nrc1799
- Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One 2012; 7:e34615; PMID:22509334; http://dx.doi.org/10.1371/journal.pone.0034615
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597-610; PMID:16136652; http://dx.doi.org/10.1038/nrg1655
- Garcia-Lacarte M, Milagro FI, Zulet MA, Martinez JA, Mansego ML. LINE-1 methylation levels, a biomarker of weight loss in obese subjects, are influenced by dietary antioxidant capacity. Redox Rep 2015; PMID:26197243.
- Nicoletti CF, Nonino CB, De Oliveira BA, Pinhel MA, Mansego ML, Milagro FI, Zulet MA, Martinez JA. DNA Methylation and Hydroxymethylation Levels in Relation to Two Weight Loss Strategies: Energy-Restricted Diet or Bariatric Surgery. Obes Surg 2015.
- Cordero P, Campion J, Milagro FI, Goyenechea E, Steemburgo T, Javierre BM, Martinez JA. Leptin and TNF-α promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem 2011; 67:463-70; PMID:21465273; http://dx.doi.org/10.1007/s13105-011-0084-4
- Hermsdorff HH, Mansego ML, Campion J, Milagro FI, Zulet MA, Martinez JA. TNF-α promoter methylation in peripheral white blood cells: relationship with circulating TNFalpha, truncal fat and n-6 PUFA intake in young women. Cytokine 2013; 64:265-71; PMID:23796695; http://dx.doi.org/10.1016/j.cyto.2013.05.028
- Warnberg J, Nova E, Moreno LA, Romeo J, Mesana MI, Ruiz JR, Ortega FB, Sjostrom M, Bueno M, Marcos A. Inflammatory proteins are related to total and abdominal adiposity in a healthy adolescent population: the AVENA Study. Am J Clin Nutr 2006; 84:505-12; PMID:16960163
- Moreno LA, Quintela I, Fleta J, Sarria A, Roda L, Giner A, Bueno M. Postprandial triglyceridemia in obese and non-obese adolescents. Importance of body composition and fat distribution. J Pediatr Endocrinol Metab 2001; 14:193-202; PMID:11305798
- Whitworth JA, Chalmers J. World health organisation-international society of hypertension (WHO/ISH) hypertension guidelines. Clin Exp Hypertens 2004; 26:747-52; PMID:15702630; http://dx.doi.org/10.1081/CEH-200032152
- Ainsworth BE, Haskell WL, Whitt MC, Irwin ML, Swartz AM, Strath SJ, O'Brien WL, Bassett DR, Jr., Schmitz KH, Emplaincourt PO, et al. Compendium of physical activities: an update of activity codes and MET intensities. Med Sci Sports Exerc 2000; 32:S498-504; PMID:10993420; http://dx.doi.org/10.1097/00005768-200009001-00009
- Ainsworth BE, Haskell WL, Herrmann SD, Meckes N, Bassett DR, Jr., Tudor-Locke C, Greer JL, Vezina J, Whitt-Glover MC, Leon AS. 2011 Compendium of Physical Activities: a second update of codes and MET values. Med Sci Sports Exerc 2011; 43:1575-81; PMID:21681120; http://dx.doi.org/10.1249/MSS.0b013e31821ece12
- Tremblay AJ, Morrissette H, Gagne JM, Bergeron J, Gagne C, Couture P. Validation of the Friedewald formula for the determination of low-density lipoprotein cholesterol compared with β-quantification in a large population. Clin Biochem 2004; 37:785-90; PMID:15329317; http://dx.doi.org/10.1016/j.clinbiochem.2004.03.008
- Castelli WP. Cholesterol and lipids in the risk of coronary artery disease–the Framingham Heart Study. Can J Cardiol 1988; 4 Suppl A:5A-10A; PMID:3179802
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985; 28:412-9; PMID:3899825; http://dx.doi.org/10.1007/BF00280883
- Navarro-Blasco I, Alvarez-Galindo JI. Selenium content of Spanish infant formulae and human milk: influence of protein matrix, interactions with other trace elements and estimation of dietary intake by infants. J Trace Elem Med Biol 2004; 17:277-89; PMID:15139390; http://dx.doi.org/10.1016/S0946-672X(04)80030-0
- Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 2007; 35:e41; PMID:17289753; http://dx.doi.org/10.1093/nar/gkm013
- Tse MY, Ashbury JE, Zwingerman N, King WD, Taylor SA, Pang SC. A refined, rapid and reproducible high resolution melt (HRM)-based method suitable for quantification of global LINE-1 repetitive element methylation. BMC Res Notes 2011; 4:565; PMID:22204640; http://dx.doi.org/10.1186/1756-0500-4-565
- Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr 1997; 65:1220S-8S; discussion 9S-31S; PMID:9094926
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010; 70:27-56; PMID:20920744; http://dx.doi.org/10.1016/B978-0-12-380866-0.60002-2
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007; 8:253-62; PMID:17363974; http://dx.doi.org/10.1038/nrg2045
- El-Maarri O, Walier M, Behne F, Van Uum J, Singer H, Diaz-Lacava A, Nusgen N, Niemann B, Watzka M, Reinsberg J, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One 2011; 6:e16252; PMID:21311577; http://dx.doi.org/10.1371/journal.pone.0016252
- Tajuddin SM, Amaral AF, Fernandez AF, Rodriguez-Rodero S, Rodriguez RM, Moore LE, Tardon A, Carrato A, Garcia-Closas M, Silverman DT, et al. Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect 2013; 121:650-6; PMID:23552396
