ABSTRACT
In mammals, expression of UBE3A is epigenetically regulated in neurons and expression is restricted to the maternal copy of UBE3A. A recent report claimed that Drosophila melanogaster UBE3A homolog (Dube3a) is preferentially expressed from the maternal allele in fly brain, inferring an imprinting mechanism. However, complex epigenetic regulatory features of the mammalian imprinting center are not present in Drosophila, and allele specific expression of Dube3a has not been documented. We used behavioral and electrophysiological analysis of the Dube3a loss-of-function allele (Dube3a15b) to investigate Dube3a imprinting in fly neurons. We found that motor impairment (climbing ability) and a newly-characterized defect in synaptic transmission are independent of parental inheritance of the Dube3a15b allele. Furthermore, expression analysis of coding single nucleotide polymorphisms (SNPs) in Dube3a did not reveal allele specific expression differences among reciprocal crosses. These data indicate that Dube3a is neither imprinted nor preferentially expressed from the maternal allele in fly neurons.
Abbreviations
| AS | = | Angelman syndrome |
| UBE3A | = | ubiquitin protein ligase E3A |
| Dube3a | = | Drosophila melanogaster homolog of UBE3A |
| UBE3A-AS | = | UBE3A antisense transcript |
| DGRP | = | Drosophila genetic reference panel |
| SNP | = | single nucleotide polymorphism |
Introduction
Angelman syndrome (AS) results from neuronal loss of maternally expressed ubiquitin protein ligase E3A (UBE3A) in neurons.Citation1 Maternal deletions of chromosome 15q11.2-q13.1 encompassing UBE3A are the most common cause of AS. However, maternally-inherited loss-of-function (LOF) point mutations in UBE3A, imprinting defects, or paternal uniparental disomy also cause AS.Citation2 The first evidence for imprinted expression of UBE3A in neurons arose from the observation that maternally derived deletions of 15q11.2-q13.1 cause ASCitation3 and that UBE3A displays maternal allele-specific expression in human brain samples.Citation4 In mice, Ube3a is expressed from the maternal allele in hippocampal, cerebellar, and olfactory bulb mitral neurons.Citation5 Ube3a is biallelically expressed in neurons born from stem cells in the adult mammalian brain and immature neurons early in development.Citation6 Ube3a expression shifts from biallelic to maternal allele specific as neurons mature.Citation7 UBE3A is biallellically expressed in human induced pluripotent stem cells and neuronal differentiation results in paternal silencing of UBE3A.Citation8 In contrast to neurons, glial cells in the mammalian brain biallelically express UBE3A.Citation6,7,9
Imprinted expression of UBE3A in neurons is a complex mechanism mediated by the expression of a UBE3A-antisense transcript (UBE3A-AS) that interferes with expression of the sense UBE3A full-length transcript. The Prader-Willi syndrome imprinting center (PWS-IC) is maternally methylated and silenced due to an upstream Angelman syndrome imprinting-center (AS-IC) that epigenetically alters the maternal PWS-IC in the female germline via a transcriptional mechanism, leaving maternal PWS-IC nonfunctional post-fertilization.Citation10 Downstream from the PWS-IC is the SNURF/SNRPN locus, and the SNRPN transcript is expressed exclusively from the paternal allele due to the unmethylated PWS-IC promoter. In mature neurons, the SNRPN transcript progresses through a cluster of snoRNAs and spliced host genes, terminating as UBE3A-AS. The expression of UBE3A-AS on the opposite strand of UBE3A interferes with UBE3A expression on the paternal allele and is the mechanism for paternal UBE3A silencing.Citation2
Although it is well-established that UBE3A is imprinted in the mature neurons of human and mouse brain, it is unclear if UBE3A is imprinted in the brains of non-mammalian species including the model invertebrate organism Drosophila melanogaster. Flies have a single UBE3A homologCitation11, Dube3a, and mutations in this gene give rise to various phenotypes independent of the parental origin of the mutation.Citation12 Recently, it was reported that Dube3a is preferentially expressed from the maternal allele in flies.Citation13 However, there is a lack of synteny between fly and human genomes at the UBE3A/Dube3a loci and there is no prior evidence for allele specific expression in flies. Given that flies do not have a documented imprinting center, differentially methylated region or UBE3A-AS, we set out to definitively determine if Dube3a is, in fact, preferentially expressed from the maternal allele in fly brain. Using behavioral analyses, electroretinograms (ERGs) and allele-specific coding single nucleotide polymorphism (SNP) gene expression studies, we found no evidence for Dube3a imprinting in flies. Both maternal and paternal Dube3a alleles are co-expressed in fly brain.
