ABSTRACT
Glycosylation is one of the most fundamental posttranslational modifications in cellular biology and has been shown to be epigenetically regulated. Understanding this process is important as epigenetic therapies such as those using DNA methyltransferase inhibitors are undergoing clinical trials for the treatment of ovarian and breast cancer. Previous work has demonstrated that altered glycosylation patterns are associated with aggressive disease in women presenting with breast and ovarian cancer. Moreover, the tumor microenvironment of hypoxia results in globally altered DNA methylation and is associated with aggressive cancer phenotypes and chemo-resistance, a feature integral to many cancers. There is sparse knowledge on the impact of these therapies on glycosylation. Moreover, little is known about the efficacy of DNA methyltransferase inhibitors in hypoxic tumors. In this review, we interrogate the impact that hypoxia and epigenetic regulation has on cancer cell glycosylation in relation to resultant tumor cell aggressiveness and chemo-resistance.
Abbreviations
| 5-AZA-dC | = | 5-AZA-2’-deoxycytidine |
| CDGs | = | congenital disorders of glycosylation |
| DNMT | = | DNA methyltransferase |
| EMT | = | epithelial to mesenchymal transition |
| FIGO | = | Fédération Internationale de Gynécologie et d'Obstétrique |
| FUTs | = | fucosyltransferases |
| GlcNAc | = | N-Acetylglucosamine |
| GnTs | = | N-Acetylglucosaminyltransferases |
| HDACs | = | histone deacetylases |
| HGSOC | = | high grade serous ovarian carcinomas |
| HIFs | = | hypoxic inducible factors |
| HRE | = | hypoxic response element |
| MAT | = | methionine adenosyl transferase |
| POFUT1 | = | polypeptide O-fucosyltransferase |
| POFUT2 | = | putative polypeptide O-fucosyltransferase |
| PTEN | = | phosphatase and tensin homolog |
| SAM | = | S-adenosyl methionine |
| sLeX | = | sialyl Lewis X |
| Siglecs | = | sialic acid-binding IgG like lectins |
Introduction
In 2012, 32.5 million people were living with cancer. Just over 14 million of those patients were diagnosed that year, a 12.8% increase from 5 years before.Citation1 The International Agency for Research on Cancer (IARC) predicts a 68% increase on these figures by 2030 if these trends persist.Citation1,2
Breast cancer
Breast cancer is the second most common cancer worldwide after lung cancer. It accounts for 25% of all female cancer incidences.Citation2 In 2012, it was estimated that 1.7 million women were diagnosed with breast cancer and, depending on the breast cancer type and stage at diagnosis, the survival rates were close to 90%.Citation1 However, because of the frequency of breast cancer occurrence, it is still the cancer that kills the highest number of women worldwide.Citation2 Breast cancers can broadly be categorized into 4 main groups, Luminal A, Luminal B, Human epidermal growth factor receptor 2 (HER2)-enriched, or Triple negative breast cancer (TNBC).Citation3 This categorization reflects receptor phenotype and the expression of certain proteins.Citation4
Luminal A is characterized by being positive for either hormone receptors or estrogen/progesterone (ER/PR), and negative for HER2. This subtype also has a low proliferation rate, determined by the presence and level of expression of the protein Ki-67.Citation5 Luminal B tumors are positive for either ER or PR and can be positive or negative for HER2, and display a higher proliferation rate. HER2-enriched tumors are positive for the HER2 receptor and may be positive or negative for either of the hormonal receptors. TNBCs are, as the name suggests, negative for all 3 (ER/PR/HER2) receptors. This phenotype accounts for 15–20% of all breast cancers and is pathologically the most aggressive.Citation6 Where Luminal A/B and HER2+ tumors can be treated with targeted therapies, such as Tamoxifen and Herceptin, TNBCs have no such receptors to target and, thus, are subject to traditional chemotherapy and/or radiation. Lehmann et al., have further categorized TNBCs based on their gene expression profile into 7 distinct subgroups, which they classified as basal-like 1 and 2, immunomodulatory, mesenchymal, mesenchymal stem-like, luminal androgen receptor, and unstable.Citation7 Although TNBCs may initially respond better to chemotherapy in comparison to other breast cancer subtypes, these patients unfortunately have a higher rate of metastasis both to visceral organs and to the brain. Moreover, the disease-free survival rate for women with TNBC with residual disease after first line therapy is about 4 months.Citation6,8
The gold standard for breast cancer screening and diagnosis is mammography in combination with biopsy and histopathological analysis. The sensitivity for detection of a mammography can be as high as 98% but this percentage is only associated with women over the age of 50 or, more precisely, those with less dense breast tissue.Citation9,10 Sensitivities of 48% have been reported in younger women with higher density breast tissue.Citation11 Another issue associated with mammography is over-diagnosis. In a paper published in the New England Journal of Medicine, Bleyer et al., estimated that 70,000 women were over-diagnosed with breast cancer in 2008.Citation12 This leads to unnecessary expense to the health system and undue anguish for the patient. A report published by the Swiss Medical Board in 2014 came to this conclusion recommending that systematic mammography screenings be phased out.Citation13 Mammography in combination with ultrasound screening has shown higher sensitivity in diagnosing breast cancer, but ultrasound sensitivity has been shown to be operator dependent.Citation14 An ideal diagnostic option would be a biological fluid (e.g., blood or urine)-based biomarker with a high specificity and sensitivity. This would mean a minimally invasive procedure for detection and a high degree of accuracy for prognosis. The biomarkers associated with breast cancer, such as cancer antigen 15–3 (CA15-3), the soluble form of the protein MUC-1, carcinoembryonic antigen (CEA), and tissue polypeptide antigen (TPA), as examples, have varying degrees of sensitivity and specificity and are of limited diagnostic and prognostic value.Citation15-17
Ovarian cancer
In 2014, an estimated 21,980 women were diagnosed with ovarian cancer. Although this number only accounts for 1.3% of newly diagnosed cancers,Citation18 ovarian cancer is the 5th cause of cancer-related deaths in women, thereby identifying it as the one of the most lethal of the gynecologic malignancies. Ovarian cancer, if diagnosed at stage I or stage II, has a 5-year-survival rate of 92.3% and 71.7%, respectively.Citation18 However, approximately 80% of women diagnosed with ovarian cancer present at a late stage (stages III and IV) and have survival rates varying between 39 and 17%, depending on the FIGO staging system.Citation19,20 Ovarian cancer is treated with a combination of surgery (optimal debulking) and chemotherapy. The majority of patients will initially respond to treatment and go into remission. However, of those that respond, 80% will develop chemo-resistance, relapse, and/or die within 5 y.Citation21 Platinum-based chemotherapeutic agents have been used in the treatment of cancer for the last 30 y. This strategy has changed little with the exception of Taxane-based therapies introduced in the early 1990s. The lack of improvement in 5-year-survival rates since 1980, compared to that of breast cancer, suggests that we are not progressing as fast as we would like or should in our clinical management of these women. Specifically, there has been a 25% increase in the 5-year-survival rates for patients diagnosed with breast cancer in the period from 1980 to 2006, while in the same period, the 5-year-survival rate for women presenting with ovarian cancer only showed a 7% increase.Citation22,23 One of the possible reasons for this is the lack of, or presence of unspecific symptoms in the early stages of this disease.
