Abstract
Infection with high-risk types of human papilloma virus (HPV) is currently the best-established prognostic marker for head and neck squamous cell carcinoma (HNSCC), one of the most common and lethal human malignancies worldwide. Clinical trials have been launched to address the concept of treatment de-escalation for HPV-positive HNSCC with the final aim to reduce treatment related toxicity and debilitating long-term impacts on the quality of life. However, HPV-related tumors are mainly restricted to oropharyngeal SCC (OPSCC) and there is an urgent need to establish reliable biomarkers for all patients at high risk for treatment failure. A patient cohort (n = 295) with mainly non-OPSCC (72.9%) and a low prevalence of HPV16-related tumors (8.8%) was analyzed by MassARRAY to determine a previously established prognostic methylation score (MS). Kaplan-Meier revealed a highly significant correlation between a high MS and a favorable survival for OPSCC (P = 0.0004) and for non-OPSCC (P<0.0001), which was confirmed for all HNSCC by multivariate Cox regression models (HR: 9.67, 95% CI [4.61–20.30], P<0.0001). Next, we established a minimal methylation signature score (MMSS), which consists of ten most informative of the originally 62 CpG units used for the MS. The prognostic value of the MMSS was confirmed by Kaplan-Meier analysis for all HNSCC (P<0.0001) and non-OPSCC (P = 0.0002), and was supported by multivariate Cox regression models for all HNSCC (HR: 2.15, 95% CI [1.36–3.41], P = 0.001). In summary, the MS and the MMSS exhibit an excellent performance as prognosticators for survival, which is not limited by the anatomical site, and both could be implemented in future clinical trials.
Introduction
Head and neck cancer is a highly heterogeneous group of malignancies originating from different anatomical sites of the upper aero-digestive tract, including oral cavity, larynx and pharynx. The vast majority of head and neck cancer cases represent as squamous cell carcinoma (HNSCC) and are attributed to tobacco and alcohol consumption as well as infection with high-risk types of the human papilloma virus (HPV).Citation1 HPV-related tumors arise mainly in the oropharynx and exhibit distinct molecular, cellular and clinical features as opposed to non-HPV-related HNSCC.Citation2-6 Most importantly, patients with HPV-related tumors show an improved treatment outcome justifying the HPV status as one of the most important biomarkers for favorable prognosis in primary and progressed HNSCC.Citation7-11 Numerous clinical trials have been launched to evaluate different concepts of treatment de-escalation for HPV-positive HNSCC patients with the aim to reduce treatment-related toxicity without raising mortality.Citation12-15 However, a substantial amount of HNSCC patients with HPV-related tumors still progress and die, and the prevalence as well as the improved therapeutic outcome of HPV-related tumors at sites other than the oropharynx remains questionable.Citation16-20 Thus, for the majority of HNSCC we still lack a reliable prognostic biomarker for treatment decision-making and to stratify patients at high risk for treatment failure under currently established treatment modalities.Citation21
Epigenetic alterations such as DNA methylation in promoters of tumor-relevant genes have gained increasing importance for our understanding of molecular principles underlying the pathogenesis of cancer.Citation22 The epigenetic plasticity makes DNA methylation patterns bona fide prognostic and therapeutic biomarkers for human cancer, including HNSCC.Citation5,23,24 In the past, we investigated differentially methylated regions (DMR) in proximal gene promoters of HPV-related and non-HPV-related OPSCC and established a methylation score (MS), which was based on DNA methylation patterns of 62 CpG units in DMR of 5 gene promoters (ALDH1A2, OSR2, GRIA4, GATA4 and IRX4).Citation25 Although a high MS was more common in HPV-related OPSCC, it served as a reliable prognostic biomarker for overall survival independent of the HPV status and initial therapy.Citation25 The close association between promoter hypermethylation and reduced gene expression suggested a critical role of affected candidate genes of the MS in HNSCC pathogenesis as well as the response to treatment. This assumption was further supported by a recent study demonstrating that loss of ALDH1A2-RAR signaling in HNSCC cell lines induces loss of cell-cell adhesion and induction of a mesenchymal-like phenotype with accelerated tumor cell motility.Citation26
The excellent prediction value concerning both HPV-related and non-HPV-related OPSCC patients raised the attractive question whether the newly identified MS could be employed as a reliable prognosticator for all HNSCC independent of the primary anatomical location, to identify patients at high risk for treatment failure and those who might benefit from de-intensified therapy.