Results
Genomic regions surrounding fly and human UBE3A are not syntenic
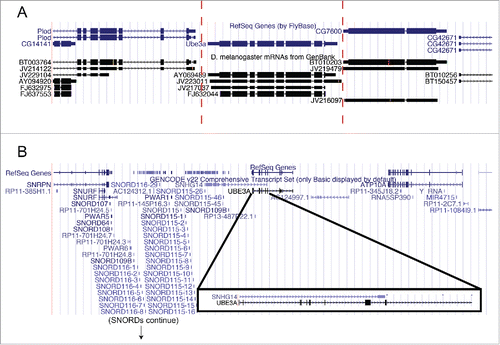
The UCSC genome browser was used to examine genomic features surrounding fly Dube3a and human UBE3A (). In flies, the 2 genes flanking Dube3a are Plod and CG7600, which have human homologs located at 7q22.1 and 8q24.13, respectively. No antisense transcript has been reported to interfere with Dube3a expression at this locus (). In contrast, the human SNHG14 (UBE3A-AS) transcript clearly overlaps with the UBE3A transcript (). Based on these data, it appears unlikely that fly Dube3a is preferentially expressed from the maternal allele as the surrounding genomic region is not syntenic to the human UBE3A region. There are no antisense transcripts detected across the Dube3a locus.
Figure 1. The genomic region surrounding UBE3A is not syntenic between flies and humans. Images from the UCSC genome browser (http://genome.ucsc.edu/) for the 10 kb region surrounding Dube3a in flies (A) and 1 Mb surrounding UBE3A in humans (B; note: the SNORD cluster has been truncated). In flies, there is no cluster of snoRNAs or the presence of SNURF/SNRPN upstream or downstream of Dube3a. There is also no RNA transcript that overlaps with Dube3a (A; red bars). In humans, note the presence of SNHG14 (UBE3A-AS) that overlaps with UBE3A transcript (B, boxed region).
Climbing behavior is not dependent upon maternal Dube3a expression
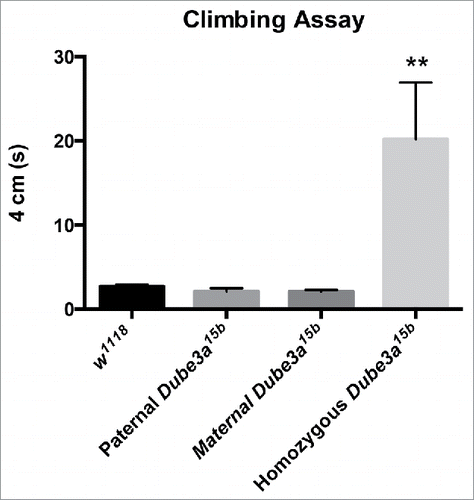
Previous work demonstrated climbing deficiencies in homozygous Dube3a15b (Dube3a null) flies, as significantly fewer Dube3a15b flies climbed 3 cm within 3 s.Citation12 Our experiments showed a significant effect of the Dube3a15b allele on climbing ability (P = 0.0021). Tukey's multiple comparisons test indicated that Dube3a15b homozygous flies took significantly longer to climb to a height of 4 cm compared to control w1118 flies, confirming previous reports of climbing defects in homozygous Dube3a15b flies. Flies that inherited this Dube3a15b allele through either the paternal or maternal germline displayed no differences in climbing behavior from controls (). These data indicate that a single copy of functional Dube3a of either paternal or maternal origin is sufficient for normal climbing behavior, and provide evidence that Dube3a is neither imprinted in flies nor preferentially expressed from the maternal allele.
Figure 2. Only Dube3a15b homozygous LOF flies display climbing deficits. At 5 d of age, flies were tested for motor function. Homozygous Dube3a15b flies had significantly impaired climbing ability compared to control w1118 flies. Paternally or maternally inherited Dube3a15b heterozygotes did not differ from controls in climbing ability (P = 0.0021). Data are presented as mean with standard error, n = 6 for all groups.