Ovarian cancer (OC) is a highly heterogeneous disease. Even though 90% of all ovarian cancers are classified as epithelial in origin, they can be further subcategorized into serous, endometrioid, mucinous, clear cell, undifferentiated carcinomas, and rare transitional cells.Citation24 However, it must be noted that malignant ovarian tumors are frequently metastatic, gastrointestinal, or uterine in origin.Citation25 Therefore, the term ovarian cancer is somewhat of a misnomer. A series of studies have led to a new paradigm for the pathogenesis and origin of epithelial ovarian cancer (EOC). The paradigm is based on a model that divides EOC into 2 broad categories, types I and II.Citation26 These tumors have distinct clinical and molecular features.Citation25 Type I tumors generally develop from benign conditions and are classified as low grade ovarian cancers. Type II tumors or high grade serous ovarian carcinomas (HGSOC) are thought to develop from what is called serous tubal intraepithelial carcinomas, which grow rapidly, are highly aggressive, and account for more than 75% of OCs.Citation27
This heterogeneity may be a possible reason for the lack of success, so far, in screening modalities for ovarian cancer. While some screening strategies have shown a degree of success, none have proven effective enough at improving overall survival when compared to traditional symptom-based screening.Citation28 Screening methods that have been explored have involved traditional imaging methods, such as ultrasound, MRI, and 18F-fluorodeoxyglucose positron emission tomography (FDG-PET). Others have involved looking at biomarkers, such as CA-125 and human epididymis protein 4. While the sensitivity and specificity of these alone proved to be low, looking at these in combination with predictive algorithms, such as the risk of ovarian cancer algorithm, has increased both sensitivity and specificity.Citation24
It is clear that ovarian and breast cancers have high degrees of variability within each disease. Within these 2 cancers, HGSOC and TNBC have proven to be the most difficult to correctly diagnose and successfully treat. The similarities do not end there. They are both associated with high relapse rates and high proportions of chemo-resistance, as stated earlier, and they also have an innately hypoxic tumor microenvironment, which has been shown to upregulate a “stem cell-like” property in both of these cancers.Citation29-31 Approximately 5% of breast cancersCitation32 and 10% of ovarian cancersCitation33 carry a BRCA1/2 mutation. These mutations lead to errors in DNA double-strand break homologous recombination repair and have shown some sensitivity to platinum-based drugs and PARP inhibitors.Citation32 However, a much higher percentage of these cancers present with errors in this pathway, which are caused by other mechanisms, such as hypermethylation, and subsequent silencing of the BRCA gene, which may be overlooked for these treatments due to lack of a BRCA mutation.Citation32 The need for highly sensitive and specific diagnostic and prognostic markers is clearly required if overall survival rates are to improve.
Glycosylation may provide some insight to the issues outlined above. Some research groups have already reported increased sensitivity and specificity of current cancer protein biomarkers by simply looking at variations in their glycoforms.Citation34,35 However, this information identifies even more questions, such as, what role these glycan alterations play in cancer biology and why there is a shift in glycosylation in this disease state.
Glycogenes may play some role in this but, from the published literature, we know that many other factors impact on the glycosylation process; therefore, the process is not gene specific (non-template driven). The two other significant contributors to an overarching understanding of cancer biology are epigenetic alterations and hypoxia. Epigenetic alterations are, in their own right, well established to occur in tumors, including breast and ovarian cancers, but, importantly, are also involved in an extensive array of pathological conditions. While these 2 topics have been the subject of numerous reviews, their potential interaction warrants investigation. In the published literature, there are data linking epigenetics and glycosylation, epigenetics and hypoxia, and hypoxia and chemo-resistance.Citation36-38 However, deciphering these interactions as a cohesive entity has enormous potential for understanding the biology not only of breast and ovarian cancer but also of tumor biology in general.
Glycosylation
Glycosylation is a posttranslational modification (PTM), whereby carbohydrate residues, or glycans, are attached to biomolecules to produce glycoconjugates, which are the primary form of PTMs for proteins and lipids. There are 2 main types of protein glycosylation: N-linked and O-linked. N-linked glycan biosynthesis takes place in the endoplasmic reticulum (ER). Glycans are attached to dolichol phosphate on the luminal side of the ER. This molecule then gets flipped to the inside of the ER for further attachment and cleavage of glycans. After folding, the glycoprotein conjugate is transported to the Golgi for further processing ().Citation39 N-linked glycans are the most abundant product of the glycosylation process.Citation40 They can be further divided based on their structures, of which there are 3: high mannose, complex, and hybrid. All 3 structures share a common core structure of 3 mannose residues attached to 2 N-Acetylglucosamine (GlcNAc) residues. Examples of these 3 classes can be seen in . The complex glycan structure is the most common class.Citation41
Figure 1. N-glycan processing. Schematic representation of the biosynthesis and processing of N-linked oligosaccharides.Citation125 Oxford nomenclature. GnT, N-Acetylglucosaminyltransferase; FucT, Fucosyltransferas; GlcT, Oligosaccharyltransferas; Lex, Lewis X epitope; sLex, Sialyl-Lex; Ley, Lewis Y. LacNAc, N-Acetyllactosamine. (Permission for use granted by Dr. Stephen A Whelan and colleagues).Citation125
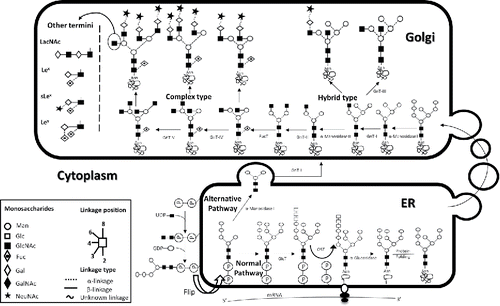
Figure 2. Glycan variations. Examples of the 3 different forms of N-linked glycans, seen on the left, linked to their N-glycosylation consensus sequence Asn-Xaa-S/T. Asn: Asparagine, Xaa: any amino acid except Proline and Serine/Threonine (S/T). (A) A high-mannose glycan (B) a complex glycan and (C) a hybrid structure. In the center are the 8 possible O-Linked structures attached to a Serine or a Threonine. On the right are the monosaccharides and linkage legends. (Variations depicted using Oxford nomenclature).
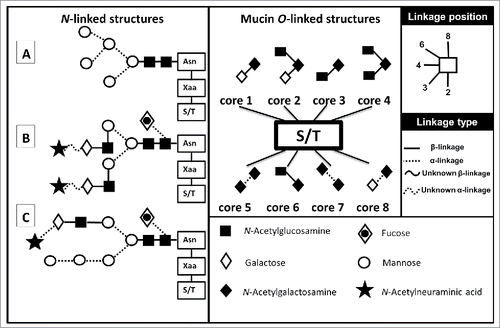
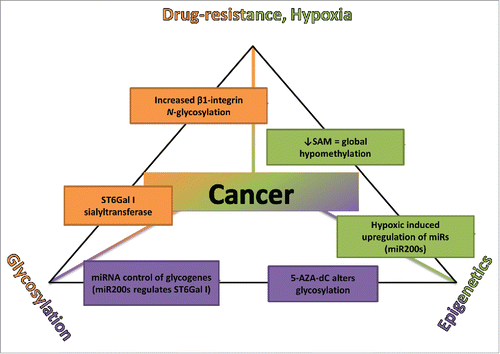
Figure 3. The Chemo-resistance-Epigenetics-Glycosylation triangle. Schematic diagram bringing together the idea that glycosylation is epigenetically regulated and that the epigenetic signature of a tumor cell is altered under hypoxic conditions conferring a drug-resistant phenotype and evidence to support this. SAM, S-adenosyl methionine; miR, microRNA; 5-AZA-dC, 5-AZA-2′-deoxyCytidine; ST6Gal I, α1–6 linkage sialyltransferase.