Results
The methylation score serves as prognostic biomarker for survival of OPSCC and non-OPSCC patients
To address the question whether the previously identified gene promoter methylation signature (ALDH1A2low, OSR2low, GATA4high, GRIA4high, IRX4high) correlates with improved outcome independent of the anatomical origin of the tumor, we conducted MassARRAY analysis with genomic DNA of a HNSCC patient cohort (n = 295) with 27.1% OPSCC and 72.9% non-OPSCC (93 laryngeal SCC, 80 oral SCC, 37 hypopharyngeal SCC and 5 nasopharygeal SCC) (Supplemental Table S1A-C). It is worth noting that there is no overlap of patients from the German study cohort (Leipzig, Germany), which were analyzed in this study as compared to those of the previous study.Citation25 During follow-up there were 107 disease-related deaths (DSS) and the median follow-up time was 29 months. MS could be determined for 220 patients and according to our previous study Citation25 2 subgroups with a MS≥3 (MS3-5, n = 110) and a MS<3 (MS0-2, n = 110) were selected for further analysis (, Supplemental Table S2A-B). There was no significant difference between both subgroups concerning all clinical or pathological features tested except for the HPV16 status (Supplemental Table S3A). As expected, we found a strong enrichment of HPV-related tumors in the MS3-5 subgroup (15% vs. 2%, P = 0.0008). Kaplan-Meier analysis revealed a favorable outcome for the MS3-5 as compared to the MS0-2 subgroup, which was highly significant for all HNSCC (p≤0.0001) as well as for OPSSC (P = 0.0004) and non-OPSCC (P≤0.0001, and Supplemental Fig. S1A-C). The effect of the MS (MS0-2 vs. MS3-5) on disease-specific survival for all patients (HR: 7.22, 95% CI [3.85–13.55], P<0.0001; Supplemental Table S3B), and different anatomical sites is summarized for subgroups with OPSCC, non-OPSCC, hypopharyngeal SCC, laryngeal SCC, and oral SCC in Fig. 2D. Multivariate Cox regression models considering most relevant risk factors (HPV status, tobacco and alcohol consumption), patient characteristics (Age, gender and OPSCC vs. non-OPSCC), pathological (TNM, staging and grading) and clinical features (first therapy) further supported the prognostic value of the MS independent of the OPSCC effect, and confirmed that MS0-2 serves as an independent prognostic biomarker for unfavorable survival (HR: 9.67, 95% CI [4.61–20.30], P<0.0001, ). No significant difference for the prognostic effect of MS was observed between OPSCC and non-OPSCC (interaction P = 0.49), different locations (interaction P = 0.28), the tumor size (T status; interaction P = 0.08), or the HPV status in the OPSCC subgroup (interaction P = 0.53. Finally, Kaplan-Meier analysis was conducted to demonstrate the effect of MS on disease-specific survival in the OPSCC patient subgroup split by HPV16 status, the non-OPSCC patient subgroup split by distant metastases status, and all patients split by tumor size (Supplemental Fig. S1D-F, Supplemental Table S4A). Distant metastasis and tumor size were selected as both variables revealed the strongest effect on disease-specific survival in univariate analysis (Supplemental Table S3B).
Figure 1. Schematic representation of the study design and association between the MS, the MMSS and the survival status of HNSCC patients. (A) The MS was evaluated based on quantitative MassARRAY data of 62 CpG units in proximal promoters of ALDH1A2, OSR2, IRX4, GRIA4 and GATA4 as described previously Citation25, and revealed informative data for 220 patients of the HNSCC cohort (n = 54 OPSCC and n = 166 non-OPSCC). The MMSS was established as a less complex signature, consisting of the 10 most informative CpG units in proximal promoters of ALDH1A2, OSR2, IRX4 and GRIA4, with data of the training cohort (n = 100; Citation25), and was applied on patients of the HNSCC cohort (n = 295 with n = 80 OPSCC and n = 215 non-OPSCC). Univariate and multivariate survival analysis was conducted to confirm prognostic effects of MS and MMSS. (B) Heatmap summarizing the MS, the MMSS and the survival status of HNSCC patients. Each column indicates a patient of the HNSCC cohort (n = 220) and the methylation status according to the established pattern ALDH1A2low, OSR2low, GATA4high, GRIA4high, IRX4high for favorable prognosis is indicated in black (congruent) or white (non-congruent). The colored bars indicate the final value for MS and the MMSS is depicted in blue (MMSS_p) or gray (MMSS_g). The survival status is indicated in gray (alive) or black (dead).
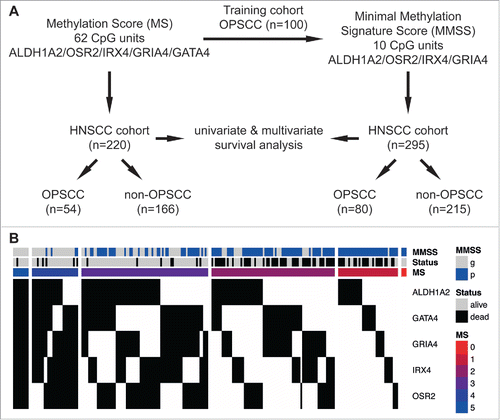
Figure 2. A high MS serves as prognostic biomarker for favorable survival in a HNSCC cohort. Kaplan-Meier plots demonstrate an improved survival of the MS3-5 (black line) as compared to the MS0-2 subgroup (red line), which is highly significant for all HNSSC patient (n = 220, A), as well as for the subgroup of patients with either OPSCC (n = 54, B) or non-OPSCC (n = 166, C). (D) The Forest plot displays the relative strength of prognostic effects as HR and 95% CI according to the methylation subgroup (MS0-2 vs. MS3-5) on disease-specific survival for all HNSCC (n = 220), and subgroup of patients with OPSCC (n = 54), non-OPSCC (n = 166), hypopharyngeal (n = 30), laryngeal (n = 69) and oral SCC (n = 63).
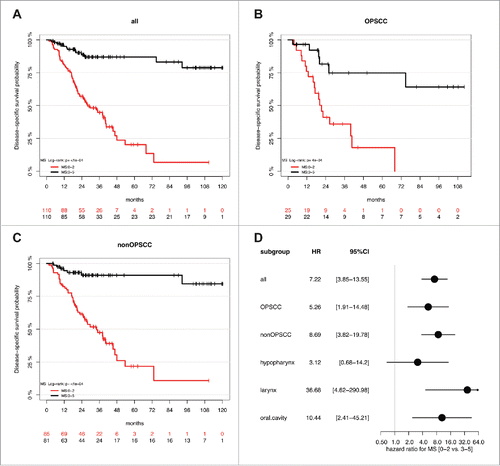
Table 1. Multivariate Cox regression model analysis for the MS (n = 211).