Synaptic transmission defects in Dube3a15b mutants are present independent of parent of origin
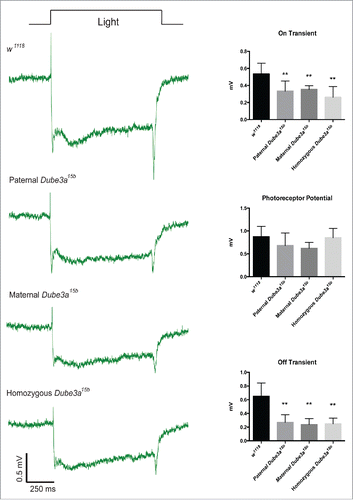
We previously demonstrated impaired synaptic transmission in Dube3a15b flies at the neuromuscular junction.Citation14 Here we investigated neuronal activity and synaptic transmission in photoreceptor neurons of the eye using ERGs. In the ERG signal, the “on” and “off” transients are indirect measures of synaptic transmission, while the photoreceptor potential is a measure of photoreceptor neuron depolarization.Citation15 Control w1118 flies displayed robust on/off transients and large photoreceptor potentials (). The Dube3a15b mutation significantly affected both on transients (P ≤ 0.0001) and off transients (P ≤ 0.0001). Tukey's post hoc multiple comparisons testing indicated that the presence of the Dube3a15b allele (either homozygous or heterozygous) reduced both on and off transients regardless of the parent of origin of the mutant allele as compared to control flies. No significant differences were observed in on/off transients among Dube3a15b allele carrying flies (heterozygous or homozygous). We observed no effect of this mutation on photoreceptor potentials (; P = 0.057). Thus, the Dube3a15b allele appears to effect synaptic transmission components of the ERG signal without interfering with photoreceptor neuron depolarization. Haploinsufficiency for Dube3a is also sufficient to decrease synaptic transmission regardless of the parental origin of the mutation, thus indicating that Dube3a is not preferentially expressed from either allele in the brain.
Figure 3. Heterozygous Dube3a15b flies have reduced on/off transients in the ERG signal independent of parent of inheritance. Control w1118 flies (n = 12) display robust on/off transients that are significantly reduced in paternal (n = 6), maternal (n = 7), and homozygous Dube3a15b (n = 11) flies (On Transient, P < 0.0001; Off transient, P < 0.0001). Note that the Dube3a15b mutation does not impair photoreceptor potentials. Data are presented as means with standard error.
Allele-specific expression of Dube3a
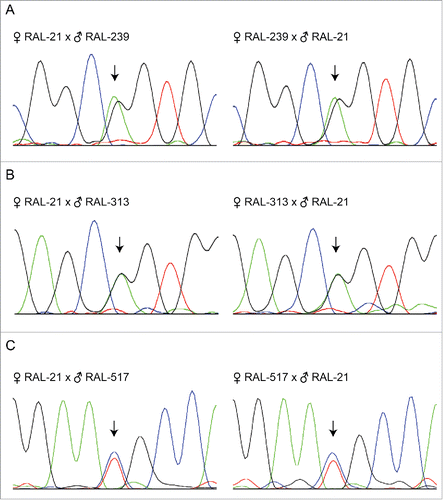
To investigate Dube3a allele specific expression at the molecular level, we performed reciprocal crosses with flies from the DGRP containing coding SNPs in Dube3a. Using the UCSC genome browser of the DGRP2 dataCitation16 (Located at: dgrp2.gnets.ncsu.edu), we selected coding SNPs located in Dube3a. Three separate DGRP lines were used; RAL-239 (G117T), RAL-313 (G78T), and RAL-517 (C171A), which each contain coding SNPs in Dube3a as compared to the reference line RAL-21. Female RAL-239 crossed to male RAL-21 and female RAL-21 crossed to male RAL-239 showed 2 similarly sized (∼50%) peaks in the chromatogram at position 117 in the Dube3a cDNA sequence (). Similar results were observed between reciprocal crosses of RAL-313 crossed to RAL-21 at position 78 (), and RAL-517 and RAL-21 at position 171 (). These data align with our previous experiments and support the conclusion that Dube3a is biallelically expressed in the fly brain, and neither imprinted nor preferentially expressed from the maternal allele.
Figure 4. Sanger sequencing chromatograms of different wild-type Dube3a coding SNPs does not indicate imprinting or preferential expression. Total fly head RNA from reciprocal crosses with different coding SNPs present in Dube3a was DNase I treated and converted to cDNA. Sanger sequencing was performed on amplified cDNA using primers flanking the SNP to identify the expression of paternally or maternally inherited Dube3a alleles. In all alleles tested, we found similar expression levels of maternally and paternally inherited alleles as indicated by 2 peaks at each SNP of the same height (arrows).