O-linked glycan biosynthesis takes place in the Golgi. O-glycans are structurally more diverse than their N-linked counterparts making them more difficult to characterize and, as a result, more difficult to classify. The majority of O-linked glycan biosynthetic pathways are not clearly defined. The best understood are the mucin type O-linked glycans. These mucin type O-glycans are covalently attached to serine/threonine protein residues via α-GalNAc, of which there are 8 possible variations in their core structure ().Citation42
Glycosylation is responsible for a wide range of biological functions, including protein folding/structure, protein trafficking, stability, biological activity, immunogenicity, receptor ligand recognition/binding, and molecule's half-life in the bloodstream. When this pathway is altered from its healthy functional state, a multitude of pathologies can occur. Aberrant glycosylation is associated with a wide group of disorders collectively known as congenital disorders of glycosylation (CDGs). Moreover, aberrant glycosylation is associated with disorders of the immune system, such as rheumatoid arthritis and IgA nephropathyCitation43 and, for many years now, is well established in cancer.Citation44
Glycosylation and cancer
There have been many observations made over the last 30 y with regards to N-glycosylation patterns and glycan profiles in various cancers, including breast and ovarian cancers.Citation45-48 Recently, large glycogene association studies were carried out. For example, Ashkani et al., analyzed the differential expression of 210 glycosylation related genes from 6 different cancer types including breast and ovarian cancer.Citation49 Using specific software and statistical analyses they were able to identify each cancer type with a greater that 95% success rate using all 210 genes.Citation49 Ashkani et al., also successfully subtyped HER2-enriched and basal-like breast cancer from luminal types.Citation49 Similar studies have been conducted by Potapenko et al., specifically looking at glycogene expression in breast cancer. They assessed a panel of 419 glycogenes and successfully correlated them with diagnosis, progression, and survival.Citation50,51 Interestingly, each gene product is involved in modifying the structure of N- and O-linked glycans.Citation49 However, the 3 most prominently featured structural modifications in the literature are alterations in branching, sialylation, and fucosylation.
Branching
Branching is the addition of N-GlcNAc to the 2 terminal core mannose residues and is carried out by a family of enzymes called N-Acetylglucosaminyltransferases (GnTs). Increased branching has been observed in many cancers, when compared to healthy patient controls and benign conditions.Citation52,53 For example, the epidermal growth factor receptor (EGFR) is a glycoprotein that has been implicated in about 30% of all epithelial cancers.Citation54 Increased branching of the N-glycans on EGFR increases the binding affinity of Galectin 3, which regulates EGFR activity.Citation55 Moreover, increased levels of Galectin 3 have been linked with metastases in many tumor types, including in breast and ovarian cancer cell lines.Citation55-57 Also, the addition of a bisecting GlcNAc, regulated by the enzyme mannosyl glycoprotein N-Acetylglucosaminyltransferase 3 (GnT III), results in increased affinity for Galectins altering the turnover of growth factor receptors and downstream signaling.Citation58
Epithelial-cadherin (E-cadherin) is an important cell adhesion molecule. Errors in, or deletion of E-cadherin function are a major contributor to cancer progression and occur in more than 70% of all human carcinomas.Citation59 GnT-V overexpression in MKN45 cells, a gastric cancer cell line, induces E-cadherin mislocalization from the cell membrane into the cytoplasm, resulting in impairment of cell to cell aggregation.Citation59
Sialylation
Sialylation is the addition of sialic acid to a terminal galactose of an N-glycan with the aid of sialyltransferases. The enzymes ST3Gal1, 3, 4, and 6 produce α2–3-linked sialic acid and ST6Gal1 forms an α1–6 linkage. There are over 50 members of the sialic acid family. However, the one found most frequently is N-Acetylneuraminic acid.Citation60 Sialic acids are usually added to terminal carbohydrates (for example galactose) of various glycans. This addition is required for many biologic processes such as cell to cell adhesion, cell migration and immune-surveillance.Citation61 However, aberrant sialylation has been associated with many disease states including cancer.Citation62,63 The sialyl Lewis X (sLeX) epitope is a tetra-saccharide that is recognized by the Selectin family of receptors present on the surface of leukocytes, platelets, and the endothelium. Binding of SLeX to these receptors aids in the extravasation and hematogenic transport of immune cells.Citation61 These mechanisms have been exploited by tumor cells and have been linked with malignancy and metastases in many cancers including colon, prostate, breast, and ovarian cancer.Citation64-67 Jandus et al., have shown that increased expression of terminal sialic acids or sialoglycans aid in immune evasion of tumor cells through binding of sialic acid-binding IgG like lectins (Siglecs) 7 and 9. These Siglecs 7 and 9 are expressed on the surface of natural killer (NK) cells and, once bound by their ligand, result in the inhibition of NK cells' cytotoxic effects.Citation68
Fucosylation
Fucosylation of N- and O-linked glycans is carried out by a family of 13 enzymes known as the fucosyltransferases (FUTs), which are numbered FUT 1-11, polypeptide O-fucosyltransferase (POFUT1), and putative polypeptide O-fucosyltransferase (POFUT2).Citation69 Depending on the linkage type and the cell type, these enzymes attach a fucose to the terminal portion of a glycan with the exception of FUT 8, which catalyzes the attachment of a fucose to the core GlcNAc bonded to the side chain of the Asparagine residue, resulting in what is known as a core fucose. Fucosylation is necessary for many biological processes. For example, Zhao et al., demonstrated that core fucosylation is essential to fetal development and growth, showing that 70% of FUT 8 knockdown mice died 3 d after birth, with the remaining neonates showing a wide array of complications, such as emphysema-like symptoms.Citation70 Lack of core fucose enhances antibody-dependent cellular cytotoxicity 50–100%.Citation71 Terminal fucose is also important for leucocyte extravasation as seen in the CDG, leukocyte adhesion deficiency II (LAD II). LAD II occurs as a result of a mutation in the SLC35C1 gene coding for the GDP-fucose transporter and is characterized by a disruption in the tethering of leucocytes to endothelial cells via selectin binding, thereby inhibiting extravasation.Citation72 While outer arm fucose is part of the Lewis and sLex antigen, fucose levels have also been linked to cancers in other ways.Citation73-77 Specifically, increased levels of various FUTs have been reported in hepatocellular carcinoma, head and neck squamous cell carcinoma, lung cancer, and prostate cancer.Citation73-76 Also a fucosylated form of α-fetoprotein, α-fetoprotein L3, was suggested as a predictive biomarker for hepatocellular carcinoma (HCC) more than 20 y ago.Citation78 It was shown to predict the onset of HCC from cirrhotic livers before the tumor was picked up by imaging techniques with a sensitivity of 73% and a specificity of 96%.