Identification of a minimal methylation signature score as prognostic biomarker in a training cohort of OPSCC patients
The data presented so far provided strong evidence for an excellent predictive value of the MS as a suitable prognosticator for all HNSCC. However, we had to exclude 75 of 295 patients (25.4%) from our cohort due to incomplete availability of quantitative DNA methylation data for all 62 CpG units, which were used to calculate the MS. We applied the LASSO method to Cox regression models in order to reduce the complexity of the MS while maintaining a high prognostic accuracy. As a training data set we used the methylation values of the previously published OPSCC cohort (n = 100) in which the MS was originally discoveredCitation25 (Supplemental Table S2C). The dichotomized methylation values provided a superior signature as compared to the untransformed CpG values (data not shown), and were used for further analysis. The final minimal methylation signature score (MMSS) consisted of 10 [ALDH1A2 (n = 1), OSR2 (n = 2), GRIA4 (n = 2) and IRX4 (n = 5)] out of the originally used 62 CpG units (Supplemental Table S2C). No CpG unit of the GATA4 promoter contributed to the MMSS. Next, patients of the training cohort were divided into 2 subgroups according to the MMSS value (MMSS_g<−0.6385 and MMSS_p>−0.6385), and the prognostic relevance was confirmed by Kaplan-Meier analysis (P<0.0001, Supplemental Fig. S2A). Moreover, the newly established MMSS and the original MS had a comparable prediction error concerning survival of patients in the training cohort (Supplemental Fig. S2B).
Prognostic value of the MMSS for survival in a HNSCC patient cohort with OPSCC and non-OPSCC
Application of the MMSS allowed the evaluation of all samples of the HNSCC patient cohort, including those for which the MS could not be calculated (Supplemental Table S2B). Similar to the MS3-5 the MMSS_g was significantly associated with HPV-related tumors but additionally with smaller tumor size (Supplemental Table S3A). Kaplan-Meier analysis revealed a favorable outcome for MMSS_g (n = 113) as compared to MMSS_p (n = 182), which was highly significant for all HNSCC (P≤0.0001) and the non-OPSCC subgroup (P = 0.0002), but did not reach statistical significance for the smaller subgroup of OPSCC (P = 0.1, ). In the subgroup of non-OPSCC, the MMSS exhibited the strongest prognostic effect for laryngeal SCC (P = 0.0007) followed by oral SCC (P = 0.05, Supplemental Fig. S3A-C). The effect of the MMSS (MMSS_p vs. MMSS_g) on disease-specific survival for all patients (HR: 2.43, 95% CI [1.56–3.80], P<0.0001; Supplemental Table S3B), and different anatomical sites is summarized for subgroups with OPSCC, non-OPSCC, hypopharyngeal SCC, laryngeal SCC, and oral SCC in . Multivariate Cox models considering co-variables as described above further supported the prognostic value of the MMSS and confirmed that the MMSS_p serves as an independent prognostic biomarker for an unfavorable survival (). Again, no significant difference for the prognostic effect of MMSS was observed between OPSCC and non-OPSCC (interaction P = 0.41), different locations (interaction P = 0.47), the HPV status in the OPSCC subgroup (interaction P = 0.99), or the M status in the non-OPSCC subgroup (P = 0.5.) Again, Kaplan-Meier analysis was conducted to demonstrate the effect of the MMSS on disease-specific survival in the OPSCC patient subgroup split by HPV16 status, the non-OPSCC patient subgroup split by distant metastases status, and all patients split by tumor size (Supplemental Fig. S3D-F, Supplemental Table S4B). It is worth noting that there was some indication for a stronger effect of both methylation signatures in patients with smaller tumor size (interaction P = 0.08/0.003 for Tumor size [T1/2 vs. T3/4] vs. MMSS or MS). However, the prognostic value of MS and MMSS was still statistically significant in patients with larger tumor size (Supplemental Tables S4A-B).
Figure 3. The MMSS_g serves as prognostic biomarker for favorable survival in a HNSCC cohort. Kaplan-Meier plots demonstrate an improved survival of the MMSS_g (black line) as compared to the MMSS_p subgroup (red line), which is highly significant for all HNSSC patient (n = 295, A), as well as for the subgroup of patients with non-OPSCC (n = 215, C). (D) The Forest plot displays the relative strength of the prognostic effects as HR and 95% CI according to the methylation subgroup (MMSS_p vs. MMSS_g) on disease-specific survival for all HNSCC (n = 295), and subgroup of patients with OPSCC (n = 80), non-OPSCC (n = 215), hypopharyngeal (n = 37), laryngeal (n = 93) and oral SCC (n = 80).
Table 2. Multivariate Cox regression model analysis for the MMSS (n = 286).
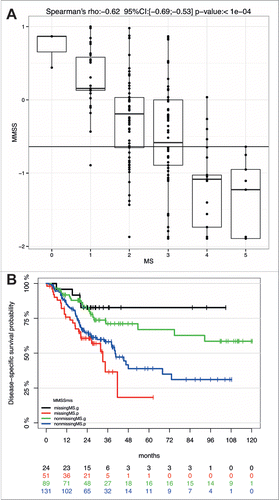
In summary, these data demonstrated the feasibility of establishing a more simplified prognosticator by selection of the most informative CpG units. However, the reduced complexity was accompanied by a decrease in the performance of the MMSS to predict survival as indicated by the smaller hazard ratio in univariate and multivariate analysis. To evaluate the difference between MS and MMSS we conducted a comparative analysis for all samples of the HNSCC patient cohort for which data for both methylations scores were available. As expected, we found a highly significant correlation between MS and MMSS values (rho = 0.62, P<0.0001) but only moderate agreement between high-risk classifications (κ = 0.45), and the most prominent discordance was detected for tumors with a MS at the cut-off point for definition of MS0-2 and MS3-5 subgroups (). Although this discordance resulted in a reduced accuracy of the MMSS to predict survival, we speculated that the MMSS represents an appropriate alternative as reliable prognosticator for tumor samples for which the MS could not be evaluated. Indeed, Kaplan-Meier analysis for the 75 HNSCC of the patient cohort with a missing MS revealed a favorable survival for the MMSS_g as compared to the MMSS_p subgroup, which was highly significant (P = 0.006, ). Interestingly, the predictive value of the MMSS was even higher for the patient subgroup (n = 75) without MS (HR: 4.0, 95% CI [1.4–11.6]) as compared to the larger subgroup (n = 220) with MS (HR: 2.1, 95% CI [1.3–3.4]).