- Tapp HS, Commane DM, Bradburn DM, Arasaradnam R, Mathers JC, Johnson IT, Belshaw NJ. Nutritional factors and gender influence age-related DNA methylation in the human rectal mucosa. Aging Cell 2013; 12:148-55; PMID:23157586; http://dx.doi.org/10.1111/acel.12030
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 2011; 6:623-9; PMID:21739720; http://dx.doi.org/10.4161/epi.6.5.15335
- Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, Vishwanatha JK, Santella RM, Cardarelli R. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr 2011; 141:1165-71; PMID:21525250; http://dx.doi.org/10.3945/jn.110.134536
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009; 10:805-11; PMID:19789556; http://dx.doi.org/10.1038/nrg2651
- Ruiz-Hernandez A, Kuo CC, Rentero-Garrido P, Tang WY, Redon J, Ordovas JM, Navas-Acien A, Tellez-Plaza M. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin Epigenetics 2015; 7:55; PMID:25984247; http://dx.doi.org/10.1186/s13148-015-0055-7
- Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 2011; 88:450-7; PMID:21457905; http://dx.doi.org/10.1016/j.ajhg.2011.03.003
- Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010; 29:3650-64; PMID:20440268; http://dx.doi.org/10.1038/onc.2010.129
- Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet 2013; 4:132; PMID:23882278
- Lind L, Penell J, Luttropp K, Nordfors L, Syvanen AC, Axelsson T, Salihovic S, Van Bavel B, Fall T, Ingelsson E, et al. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environ Int 2013; 59:456-61; PMID:23933504; http://dx.doi.org/10.1016/j.envint.2013.07.008
- White AJ, Sandler DP, Bolick SC, Xu Z, Taylor JA, DeRoo LA. Recreational and household physical activity at different time points and DNA global methylation. Eur J Cancer 2013; 49:2199-206; PMID:23473616; http://dx.doi.org/10.1016/j.ejca.2013.02.013
- Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc Nutr Soc 2012; 71:154-65; PMID:22124338; http://dx.doi.org/10.1017/S0029665111003338
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell 2011; 146:866-72; PMID:21925312; http://dx.doi.org/10.1016/j.cell.2011.08.042
- Delgado-Cruzata L, Zhang W, McDonald JA, Tsai WY, Valdovinos C, Falci L, Wang Q, Crew KD, Santella RM, Hershman DL, et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J Nutr 2015; 145:783-90; PMID:25833781; http://dx.doi.org/10.3945/jn.114.202853
- Agodi A, Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M. DAPK1 Promoter Methylation and Cervical Cancer Risk: A Systematic Review and a Meta-Analysis. PLoS One 2015; 10:e0135078; PMID:26267895; http://dx.doi.org/10.1371/journal.pone.0135078
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 2011; 6:828-37; PMID:21636973; http://dx.doi.org/10.4161/epi.6.7.16500
- Piyathilake CJ, Badiga S, Alvarez RD, Partridge EE, Johanning GL. A lower degree of PBMC L1 methylation is associated with excess body weight and higher HOMA-IR in the presence of lower concentrations of plasma folate. PLoS One 2013; 8:e54544; PMID:23358786; http://dx.doi.org/10.1371/journal.pone.0054544
- McCullough LE, Mendez MA, Miller EE, Murtha AP, Murphy SK, Hoyo C. Associations between prenatal physical activity, birth weight, and DNA methylation at genomically imprinted domains in a multiethnic newborn cohort. Epigenetics 2015; 10:597-606; PMID:25928716; http://dx.doi.org/10.1080/15592294.2015.1045181
- Perng W, Villamor E, Shroff MR, Nettleton JA, Pilsner JR, Liu Y, Diez-Roux AV. Dietary intake, plasma homocysteine, and repetitive element DNA methylation in the Multi-Ethnic Study of Atherosclerosis (MESA). Nutr Metab Cardiovasc Dis 2014; 24:614-22; PMID:24477006; http://dx.doi.org/10.1016/j.numecd.2013.11.011
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2012; 41:126-39; PMID:20846947; http://dx.doi.org/10.1093/ije/dyq154
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, Lee DH. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect 2010; 118:370-4; PMID:20064773; http://dx.doi.org/10.1289/ehp.0901131
- Campion J, Milagro FI, Goyenechea E, Martinez JA. TNF-α promoter methylation as a predictive biomarker for weight-loss response. Obesity (Silver Spring) 2009; 17:1293-7; PMID:19584886
- Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, Shi H, Zhu H, Snieder H, Wang X. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics 2013; 8:522-33; PMID:23644594; http://dx.doi.org/10.4161/epi.24506
- Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 2010; 5:e9692; PMID:20300621; http://dx.doi.org/10.1371/journal.pone.0009692
- Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimburger O, Barany P, Alvestrand A, Nordfors L, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med 2007; 261:488-99; PMID:17444888; http://dx.doi.org/10.1111/j.1365-2796.2007.01777.x