While the preceding section has talked about these modifications separately, they rarely occur independently of each other. In most of the cases referred to, altered states of branching, fucosylation, and/or sialylation can be seen simultaneously, and have been shown to vary, depending on the cancer. It may also be the case that glycan profiles are tissue specific depending on the function of the glyco-conjugates in that tissue. Abd Hamid et al., observed that, in breast cancer patients, certain glycoforms containing the sLex epitope are a more sensitive tool for staging and determining metastasis when compared to the traditional biomarker CA15-3.Citation17 Kurebayashi et al., concluded that when using sLeX in combination with CA15-3 and CEA, monitoring sensitivity in relation to breast cancer progression increased by 17%.Citation79 While it is clear that cancer alters the homeostasis of glycosylation and that there is a possible correlation between cancer type, stage, metastatic potential, and prognosis, the big question still remains unanswered: Why? One of the possible answers may be the epigenetic regulation of glycosylation.
Epigenetics
Epigenetics is the study of heritable alterations in gene expression that occur independently of modifications in the DNA nucleotide sequence. These include histone modification, remodeling of microRNA (miR), and DNA methylation (hypo/hyper). Histone modifications, such as methylation and acetylation, can either activate or repress gene transcription through alterations in the chromatin structure. miRs control the production of the gene product through binding to the mRNA 3′UTR regions, thereby inhibiting protein translation or enhancing mRNA degradation. DNA methylation primarily occurs on CpG rich regions (CpG islands), where a methyl group is covalently attached to a cytosine with the aid of DNA methyltransferases (DNMTs). All of these epigenetic alterations are regulatory steps ultimately controlling cell and tissue phenotype.Citation38,80,81 However, dysregulation of epigenetic alterations is also present in many disease states, including cancer.Citation82
Epigenetics and cancer
Global hypomethylation with site-specific hypermethylation has been repeatedly observed on the DNA of cancer cells.Citation83 Promoter hypermethylation results in gene silencing as a consequence of transcription factors being unable to bind to a promoter region of a gene. The subsequent recruitment of methyl-binding domain proteins also results in gene silencing via interactions with histone deacetylases (HDACs). The hypermethylation of genes involved in cell cycle control or tumor suppression causes these genes to be silenced and optimizes the environment for cancer initiation and progression. This makes DNMTs attractive targets for cancer therapy. Two DNMT inhibitors (DNMTi), have been approved for treatment of hematological malignancies: 5-AZAcytidine (Vidaza), approved by the FDA in 2004, and 5-AZA-2′-deoxycytidine (Dacogen/5-AZA-dC), approved by the FDA in 2006.Citation84 Moreover, 5-AZA-dC is currently in clinical trials for the treatment of various other cancers arising in the lung, liver, breast, and ovary.Citation85 While there are promising results using this therapy, 5-AZA-dC is a pan DNMTi causing DNA hypomethylation on all CpG rich genes, even those that may require silencing for healthy physiological function to take place. While the mechanism of action for this inhibitor is well documented, how 5-AZA-dC affects the whole cell is unclear and may be cell type specific. This theory is supported by results produced by Ari et al., who treated 2 breast cancer cell lines, MCF-7 (non-metastatic cell line) and MDA-MB-231 (TNBC metastatic cell line), with 5-AZA-dC. These treatments reduced the invasion capacity of MDA-MB-231 but increased the invasion capacity of the non-invasive MCF-7. This treatment also reduced viability in both cell lines but only induced apoptosis in MCF-7s. This suggests that in the MDA-MB-231 cells the apparent reduction in viability may be due to necrosis because of impaired apoptotic pathway(a).Citation86
Epigenetics and glycosylation
Epigenetic regulation of glycosylation is a relatively new emerging concept but with enough studies completed to highlight its importance.Citation36,48,87 There are about 25,000 protein coding genes in the human genome but when we compare this to other eukaryotes, the variation in gene number is not large enough to account for the vastness in our complexity. Lauc et al., proposed that the introduction of glycosylation heralded the “third revolution in evolution,” owing to our intricacy.Citation88 Horvat et al., have demonstrated that treatment with DNA methyltransferase inhibitors and histone deacetylase inhibitors in HeLa cells altered their glycan profile.Citation87 Moreover, Agrawal et al., identified miRs in the NCI-60 cancer cell set that controlled glycogene transcripts, drawing a correlation between altered epigenetic status, glycosylation, and cell phenotype.Citation36 This would suggest there is a strong link between epigenetics, glycosylation, and cancer.
Epigenetics, glycosylation, and cancer
In a study by Chakraborty et al., macrophage–melanoma hybrid cells treated with 5-AZA-dC, resulted in downregulation of GnT-V,Citation89 an enzyme which catalyzes the transfer of a β1,6-linked GlcNAc to either of the outer mannose of the N-glycan core, thereby increasing branching.Citation89 Importantly, increased branching has frequently been shown to be favorable for metastasis,Citation52 and so GnT-V was termed a “master regulator of metastasis.”Citation89 The reduced expression of GnT-V mRNA subsequently resulted in a reduction of β1,6-branched glycans.Citation89 Reduced glycosylation of the lysosome-associated membrane protein-1 (LAMP-1), a major substrate for GnT-V was also demonstrated. Results showing transcriptional regulation of the GnT-V gene (MGAT5) may have been as an indirect consequence of upregulation of nm23, a metastasis suppressor protein that negatively regulates MGAT5 transcription.Citation89 This increase in transcription could be a result of CpG island demethylation at the nm23 promoter region.Citation89
Conversely, Saldova et al., demonstrated an increase in GnT-V mRNA expression following 5-AZA-dC treatment in the ovarian cancer cell line OVCAR3. They also showed increased levels of branching and sialylation of N-glycans, which, as was discussed earlier, would also favor metastases.Citation48 Similar results were shown by Chachadi et al., in that 5-AZA-dC treatment enhanced favorable metastatic factors. These factors included an increase in the production of the sLex epitope on the MUC1 protein in HCT15 colon cancer cells and an increase in E-selectin binding of these cells, which, as stated earlier, is the first step in the extravasation of leukocytes under physiological conditions.Citation90 While Chakraborty et al., and Saldova et al.,'s results are contradictory, they may be explained by differential methylation profiles unique to each cell line, identifying the consequences of 5-AZA-dC treatment as cell line-specific. These papers not only highlight the role that epigenetics (methylation) plays in glycosylation patterns, but also the need for a more in-depth look at the effects of 5-AZA-dC on glycosylation patterns in various tumor tissues, and on tumors of varying grade and stage.
Drug resistance in cancer
Drug resistance still remains one of the biggest clinical challenges to overcome in relation to cancer therapy. Cancers develop resistance for a number of reasons; some of the main ones are highlighted in .
Table 1. Examples of drug resistant mechanisms in tumors
Recently published data from 2 individual studies have produced some interesting results on chemo-resistance in ovarian cancer. Weiner-Gorzel et al., demonstrated that increased expression of the microRNA miR-433 induces senescence and, subsequently, paclitaxel resistance in ovarian cancer.Citation91 Data published just a few months later shows that depletion of the growth factor receptor-bound protein 2 (GRB2) aids in tumor progression/survival and is a marker of poor prognosis in women with ovarian cancer.Citation92 GRB2 is involved in the PI3K/PTEN/Akt pathway and its precursor mRNA is one of the targets of miR-433.