Figure 4. The MMSS reliably predicts survival of HNSCC patients with a missing MS. (A) Comparative analysis of MS and MMSS values for all HNSCC patients (n = 220) for which data for both methylation scores were available. (B) The Kaplan-Meier plot demonstrates the prognostic value of the MMSS_g to predict improved survival of HNSCC patient with available MS (n = 89, green line) as well as those with a missing MS (n = 24, black line). The MMSS_p is associated with an unfavorable survival, which is displayed for HNSCC patients with an available MS (blue line) and those with a missing MS (red line).
Discussion
Head and neck carcinogenesis is a multistep and multifactorial process, encompassing a number of genetic and epigenetic events.Citation4,5,24 Despite advances in our knowledge on the pathogenesis and implementation of multimodal treatment the 5-year survival rate for advanced HNSCC remains discouraging. Moreover, multimodal therapy is characterized by high toxicity and treatment-related mortality resulting in severe loss of life quality for HNSCC patients. Accordingly, treatment de-intensification is an attractive and intensively studied issue in the field of head and neck oncology, with a focus on HPV-related HNSCC.Citation12,13,27 However, HPV-related tumors have been identified predominantly in the subgroup of OPSCC,Citation2 and reliable biomarkers are still missing to identify HNSCC patients with a non-HPV-related tumor, who might benefit from de-intensified therapy.
In the past, we monitored HPV-related alterations in the DNA methylome of OPSCC patients and established a MS, consisting of gene promoter methylation patterns for ALDH1A2, OSR2, GATA4, GRIA4 and IRX4 as a reliable prognosticator for PFS and OS.Citation25 Strikingly, the prognostic value of the MS was independent of the HPV status and had an excellent performance to predict the clinical outcome of non-HPV-related OPSCC patients. Based on this encouraging finding our objective was to proof the concept whether the MS could be applicable as a prognosticator for all HNSCC patients independent of the anatomical origin of the primary tumor. We determined the MS for a HNSCC patient cohort with no overlap to our previous study,Citation25 which was characterized by a low HPV prevalence and comprised tumors from various anatomical sites, including oral cavity, hypopharynx, larynx and oropharynx. Kaplan-Meier analysis revealed a highly significant correlation between the MS and clinical outcome irrespective of the anatomical site, confirming its reliable and robust predictive value. Despite these promising results, the relative high number of CpG units, which have to be used to calculate the MS turned out to be a limitation for the potential translation into clinical practice.
We used the LASSO method to Cox regression models to select the most informative CpG units for predicting overall survival of HNSCC patients and to establish a less complex prognostic signature. LASSO is a popular method for regression modeling with a large number of potential prognostic features, because it can perform automatic feature selection in a manner that results in signatures with generally good prognostic performance.Citation28 The LASSO method has been extended to the Cox model for survival analysis and has been successfully applied for the purpose of building sparse signatures for survival prognosis in many application areas including oncology.Citation29-31
We initially applied the approach to the same training cohort, which was previously employed for MS establishment,Citation25 and obtained a substantial reduction of required CpG units. The resulting MMSS finally consisted of only 10 CpG units located within the promoters of ALDH1A2, OSR2, GRIA4 and IRX4, omitting GATA4, which previously contributed to the MS. The predictive value of the MMSS could be confirmed with the HNSCC cohort, demonstrating the feasibility of our approach to reduce the complexity of the original MS while maintaining a high prognostic accuracy. However, we cannot completely exclude the possibility that omitting GATA4 affected at least in part the performance of the MMSS in the validation cohort enriched for HPV-negative tumors at anatomical sites different from OPSCC. It is conceivable that the expression and function of GATA4 is less relevant for the clinical outcome of HPV-positive OPSCC representing 32% of patients in the training but less than 9% in the validation cohort.
It is worth noting that both the original MS and the newly established MMSS display an excellent performance for the subgroup of laryngeal SCC. Although the underlying clinical and/or molecular features that are related to these strong prognostic effects remain to be addressed, our data could be relevant for the debate on strategies for functional organ preservation in patients with potentially resectable laryngeal SCC. In the past, several large randomized clinical trials investigated definitive radiotherapy with or without adjuvant chemotherapy or sequential chemotherapy administration, in the form of induction chemotherapy followed by radiotherapy or chemoradiation to address the issue of adequate treatment.Citation32-36 In this context, it will be interesting to address the question, whether the MS and the MMSS might be informative to stratify patients with advanced laryngeal SCC and identify those, who will benefit from treatment strategies with the aim of functional organ preservation. Another relevant issue concerning the translation of our findings into future clinical trials is the technology, which might be best suited to determine the MS or the MMSS. In our study, the MassARRAY technology was used that is well established for automated and routine analyses of DNA methylation patterns in basic and clinical research laboratories. However, the preferred technology in almost all routine clinical laboratories is bisulfite sequencing using the pyro-sequencer, which provides highly concordant results with MassARRAY data.Citation37,38 Both technologies are highly accepted in the epigenetics community and share comparable assay design and financial costs.