Discussion
Our behavioral and molecular analyses indicate that neural expression of Dube3a alleles is independent of parental origin. Using the same Dube3a15b allele as Wu et al., we confirmed climbing deficiency in Dube3a homozygous LOF flies.Citation12 Here we demonstrated that paternally- or maternally-deficient Dube3a15b flies do not display climbing difficulties, indicating that one functional copy of Dube3a is sufficient for normal motor function independent of parent of origin. Our lab previously demonstrated synaptic transmission deficits at the neuromuscular junction in Dube3a15b homozygous fliesCitation14, and we confirmed synaptic transmission deficits here using ERGs. In our previous work at the 3rd instar neuromuscular junction, we reported that excitatory junction potentials decreased in amplitude faster in Dube3a LOF flies in comparison with controls,Citation14 possibly due to a depletion of synaptic vesicles in the readily releasable pool.Citation17 Here we show reductions in on/off transients in the ERG signal, which may also be due to a depletion of synaptic vesicles. Regardless, on/off transients were reduced in Dube3a homozygous and reciprocal heterozygous Dube3a15b flies, supporting the conclusion that Dube3a is neither imprinted nor preferentially expressed and that Dube3a haploinsufficiency is sufficient to cause synaptic transmission defects. The strongest piece of evidence indicating that Dube3a is not imprinted in flies comes from sequencing several different wild-type alleles of Dube3a. Reciprocal crosses for 3 separate wild-type Dube3a alleles indicated similar, if not identical, expression of both maternally- and paternally-inherited alleles.
In contrast to Chakraborty et al.Citation13, we found no evidence that Dube3a is imprinted or preferentially expressed from the maternal allele in Drosophila melanogaster. On the other hand, our data is compatible with the previously reported learning deficits in Dube3a maternally-deficient flies since we found that Dube3a haploinsufficiency results in ERG on/off transient deficiencies regardless of parental origin. As the UBE3A imprinting mechanism in mature mammalian neurons is complex and similar PWS/AS-ICR or antisense transcripts have not been detected in Drosophila, our behavioral, electrophysiological, and Dube3a allele-specific expression data suggest that Dube3a is neither imprinted nor preferentially expressed in fruit flies.
Materials and methods
Drosophila stocks
All flies were maintained at 25°C on a 12-hour light/dark cycle and raised on standard corn meal media (Bloomington Stock Center: http://tinyurl.com/zs5tpbf). Dube3a15b is a null LOF allele that produces a truncated transcript that does not make functional Dube3a protein.Citation11 w1118 flies were used as controls. The following Drosophila Genetic Reference Panel (DGRP) linesCitation16 were also obtained from the Bloomington Drosophila Stock Center: RAL-21, RAL-239, RAL-313, and RAL-517 (stock numbers 28122, 28161, 25180, and 25197, respectively).
Genome comparisons
Genomic regions surrounding Dube3a in Drosophila melanogaster were compared to human UBE3A using the UCSC genome browser (http://genome.ucsc.edu/).Citation18 We used fly BDGP Release 6 + ISO1 MT/dm6 assembly and human GRCh38/hg38 assembly for comparisons.
Climbing assay
Climbing assays were performed as previously described.Citation19 In brief, 3 flies of each genotype at 5 d post eclosure were transferred to empty vials and allowed 1 min to acclimate. Flies were tapped to the bottom of the vial and the time was measured for the first fly to climb past a 4 cm vertical mark. Three trials were averaged per group of flies with 1 min intervals.
Electroretinogram (ERG)
ERGs were performed as previously describedCitation20 with modifications. Flies were immobilized on ice for 2 min, transferred to a glass slide, and held in place with dental wax. Tungsten wire electrodes were gently inserted into the right eye (recording electrode) and abdomen (ground electrode). Data was recorded using a Model 1800 Microelectrode AC Amplifier (A-M Systems), digitized with a Micro3 1401 digitizer (Cambridge Electronic Design), and analyzed with Spike2 software (Cambridge Electronic Design). Fly eyes were stimulated using 1-s light pulses from a 5 mm white LED (Radioshack).
Statistical analysis
Significance testing was performed by 1-way ANOVA analysis. All statistics were performed using Prism version 6.0 (GraphPad).