Glycosylation has also been implicated in drug resistance. Lesniak et al., looked at the role of the β1-integrin in HER-2 positive luminal breast cancer resistance to Trastuzumab.Citation93 They found that a higher molecular weight integrin coincided with epithelial to mesenchymal transition (EMT). They showed that the higher molecular weight was due to N-glycosylation of the β1-integrin. They also demonstrated that antagonization of the higher molecular weight isoform with the monoclonal antibody (mAb) AIIB2 restored the cells to an epithelial phenotype. This mAb has also been shown to increase the sensitivity to Trastuzumab and Lapatinib.Citation94 Schultz et al., subsequently demonstrated that ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian cancer cells.Citation95 While ST6Gal-I activity on efficacy of cisplatin is not yet understood, possible mechanisms suggested for this were inhibition of the Fas receptor internalization and subsequent inhibition of this apoptotic pathway or mislocalization of transporter proteins due to aberrant glycosylation.
Hypoxia
Another element associated with tumor progression is hypoxia. As the tumor grows in size, so does its energy demand and its need for oxygen. When the oxygen demand outweighs the availability of oxygen in the tumor, it becomes hypoxic. Hockel et al., proposed that tumor hypoxia is represented by a partial oxygen pressure (pO2) of 8–10 mmHg (∼1% of pO2 at sea level, 760 mmHg). However, normal pO2 (physoxia) varies depending on the tissue type; so, for example, physoxia in brain tissue is reported to average 34.5 mmHg (4.5%), while physoxia in the lung has been reported to be 43 mmHg (5.6%).Citation96,97 This alters the pO2 at which hypoxic conditions occur and should be taken into consideration in in vitro experiments in which cell lines are usually incubated at a pO2 of 19.95%.Citation98
Hypoxia and chemo-resistance
The association between hypoxia and chemo-resistance has been widely published. Hypoxic conditions in the tumor microenvironment activate hypoxic inducible factors (HIFs). HIF-1α is a transcription factor that recognizes a sequence present in numerous genes called the hypoxic response element (HRE). The binding of HIF-1α to these elements regulates the transcription of the gene producing or reducing factors, which will promote cell survival under hypoxic conditions. Some of these include the upregulation of growth factors, such as vascular endothelial growth factors and fibroblast growth factor, which promote angiogenesis and cell survival, respectively.Citation37,99 Hypoxia also induces the MDR1 gene, which codes for the permeability glycoprotein, a transmembrane protein that transports a wide variety of exogenous compounds out of the cell, thereby maintaining homeostasis.Citation100 All of these factors combine to create an aggressive and possibly metastatic phenotype.Citation100,101 There is also the opinion that hypoxia induces epigenetic alterations, which aid in cell survival and will be discussed in the following section.Citation102
Hypoxia and epigenetic regulation
Watson et al., demonstrated epigenetic signatures that promoted and maintained a hypoxia-adapted cellular phenotype with a potential role in prostate cancer development.Citation38
Shahrzad et al., published data that showed that the hypoxic conditions observed in tumors resulted in the global DNA hypomethylation we associate with cancer.Citation103 While they alluded to the possible mechanism underlying this altered state, a paper published in the same year by Liu et al., gave a more comprehensive overview of the possible mechanism of hypomethylation in hepatocellular carcinoma (HCC). Specifically, the enzyme methionine adenosyl transferase (MAT) catalyzes the production of S-adenosyl methionine (SAM), which is a methyl donor substrate used in many cellular processes, including DNA and histone methylation. MAT is encoded by 2 genes in mammals, MAT1A and MAT2A. A switch from the MAT1A to the MAT2A protein is frequently seen in HCC and is associated with a downregulation of SAM.Citation104 In addition to its role in DNA methylation, SAM has also been shown to regulate hepatocyte growth and differentiation.Citation105 Interestingly, Liu and colleagues have shown that MAT2A has a HRE in its promoter region, which binds HIF-1α, which then upregulates transcription of MAT2A.Citation106
Hypoxia also regulates the levels of some miRs. Specifically, Kulshreshtha et al., showed the induction of numerous miRs in colon and breast cancer cell lines grown under hypoxic conditions, including miR-210, which available data suggests is one of the most consistently activated miRs in response to hypoxia.Citation107 miR-210 inhibition of vacuole membrane protein 1, a stress-induced protein that promotes the formation of intracellular vacuoles followed by cell death, is involved in autophagy and has been shown to mediate metastasis in breast and kidney cancer and HCC cells.Citation108,109 In addition, hypoxia-induced expression of miR-424 in human endothelial cells has been shown to result in the decline of cullin 2 (CUL2), a core protein of the cullin-RING ligase system used to target proteins for degradation, which potentially stabilizes HIF1α.Citation110 Certain miRs, such as miR-21, have been reported to upregulate the production of HIF-1α in prostate cancer cell lines through the inhibition of phosphatase and tensin homolog (PTEN) and the activation of the Akt and ERK signaling pathways.Citation111 The miR-200 family of RNAs has also been identified to play a vital role in regulating cell polarity and migration and, consequently, the epithelial to mesenchymal transition. Chan et al., demonstrated that hypoxia downregulated miR-200b, one of the 5 miR-200 family members, resulting in an increased angiogenic response.Citation112 Interestingly, Agrawal and colleagues have shown that this family of miR-200s targets various glycogenes, including α-L-fucosidase-2 (FUC2), which is involved in the hydrolysis of terminal α-1,2-L-fucose residues from glycoproteins and α-2,6-sialyltransferase (ST6GALNAC), whose protein product catalyzes the addition of sialic acid to terminal galactose, a feature upregulated in many cancers.Citation36
The Chemo-resistance-Epigenetics-Glycosylation triangle
As evidenced above, there is a clear link between (a) chemo-resistance and hypoxia; (b) hypoxia and epigenetics; and (c) glycosylation and epigenetics (). A possible conclusion to draw is that there is a link between hypoxia and the regulation of glycosylation and, following from this, between glycosylation and chemo-resistance. There have been several papers published looking at the effect of hypoxia on cancer associated carbohydrate antigens in colon cancer. They specifically noted how HIF increased the expression of sialyl Lewis A and sialyl Lewis X.Citation113-115 Sialyl Lewis A is structurally and functionally similar to sialyl Lewis X, apart from some variation in glycosidic bonds. While these results are informative, they focused on 2 carbohydrate antigens and not the whole cell glycome. As reviewed earlier, the miR200 family has been linked to hypoxia and, with the regulation of certain glycogenes.Citation36 Also, hypoxia has been shown to alter DNA methylation,Citation102 another epigenetic mechanism whose change in state affects glycosylation.Citation48 As glycosylation is non-template driven, there are numerous factors that can alter this process, such as the efficiency of sugar nucleotide transporters, the concentration of glycosyltransferase enzymes in the endoplasmic reticulum or Golgi, and the availability of sugar nucleotides. Two of the hallmarks of a cancer cell are altered cellular metabolism and increase in glucose uptake for glycolysis. This switch from oxidative phosphorylation to glycolysis has been shown to induce HIF-1 activation independent of hypoxia.Citation116 This switch could possibly alter glycosylation through increased availability of sugar nucleotides, HIF regulation of glycogenes, and HIF regulation of epigenetic controls.