The presented data also raises the question whether the MS and MMSS are associated with the CpG island methylator phenotype (CIMP), which was originally discovered in colorectal cancer and has been reported among a wide variety of human malignancies, including HNSCC.Citation39,40 However, marker gene panels to define CIMP have been largely inconsistent among studies limiting the final interpretation of most published data and their clinical relevance. So far, only few studies confirmed the existence of CIMP in HNSCC patients based on a high-throughput methylation approach. As an example, Lechner and colleagues discovered an enrichment of hypermethylated regions in HPV-positive HNSCC many of those mapped to CpG islands suggesting a possible association between HPV and CIMP.Citation41 Interestingly, HPV-positive tumors with CIMP had a poor clinical outcome exhibiting significantly shorter survival. An association between CIMP and poor prognosis was also reported for patients with oral cancer, though CIMP was not an independent factor in predicting prognosis.Citation42 It might be worth addressing a potential link between the MS or MMSS with CIMP in larger cohorts of HNSCC patients for which genome-wide methylation data are available.
In summary, the present study confirms the predictive power of the previously established MS as robust prognosticator for all HNSCC patients, including non-OPSCC or non-HPV-related tumors. This finding is of particular clinical relevance as we still lack appropriate prognostic biomarkers for this large subgroup of HNSCC patients, which represents the most lethal cases. Furthermore, we improved the applicability of the MS by reducing the number of informative CpG units, and provide compelling evidence that the MMSS serves as an alternative tool for tumor samples for which the MS could not be evaluated. However, the translation of our findings from bench to bedside also demands a better understanding of the complex interactions between alterations in the DNA methylome and the mutational landscape, both of which have a major impact on the pathogenesis and most likely clinical outcome of HNSCC patients.Citation4 The expected data of such a challenging endeavor will highlight critical molecular mechanisms of treatment failure and pave the way to implement epigenetic patterns into future biomarker-driven clinical trials.
Materials and Methods
Patient samples
Snap-frozen biopsies from HNSCC patients were collected between 2003 and 2012 at initial diagnosis presented in the Ear-Nose-Throat Units of Treviso, Mirano and Trieste (Italian cohort, n = 115), and from HNSCC patients treated at the University Hospital Leipzig between 2009 and 2012 (German cohort, n = 180). Tumor samples of the German cohort were not analyzed in our previous study.Citation25 Clinical and demographic data of individual HNSCC patients as well as an overview of both cohorts used in this study are given in Supplemental Table S1A-C. The study was performed according to the ethical standards of the Declaration of Helsinki after approval by the local institutional review boards (345/AULSS9, 421/AULSS9, 201-10-12072010 and 202-10-12072010).
Genomic DNA isolation
For the Italian cohort, 2–10 mg of 16 μm tissue cryosections were homogenized in liquid nitrogen and stored at −80°C. Hematoxylin and eosin (HE) staining of adjacent sections indicated a tumor cell content of at least 70%. Genomic DNA was extracted using the Magna Pure kit (Roche Applied Science, Mannheim, Germany). Total RNA was isolated using the RNeasy mini kit including the on-column DNase I digestion according to the manufacturer's instructions (Qiagen, Hilden, Germany). Quantity and quality of DNA was determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
For the German cohort, samples were put into TRIzol™ (Life technologies, Carlsbad) and stored at −80°C. The frozen samples in TRIzol® were homogenized for 80 sec at 4000 rpm using the Minilys benchtop homogenizer (Peqlab Biotechnologie GmbH Erlangen, Germany) followed by isolation of both RNA and DNA.Citation43 The aqueous phase was used for RNA preparation by use of the Ambion Pure Link™ RNA Mini Kit (Life technologies) including the on-column DNase I digestion (Qiagen, Hilden, Germany) according the manufacturer's instructions. Genomic DNA was prepared from the organic phase according to the standard TRIzol® protocol. Quantity and quality of DNA and RNA were determined using the NanoDrop 2000 spectrophotometer.
HPV status
The HPV DNA and viral RNA status were determined by multiplex HPV genotyping and viral transcript analysis as previously described.Citation44 Tumors were classified into 3 subgroups: DNA-negative (DNA-), DNA-positive but RNA-negative (DNA+/RNA-), and DNA-positive and RNA-positive (DNA+/RNA+; Supplemental Table S1A-C). DNA- or DNA+/RNA- tumors were classified as non-HPV-related and DNA+/RNA+ tumors as HPV-related.
Quantitative DNA methylation analysis
Quantification of DNA methylation was determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY) as previously described,Citation25 and the original data are given in Supplemental Table S2A. The genomic position and orientation of the amplicons as well as the gene promoter-specific primer sequences and relevant Agilent probes and genomic position are listed in Supplemental Table S5A-B. Heatmaps were generated to visualize DNA methylation levels for each CpG unit or averages of amplicons using R/Bioconductor library gplots version 3.1/2.14. The methylation score (MS) was calculated as previously described considering a total of 62 CpG units.Citation25 In brief, the average methylation value of all CpG units in each of the five amplicons was calculated. Samples were divided into 2 subgroups regarding high and low DNA methylation levels based on the median values of the training cohort.Citation25 A combined MS was obtained for 220 of the 295 patients, and 2 subgroups with a MS≥3 (MS3-5, n = 110) or a MS<3 (MS0-2, n = 110) were defined for further analysis.Citation25
Minimal methylation signature score
The minimal methylation signature score (MMSS) was established on the original training cohort Citation25 applying LASSO-penalized Cox regression models,Citation45 using either the untransformed (continuous) values of individual CpG units or the dichotomized values of individual CpG units with the median value as cut-off. The optimal LASSO penalty parameters were determined by maximizing the 10-fold cross-validated partial log-likelihood. The predictive accuracies of the prognostic models were quantified with prediction error curves based on the time-dependent Brier score.Citation46 The 0.632+ bootstrap subsampling method Citation47,48 was used to adjust prediction error estimates in the training cohort (Supplemental Fig. S2B). The final MMSS for OS was based on 10 CpG units and was calculated by the following equation using dichotomized CpG variables:
MMSS = 0.844 ALDH1A2_CpG2 + 0.132 OSR2_CpG6 + 0.022 OSR2_CpG10 – 0.019 GRIA4_CpG2.3 – 0.790 GRIA4_CpG34.35.36 – 0.233 IRX4_CpG1 – 0.104 IRX4_CpG3 – 0.215 IRX4_CpG7 – 0.100 IRX4_CpG12.13.14.15 – 0.433 IRX4_CpG22.