Allele-specific Dube3a sequencing
Using the DGRP and the UCSC Genome browser of DGRP2 data (dgrp2.gnets.ncsu.eduCitation16), we selected 3 lines with single nucleotide polymorphisms (SNPs) in coding exons of Dube3a. RAL-21 was used as a reference line, and lines RAL-313, RAL-239, and RAL-517 harbored G78T, G117T, and C171A SNPs, respectively. We performed reciprocal crosses and extracted RNA from heads for cDNA synthesis. Totals of 20–30 fly heads from each cross were removed and homogenized in TRI Reagent Solution (Ambion). Total RNA was extracted using Directzol RNA MiniPrep Plus (Zymo Research Corp) according to the manufacturer's instructions, which include a DNAse step to remove genomic DNA. RNA was converted to cDNA using Transcriptor First Strand cDNA Synthesis Kit (Roche). A 950 bp fragment of Dube3a encompassing our chosen SNPs was PCR amplified with Phusion polymerase (Thermofisher Scientific) using the following forward and reverse primers, respectively: 5′-ATGAACGGTGGCGGGG-3′, and 5′-CGCTGCTTTGGGATGAACAC-3′. PCR products were sent for Sanger sequencing (GENEWIZ) using the following sequencing primer: 5′-AATGAAGCTCTTGCCCAGTC-3′. Sequence data was analyzed using CodonCode Aligner (CodonCode Corporation).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by a pre-doctoral fellowship to KAH from the Duplication 15q Alliance and the UTHSC Neuroscience Institute. MSL was supported by the Dorothy/Daniel Gerwin Parkinson Research Fund, and National Institutes of Health grants R01 NS069936 and R21 GM118962. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
References
- Kishino T, Lalande M Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 1997; 15:70-3; PMID:8988171; http://dx.doi.org/10.1038/ng0197-70
- LaSalle JM, Reiter LT, Chamberlain SJ. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics 2015; 7:1213-28; PMID:26585570; http://dx.doi.org/10.2217/epi.15.70
- Knoll JH, Nicholls RD, Magenis RE, Graham JM Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 1989; 32:285-90; PMID:2564739; http://dx.doi.org/10.1002/ajmg.1320320235
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 1997; 17:14-15; PMID:9288088; http://dx.doi.org/10.1038/ng0997-14
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997 Sep; 17(1):75-8; PMID:9288101
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet 2008; 17:111-8; PMID:17940072; http://dx.doi.org/10.1093/hmg/ddm288
- Judson MC, Sosa-Pagan JO, Del Cid WA, Han JE, Philpot BD. Allelic specificity of Ube3a expression in the mouse brain during postnatal development. J Comp Neurol 2014; 522:1874-96; PMID:24254964; http://dx.doi.org/10.1002/cne.23507
- Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci U S A 2010; 107:17668-73; PMID:20876107; http://dx.doi.org/10.1073/pnas.1004487107
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet 2003; 12:837-47; PMID:12668607; http://dx.doi.org/10.1093/hmg/ddg106
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 2013; 368:20110336; PMID:23166397; http://dx.doi.org/10.1098/rstb.2011.0336
- Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet 2006; 15:2825-35; PMID:16905559; http://dx.doi.org/10.1093/hmg/ddl225
- Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, Fischer JA. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci U S A 2008; 105:12399-404; PMID:18701717; http://dx.doi.org/10.1073/pnas.0805291105
- Chakraborty M, Paul BK, Nayak T, Das A, Jana NR, Bhutani S. The E3 ligase ube3a is required for learning in Drosophila melanogaster. Biochem Biophys Res Commun 2015; 462:71-7; PMID:25935478; http://dx.doi.org/10.1016/j.bbrc.2015.04.110
- Valdez C, Scroggs R, Chassen R, Reiter LT. Variation in Dube3a expression affects neurotransmission at the Drosophila neuromuscular junction. Biol Open 2015; 4:776-82; PMID:25948754; http://dx.doi.org/10.1242/bio.20148045
- Ugur B, Chen K, Bellen HJ. Drosophila tools and assays for the study of human diseases. Dis Model Mech 2016; 9:235-44; PMID:26935102; http://dx.doi.org/10.1242/dmm.023762
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The drosophila melanogaster genetic reference panel. Nature 2012; 482:173-8; PMID:22318601; http://dx.doi.org/10.1038/nature10811
- Desai-Shah M, Cooper RL. Different mechanisms of Ca2+ regulation that influence synaptic transmission: comparison between crayfish and Drosophila neuromuscular junctions. Synapse 2009; 63:1100-21; PMID:19650116; http://dx.doi.org/10.1002/syn.20695
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996-1006. Article published online before print in May 2002 ; PMID:12045153; http://dx.doi.org/10.1101/gr.229102.
- Hatfield I, Harvey I, Yates ER, Redd JR, Reiter LT, Bridges D. The role of TORC1 in muscle development in Drosophila. Sci Rep 2015; 5:9676; PMID:25866192; http://dx.doi.org/10.1038/srep09676
- Vilinsky I, Johnson KG. Electroretinograms in Drosophila: a robust and genetically accessible electrophysiological system for the undergraduate laboratory. J Undergrad Neurosci Educ 2012; 11:A149-57; PMID:23494679