Further elucidating the role that hypoxia plays in glycosylation and the mechanism by which epigenetic alterations, occurring as a result of hypoxia, affect the type of glycan profile produced by the tumor cell or the effect a tumor has on the whole serum glycome, may be a new avenue for diagnosis, prognosis, and efficient treatment selection. Gornik et al., have carried out a study on human serum producing results that highlight the fact that glycans have good temporal stability and intra-individuality.Citation117 As mentioned earlier, many studies have demonstrated that glycans vary with many disease states and good temporal stability means glycans may be used to track disease progression and treatment response and/or the development of chemo-resistance. These markers would improve the efficacy of treatment and potentially reduce the cost to the patient and the health care system. Glycosylation has been credited for the evolution of multicellular organisms and for our complexity.Citation118 Appreciating that cancer is a complex disease, it is logical to assume that changes in this complexity would serve as a good biomarker for determining specific aspects of a disease state or, more precisely, cancer stage, prognosis, progression, and treatment options. For example, one possible mechanism involved in a cancer metastasizing is detachment of the cell from the tumor tissue (EMT), intravasation, transportation to different site, attachment to endothelial cells, and extravasation and specific glycans are necessary for these processes.Citation119
Conclusion
While glycosylation studies are producing exciting new results and revealing new possibilities for treatment, prognosis, and determining progression of cancer and other diseases, glycosylation cannot be viewed as a singular entity. In a review published by Zoldoš et al., in 2013 they emphasize the need for “multi-omics” data analysis to further advance our understanding of the transition from healthy physiology to a disease state.Citation120 Using high-throughput glycan analysis technology developed in NIBRT, we, together with our colleagues, published the results of the first genome-wide association study (GWAS) of the human IgG N-glycome.Citation121 In this study, we identified 9 genes that reach the GWAS P-value threshold for significance. Surprisingly, only 4 of those 9 genes were directly involved in N-glycosylation (ST6GALI, B4GALT1, FUT8, and MGAT3). The other 5 genes had a stronger association to IgG N-glycosylation than the 4 previously mentioned. This suggests a more complex level of N-glycosylation regulation for this protein (IgG) than previously thought. These five genes have also been linked with autoimmune diseases and neoplasia.Citation121 This study subsequently went on to identify possible plasma biomarkers for systemic lupus erythematous (SLE), confirming the power of multiple –omic studies and paving the way for other GWA studies of this type.
A study carried out by Haakensen et al., also emphasizes the increased benefit of using multiple –omics and shows how this type of analysis would be a more powerful tool than looking at individual aspects of a biological system.Citation122 They showed a significant association of serum glycans with various aspects of breast cancer, such as survival, tumor size, and aggressiveness. They also significantly correlated glycan structures to mRNA transcripts that are associated with replication, repair, metabolism, adhesion, integrins, and angiogenesis. In agreement with Abd Hamid et al.,Citation17 and Saldova et al.Citation48, they concluded that the serum glycome changes cannot all be accounted for by the alterations in glycans on glycoproteins secreted by the tumor but are chiefly the result of the biological system responding to the tumor, since the changes are found mainly on liver proteins, particularly glycans, involved in the acute phase proteins or on immunoglobulin.Citation122
The take home message here is twofold: 1) Epigenetic therapies, such as 5-AZA-dC (Decitabine), are increasingly being trialed in women presenting with ovarian and breast cancer.Citation123,124 However, little is known of the impact of these therapies on processes such as glycosylation, which are known to be epigenetically regulated. In addition, as stated earlier, published data has shown that epigenetic therapies can in fact increase glycan structure, which might potentially facilitate tumor progression and metastasis. The question is whether these specific structures could be used as markers of progression and prognosis; 2) Hypoxia is an innate tumor microenvironmental state, which has been shown to alter the epigenome and enhance drug resistance. Data has also been published on how altered glycosylation aids in drug resistance. Is it possible that hypoxia alters glycosylation through epigenetic regulation to promote tumor survival and drug resistance? Continued research into these areas is warranted.
Cancer is a disease state that occurs as a result of multiple genetic and epigenetic alterations; yet, we see similar glycan alterations across a wide range of cancers. This is because a single glycan alteration can reflect many genetic and epigenetic changes. While we feel multiple –omics analysis of breast and ovarian cancer is the way forward, glycans on their own can be a very powerful tool, for example, as targets for mAb therapies and in providing useful information when determining breast and ovarian cancer pathogenesis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The authors acknowledge funding from Science foundation Ireland Starting Investigator Research grant (SFI SIRG) under grant number 13/SIRG/2164.
ORCID
Radka Saldova http://orcid.org/0000-0001-5085-5080
References
- IARC. Globocan 2012. Lyon: 2014. [accessed 2015 Jan 27]. globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- IARC CRUK. World Cancer Factsheet. London: 2014. [accessed 2015 Jan 27]. http://publications.cancerresearchuk.org/downloads/Product/CS_REPORT_WORLD.pdf.
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature 2000; 406(6797):747-52; PMID:10963602; http://dx.doi.org/10.1038/35021093
- Chikarmane S, Tirumani S, Howard S, Jagannathan J, DiPiro P. Metastatic patterns of breast cancer subtypes: What radiologists should know in the era of personalized cancer medicine. Clin Radiol 2015. 70(1):1-10; PMID:25300558; http://dx.doi.org/10.1016/j.crad.2014.08.015
- Kumar V, Abbas AK, Fausto N, Aster JC. Robbins and Cotran Pathologic Basis of Disease, Professional Edition: Expert Consult-Online: Elsevier Health Sciences; 2009
- Ismail-Khan R, Bui MM. A review of triple-negative breast cancer. Cancer control: journal of the moffitt cancer center. 2010; 17(3):173-6; PMID:20664514
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121(7):2750; PMID:21633166; http://dx.doi.org/10.1172/JCI45014
- Jiao Q, Wu A, Shao G, Peng H, Wang M, Ji S, Liu P, Zhang J. The latest progress in research on triple negative breast cancer (TNBC): risk factors, possible therapeutic targets and prognostic markers. J Thorac Dis 2014; 6(9):1329-35; PMID:25276378; http://dx.doi.org/10.3978/j.issn.2072-1439.2014.08.13
- dos Anjos Pultz B, da Luz FAC, de Faria PR, Oliveira APL, de Araújo RA, Silva MJB. Far beyond the usual biomarkers in breast cancer: a review. J Cancer 2014; 5(7):559; PMID:25057307; http://dx.doi.org/10.7150/jca.8925