The MMSS was dichotomized using the median value of the training cohort, which proved almost identical with the “optimal cut-point” in the training cohort based on maximally selected log-rank statistics Citation49 (Supplemental Fig. S2C). MMSS values smaller than the cut-point were considered to predict a good prognosis (MMSS_g, n = 113) and values larger than the cut-point were considered to predict a poor prognosis (MMSS_p, n = 182) in the HNSCC cohort. All statistical analyses were performed with the R/Bioconductor software environment (version 3.1/2.14) using add-on R packages glmnet (version 1.9), pec (version 2.2.9) and maxstat (version 0.7.20).
Survival functions and statistics
Disease-specific survival (DSS) was determined as time interval from registration date until date of tumor-related death, censoring patients, who were alive at last follow-up or died due to non-tumor-related deaths. Distribution of DSS times was estimated by the Kaplan-Meier method. Differences between subgroups were determined by Log-rank test. Univariable and multivariable Cox regression models were used to assess the prognostic value in terms of hazard ratios and 95% confidence intervals. Interaction term was tested to assess heterogeneity of MS/MMSS effect between subgroups/locations. Forest plots were used to display hazard ratios of MS/MMSS across locations. Median follow-up time was estimated by reverse Kaplan-Meier method. Fisher's exact test was used to test association between categorical variables. Spearman's correlation coefficient between quantitative MS and MMSS was calculated. Agreement between dichotomized scores was calculated with Cohen's kappa. All tests were 2-sided. P-values below 0.05 were considered statistically significant. All analyses were carried out with software R 3.1.
R/Bioconductor software codes
Analysis has been performed with knitr (R+Latex). Original knitr.Rnw files and the extracted R Codes from the .Rnw file via ‘purl’ as well as calls for prediction error curves and convenience R functions are available upon request.
Disclosure of Potential Conflicts of Interest
Gunnar Wichmann received funding for translational research projects on “Pharmacological drug interaction of cilengitide in treatment of head and neck squamous cell carcinoma (HNSCC) ex vivo” from Merck KGaA and on “Efficacy of vismodegib (GDC -0449) on HNSCC ex vivo” from Roche Pharma AG. Michael Pawlita has received research funding through cooperation contracts of the DKFZ with Roche and Qiagen. He is also an inventor on patents owned by the DKFZ in the field of molecular HPV diagnostics. All other authors declare no conflict of interest.
Supplementary_Datas.zip
Download Zip (1.6 MB)Acknowledgments
This work was supported in part by the Helmholtz Foundation and funds from the German Cancer Research Consortium (to C.P. and D.W.), the European Commission grant HPV-AHEAD (FP7-HEALTH-2011-282562 to M.P.), the German Research Foundation (HE-5760/3-1 to J.H.), Lega Italiana per la Lotta contro i Tumori (LILT to A.D.M.), funding via projects on “Pharmacological drug interaction of cilengitide in treatment of head and neck squamous cell carcinoma (HNSCC) ex vivo” from Merck KGaA and on “Efficacy of vismodegib (GDC -0449) on HNSCC ex vivo” from Roche PharmaAG (to G.W.).
References
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011; 11:9-22; PMID:21160525; http://dx.doi.org/10.1038/nrc2982
- Ndiaye C, Mena M, Alemany L, Arbyn M, Castellsague X, Laporte L, Bosch FX, de Sanjose S, Trottier H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol 2014; 15:1319-31; PMID:25439690; http://dx.doi.org/10.1016/S1470-2045(14)70471-1
- Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, Roberts S. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck 2013; 35:747-55; PMID:22267298; http://dx.doi.org/10.1002/hed.22015
- Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015; 517:576-82; PMID:25631445; http://dx.doi.org/10.1038/nature14129
- Kostareli E, Holzinger D, Hess J. New concepts for translational head and neck oncology: lessons from HPV-related oropharyngeal squamous cell carcinomas. Front Head Neck Cancer 2012; 2:1-10
- Hayes DN, Van Waes C, Seiwert TY. Genetic landscape of human papillomavirus-associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol: Off J Am Soc Clin Oncol 2015; 33:3227-34; PMID:26351353; http://dx.doi.org/10.1200/JCO.2015.62.1086
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. New Engl Med 2010; 363:24-35; PMID:20530316; http://dx.doi.org/10.1056/NEJMoa0912217
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol: Off J Am Soc Clin Oncol 2011; 29:4294-301; PMID:21969503; http://dx.doi.org/10.1200/JCO.2011.36.4596
- Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol 2015; 12:11-26; PMID:25403939; http://dx.doi.org/10.1038/nrclinonc.2014.192
- Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, Soulieres D, Trotti A, Avizonis V, Ridge JA, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol: Off J Am Soc Clin Oncol 2014; 32:3365-73; PMID:24958820; http://dx.doi.org/10.1200/JCO.2014.55.1937
- Sacco AG, Cohen EE. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol: Off J Am Soc Clin Oncol 2015; 33:3305-13; PMID:26351341; http://dx.doi.org/10.1200/JCO.2015.62.0963
- Masterson L, Moualed D, Liu ZW, Howard JE, Dwivedi RC, Tysome JR, Benson R, Sterling JC, Sudhoff H, Jani P, et al. De-escalation treatment protocols for human papillomavirus-associated oropharyngeal squamous cell carcinoma: a systematic review and meta-analysis of current clinical trials. Eur J Cancer 2014; 50:2636-48; PMID:25091798; http://dx.doi.org/10.1016/j.ejca.2014.07.001
- Mirghani H, Amen F, Blanchard P, Moreau F, Guigay J, Hartl DM, Lacau St Guily J. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: ongoing trials, critical issues and perspectives. Int J Cancer 2015; 136:1494-503; PMID:24622970; http://dx.doi.org/10.1002/ijc.28847
- Psyrri A, Rampias T, Vermorken JB. The current and future impact of human papillomavirus on treatment of squamous cell carcinoma of the head and neck. Ann Oncol: Off J Euro Soc Med Oncol / ESMO 2014; 25:2101-15; PMID:25057165; http://dx.doi.org/10.1093/annonc/mdu265
- Bhatia A, Burtness B. Human papillomavirus-associated oropharyngeal cancer: defining risk groups and clinical trials. J Clin Oncol: Off J Am Soc Clin Oncol 2015; 33:3243-50; PMID:26351343; http://dx.doi.org/10.1200/JCO.2015.61.2358
- Young RJ, Urban D, Angel C, Corry J, Lyons B, Vallance N, Kleid S, Iseli TA, Solomon B, Rischin D. Frequency and prognostic significance of p16(INK4A) protein overexpression and transcriptionally active human papillomavirus infection in laryngeal squamous cell carcinoma. Br J Cancer 2015; 112:1098-104; PMID:25688737; http://dx.doi.org/10.1038/bjc.2015.59
- Lassen P, Primdahl H, Johansen J, Kristensen CA, Andersen E, Andersen LJ, Evensen JF, Eriksen JG, Overgaard J. Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol: J Euro Soc Therap Radiol Oncol 2014; 113:310-6; PMID:25544647; http://dx.doi.org/10.1016/j.radonc.2014.11.032
- Combes JD, Franceschi S. Role of human papillomavirus in non-oropharyngeal head and neck cancers. Oral Oncol 2014; 50:370-9; PMID:24331868; http://dx.doi.org/10.1016/j.oraloncology.2013.11.004
- Salazar CR, Smith RV, Garg MK, Haigentz M Jr., Schiff BA, Kawachi N, Anayannis N, Belbin TJ, Prystowsky MB, Burk RD, et al. Human papillomavirus-associated head and neck squamous cell carcinoma survival: a comparison by tumor site and initial treatment. Head Neck Pathol 2014; 8:77-87; PMID:24002971; http://dx.doi.org/10.1007/s12105-013-0486-4
- Halec G, Holzinger D, Schmitt M, Flechtenmacher C, Dyckhoff G, Lloveras B, Hofler D, Bosch FX, Pawlita M. Biological evidence for a causal role of HPV16 in a small fraction of laryngeal squamous cell carcinoma. Br J Cancer 2013; 109:172-83; PMID:23778529; http://dx.doi.org/10.1038/bjc.2013.296
- Dillon MT, Harrington KJ. Human papillomavirus-negative pharyngeal cancer. J Clin Oncol: Off J Am Soc Clin Oncol 2015; 33:3251-61; PMID:26351347; http://dx.doi.org/10.1200/JCO.2015.60.7804
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev enet 2013; 14:765-80; PMID:24105274; http://dx.doi.org/10.1038/nrg3554
- Arantes LM, de Carvalho AC, Melendez ME, Carvalho AL, Goloni-Bertollo EM. Methylation as a biomarker for head and neck cancer. Oral Oncol 2014; 50:587-92; PMID:24656975; http://dx.doi.org/10.1016/j.oraloncology.2014.02.015
- van Kempen PM, Noorlag R, Braunius WW, Stegeman I, Willems SM, Grolman W. Differences in methylation profiles between HPV-positive and HPV-negative oropharynx squamous cell carcinoma: a systematic review. Epigenetics: Off J DNA Methylation Soc 2014; 9:194-203; PMID:24169583; http://dx.doi.org/10.4161/epi.26881
- Kostareli E, Holzinger D, Bogatyrova O, Hielscher T, Wichmann G, Keck M, Lahrmann B, Grabe N, Flechtenmacher C, Schmidt CR, et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J Clin Investigat 2013; 123:2488-501; PMID:23635773; http://dx.doi.org/10.1172/JCI67010
- Seidensaal K, Nollert A, Feige AH, Muller M, Fleming T, Gunkel N, Zaoui K, Grabe N, Weichert W, Weber KJ, et al. Impaired aldehyde dehydrogenase 1 subfamily member 2A-dependent retinoic acid signaling is related with a mesenchymal-like phenotype and an unfavorable prognosis of head and neck squamous cell carcinoma. Mol Cancer 2015; 14:204; PMID:26634247; http://dx.doi.org/10.1186/s12943-015-0476-0
- O'Sullivan B, Huang SH, Siu LL, Waldron J, Zhao H, Perez-Ordonez B, Weinreb I, Kim J, Ringash J, Bayley A, et al. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J Clin Oncol: Off J Am Soc Clin Oncol 2013; 31:543-50; PMID:23295795; http://dx.doi.org/10.1200/JCO.2012.44.0164