- Drukteinis JS, Mooney BP, Flowers CI, Gatenby RA. Beyond Mammography: New frontiers in breast cancer screening. Am J Med 2013; 126(6):472-9; PMID:23561631; http://dx.doi.org/10.1016/j.amjmed.2012.11.025
- Mandelson MT, Oestreicher N, Porter PL, White D, Finder CA, Taplin SH, White E. Breast density as a predictor of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst 2000; 92(13):1081-7; PMID:10880551; http://dx.doi.org/10.1093/jnci/92.13.1081
- Bleyer A, Welch HG. Effect of three decades of screening mammography on breast-cancer incidence. N Eng J Med 2012; 367(21):1998-2005; PMID:23171096; http://dx.doi.org/10.1056/NEJMoa1206809
- Biller-Andorno N, Jüni P. Abolishing mammography screening programs? A view from the Swiss medical board. N Eng J Med 2014; 370(21):1965-7; PMID:24738641; http://dx.doi.org/10.1056/NEJMp1401875
- Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 2013; 8(1):3; PMID:23301032; http://dx.doi.org/10.1371/journal.pone.0053141
- Pedersen AC, Sørensen PD, Jacobsen EH, Madsen JS, Brandslund I. Sensitivity of CA 15-3, CEA and serum HER2 in the early detection of recurrence of breast cancer. Clin Chem Lab Med 2013; 51(7):1511-9; PMID:23403727; http://dx.doi.org/10.1515/cclm-2012-0488
- Pierce A, Saldova R, Abd Hamid UM, Abrahams JL, McDermott EW, Evoy D, Duffy MJ, Rudd PM. Levels of specific glycans significantly distinguish lymph node-positive from lymph node-negative breast cancer patients. Glycobiology 2010; 20(10):1283-8; PMID:20581008; http://dx.doi.org/10.1093/glycob/cwq090
- Abd Hamid UM, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Pardo M, Antrobus R, Chapman CJ, Zitzmann N, et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology 2008; 18(12):1105-18; PMID:18818422; http://dx.doi.org/10.1093/glycob/cwn095
- SEER. SEER Stat Fact Sheets: Ovary Cancer 2014. NIH [accessed 2015 Jan 27]. Available from: http://seer.cancer.gov/statfacts/html/ovary.html
- Society AC. Ovarian Cancer: American Cancer Society; 2015 [updated 2015 May 8, cited 2015 Nov 4]. Available from: http://www.cancer.org/cancer/ovariancancer/detailedguide/ovarian-cancer-survival-rates
- Thalhammer T, Mungenast F. Estrogen biosynthesis and action in ovarian cancer. Frontiers Endocrinol 2014; 5:192; PMID:25429284; http://dx.doi.org/10.3389/fendo.2014.00192
- Patel S, Kumar L, Singh N. Metformin and epithelial ovarian cancer therapeutics. Cell Oncol 2015; 38(5):365-75; PMID:26266765; http://dx.doi.org/10.1007/s13402-015-0235-7
- DeSantis C, Siegel R, Jemal A. Breast cancer facts and figures Atlanta: American Cancer Society, Inc. 2013–2014. Atlanta: American Cancer Society, Inc. 2013
- Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS) Atlanta, GA: American Cancer Society, 2012. [accessed 2015 Jan 27]. http://uacc.arizona.edu/sites/default/files/acs_2012.pdf
- Cohen JG, White M, Cruz A, Farias-Eisner R. In 2014, can we do better than CA125 in the early detection of ovarian cancer? World J Biol Chem 2014; 5(3):286-300; PMID:25225597; http://dx.doi.org/10.4331/wjbc.v5.i3.286
- Bowtell DDL. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer 2010; 10(11):803-8; PMID:20944665; http://dx.doi.org/10.1038/nrc2946
- Shih IM, Kurman RJ. Ovarian Tumorigenesis: A proposed model based on morphological and molecular genetic analysis. Am J Pathol 2004; 164(5):1511-8; PMID:15111296; http://dx.doi.org/10.1016/S0002-9440(10)63708-X
- Kurman RJ, Shih IM. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—Shifting the paradigm. Hum Pathol 2011; 42(7):918-31; PMID:21683865; http://dx.doi.org/10.1016/j.humpath.2011.03.003
- Vine MF, Calingaert B, Berchuck A, Schildkraut JM. Characterization of prediagnostic symptoms among primary epithelial ovarian cancer cases and controls. Gynecol Oncol 2003; 90(1):75-82; PMID:12821345; http://dx.doi.org/10.1016/S0090-8258(03)00175-6
- Liang D, Ma Y, Liu J, Trope C, Holm R, Nesland J, Suo Z. The hypoxic microenvironment upgrades stem-like properties of ovarian cancer cells. BMC Cancer 2012; 12(1):201; PMID:22642602; http://dx.doi.org/10.1186/1471-2407-12-201
- Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, Clouthier SG, Wicha MS. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci 2012; 109(8):2784-9; PMID:22308314; http://dx.doi.org/10.1073/pnas.1018866109
- O'Reilly EA, Gubbins L, Sharma S, Tully R, Guang MHZ, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, et al. The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clin 2015; 3:257-75; PMID:26676166; http://dx.doi.org/10.1016/j.bbacli.2015.03.003
- Jacot W, Theillet C, Guiu S, Lamy PJ. Targeting triple-negative breast cancer and high-grade ovarian carcinoma: refining BRCAness beyond BRCA1/2 mutations? Future Oncol 2015; 11(4):557-9; PMID:25686112; http://dx.doi.org/10.2217/fon.14.268
- Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, Lambrechts D, Despierre E, Barrowdale D, McGuffog L, et al. Association between brca1 and brca2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012; 307(4):382-9; PMID:22274685; http://dx.doi.org/10.1001/jama.2012.20
- Llop E, Ferrer-Batallé M, Barrabés S, Guerrero PE, Ramírez M, Saldova R, Rudd PM, Aleixandre RN, Comet J, de Llorens R, et al. Improvement of prostate cancer diagnosis by detecting PSA glycosylation-specific changes. Theranostics. 2016; 6(8):1190-204; PMID:27279911; http://dx.doi.org/10.7150/thno.15226
- Kumada T, Toyoda H, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, Kanamori A, Tanaka J, Kagebayashi C, Satomura S. High-sensitivity Lens culinaris agglutinin-reactive α-fetoprotein assay predicts early detection of hepatocellular carcinoma. J Gastroenterol 2014; 49(3):555-63; PMID:24057163; http://dx.doi.org/10.1007/s00535-013-0883-1
- Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu KL, Liang Y, et al. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci U S A 2014; 111(11):4338-43; PMID:24591635; http://dx.doi.org/10.1073/pnas.1321524111
- Li S, Payne S, Wang F, Claus P, Su Z, Groth J, Geradts J, de Ridder G, Alvarez R, Marcom PK, et al. Nuclear basic fibroblast growth factor regulates triple-negative breast cancer chemo-resistance. Breast Cancer Res 2015; 17(1):1-16; PMID:26141457; http://dx.doi.org/10.1186/s13058-014-0509-4
- Watson JA, Watson CJ, McCrohan AM, Woodfine K, Tosetto M, McDaid J, Gallagher E, Betts D, Baugh J, O'Sullivan J, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum Mol Genetics 2009; 18(19):3594-604; PMID:19584087; http://dx.doi.org/10.1093/hmg/ddp307
- Larkin A, Chang MM, Whitworth GE, Imperiali B. Biochemical evidence for an alternate pathway in N-linked glycoprotein biosynthesis. Nat Chem Biol 2013; 9(6):367-73; PMID:23624439; http://dx.doi.org/10.1038/nchembio.1249
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 2002; 298(5599):1790-3; PMID:12459590; http://dx.doi.org/10.1126/science.298.5599.1790
- Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol 2012; 8(5):434-6; PMID:22446837; http://dx.doi.org/10.1038/nchembio.921
- Bill RM, Revers L, Wilson I. Protein Glycosylation: Springer Science & Business Media; 2012
- Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol 2008; 8(11):874-87; PMID:18846099; http://dx.doi.org/10.1038/nri2417
- Hakomori SI. Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. In: George FVW, George K, editors. Advances in cancer research: Academic Press; 1989; 52:p. 257-331.
- Drake RR, Jones EE, Powers TW, Nyalwidhe JO. Chapter Ten - altered glycosylation in prostate cancer. In: Richard RD, Lauren EB, editors. Advances in Cancer Research: Academic Press; 2015; 126:p. 345-82.