- Bovelstad HM, Nygard S, Storvold HL, Aldrin M, Borgan O, Frigessi A, Lingjaerde OC. Predicting survival from microarray data–a comparative study. Bioinformatics 2007; 23:2080-7; PMID:17553857; http://dx.doi.org/10.1093/bioinformatics/btm305
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122:3616-27; quiz 99; PMID:24030381; http://dx.doi.org/10.1182/blood-2013-08-518886
- Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol 2013; 14:1295-306; PMID:24239208; http://dx.doi.org/10.1016/S1470-2045(13)70491-1
- Yuan Y, Van Allen EM, Omberg L, Wagle N, Amin-Mansour A, Sokolov A, Byers LA, Xu Y, Hess KR, Diao L, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol 2014; 32:644-52; PMID:24952901; http://dx.doi.org/10.1038/nbt.2940
- Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. New Engl J Med 2003; 349:2091-8; PMID:14645636; http://dx.doi.org/10.1056/NEJMoa031317
- Urba S, Wolf G, Eisbruch A, Worden F, Lee J, Bradford C, Teknos T, Chepeha D, Prince M, Hogikyan N, et al. Single-cycle induction chemotherapy selects patients with advanced laryngeal cancer for combined chemoradiation: a new treatment paradigm. J Clin Oncol: Off J Am Soc Clin Oncol 2006; 24:593-8; PMID:16380415; http://dx.doi.org/10.1200/JCO.2005.01.2047
- Pointreau Y, Garaud P, Chapet S, Sire C, Tuchais C, Tortochaux J, Faivre S, Guerrif S, Alfonsi M, Calais G. Randomized trial of induction chemotherapy with cisplatin and 5-fluorouracil with or without docetaxel for larynx preservation. J Natl Cancer Inst 2009; 101:498-506; PMID:19318632; http://dx.doi.org/10.1093/jnci/djp007
- Lefebvre JL, Rolland F, Tesselaar M, Bardet E, Leemans CR, Geoffrois L, Hupperets P, Barzan L, de Raucourt D, Chevalier D, et al. Phase 3 randomized trial on larynx preservation comparing sequential vs alternating chemotherapy and radiotherapy. J Natl Cancer Inst 2009; 101:142-52; PMID:19176454; http://dx.doi.org/10.1093/jnci/djn460
- Lefebvre JL, Pointreau Y, Rolland F, Alfonsi M, Baudoux A, Sire C, de Raucourt D, Malard O, Degardin M, Tuchais C, et al. Induction chemotherapy followed by either chemoradiotherapy or bioradiotherapy for larynx preservation: the TREMPLIN randomized phase II study. J Clin Oncol: Off J Am Soc Clin Oncol 2013; 31:853-9; PMID:23341517; http://dx.doi.org/10.1200/JCO.2012.42.3988
- Claus R, Lucas DM, Ruppert AS, Williams KE, Weng D, Patterson K, Zucknick M, Oakes CC, Rassenti LZ, Greaves AW, et al. Validation of ZAP-70 methylation and its relative significance in predicting outcome in chronic lymphocytic leukemia. Blood 2014; 124:42-8; PMID:24868078; http://dx.doi.org/10.1182/blood-2014-02-555722
- Claus R, Wilop S, Hielscher T, Sonnet M, Dahl E, Galm O, Jost E, Plass C. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics: Off J DNA Methylation Soc 2012; 7:772-80; PMID:22647397; http://dx.doi.org/10.4161/epi.20299
- Hughes LA, Melotte V, de Schrijver J, de Maat M, Smit VT, Bovee JV, French PJ, van den Brandt PA, Schouten LJ, de Meyer T, et al. The CpG island methylator phenotype: what's in a name? Cancer Res 2013; 73:5858-68; PMID:23801749; http://dx.doi.org/10.1158/0008-5472.CAN-12-4306
- Suzuki H, Yamamoto E, Maruyama R, Niinuma T, Kai M. Biological significance of the CpG island methylator phenotype. Biochem Biophys Res Commun 2014; 455:35-42; PMID:25016183; http://dx.doi.org/10.1016/j.bbrc.2014.07.007
- Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, Thirlwell C, Dibra HK, Jay A, Butcher L, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med 2013; 5:15; PMID:23419152; http://dx.doi.org/10.1186/gm419
- Jithesh PV, Risk JM, Schache AG, Dhanda J, Lane B, Liloglou T, Shaw RJ. The epigenetic landscape of oral squamous cell carcinoma. Br J Cancer 2013; 108:370-9; PMID:23287992; http://dx.doi.org/10.1038/bjc.2012.568
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 1993; 15:532-4, 6–7
- Holzinger D, Schmitt M, Dyckhoff G, Benner A, Pawlita M, Bosch FX. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res 2012; 72:4993-5003; PMID:22991302; http://dx.doi.org/10.1158/0008-5472.CAN-11-3934
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997; 16:385-95; PMID:9044528; http://dx.doi.org/10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3
- Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med 1999; 18:2529-45; PMID:10474158; http://dx.doi.org/10.1002/(SICI)1097-0258(19990915/30)18:17/18<2529::AID-SIM274>3.0.CO;2-5
- Binder H, Schumacher M. Adapting prediction error estimates for biased complexity selection in high-dimensional bootstrap samples. Stat Appl Genet Mol Biol 2008; 7:Article12; PMID:18384265; http://dx.doi.org/10.2202/1544-6115.1346
- Efron B, Tibshirani R. Improvements on cross-validation: the 632+ bootstrap method. J Am Stat Assoc 1997; 92:548-60
- Lausen B, Hothorn T, Bretz F, Schumacher M. Assessment of optimal selected prognostic factors. Biometrical J 2004; 46:364-74; http://dx.doi.org/10.1002/bimj.200310030