- Saldova R, Reuben JM, Abd Hamid UM, Rudd PM, Cristofanilli M. Levels of specific serum N-glycans identify breast cancer patients with higher circulating tumor cell counts. Ann Oncol 2011; 22(5):1113-9; PMID:21127012; http://dx.doi.org/10.1093/annonc/mdq570
- Reticker-Flynn NE, Bhatia SN. Aberrant glycosylation promotes lung cancer metastasis through adhesion to galectins in the metastatic niche. Cancer Discov 2015; 5(2):168-81; PMID:25421439; http://dx.doi.org/10.1158/2159-8290.CD-13-0760
- Saldova R, Dempsey E, Perez-Garay M, Marino K, Watson JA, Blanco-Fernandez A, Struwe WB, Harvey DJ, Madden SF, Peracaula R, et al. 5-AZA-2′-deoxycytidine induced demethylation influences N-glycosylation of secreted glycoproteins in ovarian cancer. Epigenetics 2011; 6(11):1362-72; PMID:22086115; http://dx.doi.org/10.4161/epi.6.11.17977
- Ashkani J, Naidoo KJ. Glycosyltransferase gene expression profiles classify cancer types and propose prognostic subtypes. Scientific Reports 2016; 6:26451; PMID:27198045; http://dx.doi.org/10.1038/srep26451
- Potapenko IO, Luders T, Russnes HG, Helland A, Sorlie T, Kristensen VN, Nord S, Lingjærde OC, Børresen-Dale AL, Haakensen VD. Glycan-related gene expression signatures in breast cancer subtypes; relation to survival. Mol Oncol 2015; 9(4):861-76; PMID:25655580; http://dx.doi.org/10.1016/j.molonc.2014.12.013
- Potapenko IO, Haakensen VD, Luders T, Helland A, Bukholm I, Sorlie T, Kristensen VN, Lingjaerde OC, Børresen-Dale AL. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol Oncol 2010; 4(2):98-118; PMID:20060370; http://dx.doi.org/10.1016/j.molonc.2009.12.001
- Mehta A, Norton P, Liang H, Comunale MA, Wang M, Rodemich-Betesh L, Koszycki A, Noda K, Miyoshi E, Block T. Increased levels of tetra-antennary N-linked glycan but not core fucosylation are associated with hepatocellular carcinoma tissue. Cancer Epidemiol Biomarkers Prev 2012; 21(6):925-33; PMID:22490318; http://dx.doi.org/10.1158/1055-9965.EPI-11-1183
- Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res 2012; 11(4):2282-300; PMID:22304416; http://dx.doi.org/10.1021/pr201070k
- Huang WR, Fan XX, Tang X. SiRNA targeting EGFR effectively prevents posterior capsular opacification after cataract surgery. Mol Vision 2011; 17:2349-55; PMID:21921987
- Nangia-Makker P, Balan V, Raz A. Galectin-3-binding and metastasis. Methods Mol Biol 2012; 878:251-66; PMID:22674139; http://dx.doi.org/10.1007/978-1-61779-854-2_17
- Lin TW, Chang HT, Chen CH, Chen CH, Lin SW, Hsu TL, Wong CH. Galectin-3 binding protein and Galectin-1 interaction in breast cancer cell aggregation and metastasis. J Am Chem Soc 2015; 137(30):9685-93; PMID:26168351; http://dx.doi.org/10.1021/jacs.5b04744
- Hossein G, Keshavarz M, Ahmadi S, Naderi N. Synergistic effects of PectaSol-C modified citrus pectin an inhibitor of Galectin-3 and paclitaxel on apoptosis of human SKOV-3 ovarian cancer cells. Asian Pac J Cancer Prev 2013; 14(12):7561-8; PMID:24460334; http://dx.doi.org/10.7314/APJCP.2013.14.12.7561
- Miwa HE, Song Y, Alvarez R, Cummings RD, Stanley P. The bisecting GlcNAc in cell growth control and tumor progression. Glycoconj J 2012; 29(8-9):609-18; PMID:22476631; http://dx.doi.org/10.1007/s10719-012-9373-6
- Pinho SS, Figueiredo J, Cabral J, Carvalho S, Dourado J, Magalhaes A, Gärtner F, Mendonfa AM, Isaji T, Gu J, et al. E-cadherin and adherens-junctions stability in gastric carcinoma: functional implications of glycosyltransferases involving N-glycan branching biosynthesis, N-acetylglucosaminyltransferases III and V. Biochim Biophys Acta 2013; 3(700):2690-700; PMID:23671930; http://dx.doi.org/10.1016/j.bbagen.2012.10.021
- Varki A, Schauer R. Sialic acids. In Varki A, Cummings RD, Esko JD, et al., editors. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 14.
- Higai K, Suzuki C, Imaizumi Y, Xin X, Azuma Y, Matsumoto K. Binding affinities of NKG2D and CD94 to sialyl Lewis X-expressing N-glycans and heparin. Biol Pharmaceutical Bulletin 2011; 34(1):8-12; PMID:21212510; http://dx.doi.org/10.1248/bpb.34.8
- Büll C, Stoel MA, den Brok MH, Adema GJ. Sialic acids sweeten a tumor's life. Cancer Res 2014; 74(12):3199-204; PMID:24830719; http://dx.doi.org/10.1158/0008-5472.CAN-14-0728
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015; 15(9):540-55; PMID:26289314; http://dx.doi.org/10.1038/nrc3982
- Barthel SR, Wiese GK, Cho J, Opperman MJ, Hays DL, Siddiqui J, Pienta KJ, Furie B, Dimitroff CJ. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc Natl Acad Sci U S A 2009; 106(46):19491-6; PMID:19889975; http://dx.doi.org/10.1073/pnas.0906074106
- Saldova R, Wormald MR, Dwek RA, Rudd PM. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis Markers 2008; 25(4-5):219-32; PMID:19126966; http://dx.doi.org/10.1155/2008/601583
- Julien S, Ivetic A, Grigoriadis A, QiZe D, Burford B, Sproviero D, Picco G, Gillett C, Papp SL, Schaffer L, et al. Selectin ligand sialyl-Lewis x antigen drives metastasis of hormone-dependent breast cancers. Cancer Res 2011; 71(24):7683-93; PMID:22025563; http://dx.doi.org/10.1158/0008-5472.CAN-11-1139
- Burdick MM, McCaffery JM, Kim YS, Bochner BS, Konstantopoulos K. Colon carcinoma cell glycolipids, integrins, and other glycoproteins mediate adhesion to HUVECs under flow. Am J Physiol Cell Physiol 2003; 284(4):C977-87; PMID:12477667; http://dx.doi.org/10.1152/ajpcell.00423.2002
- Jandus C, Boligan KF, Chijioke O, Liu H, Dahlhaus M, Demoulins T, Christoph S, Marc W, Robert EH, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest 2014; 124(4):1810-20; PMID:24569453; http://dx.doi.org/10.1172/JCI65899
- Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology 2003; 13(7):41R-53R; PMID:12651883; http://dx.doi.org/10.1093/glycob/cwg054
- Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, et al. Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci USA 2005; 102(44):15791-6; PMID:16236725; http://dx.doi.org/10.1073/pnas.0507375102
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 2003; 278(5):3466-73; PMID:12427744; http://dx.doi.org/10.1074/jbc.M210665200
- Luhn K, Wild MK. Human deficiencies of fucosylation and sialylation affecting selectin ligands. Semin Immunopathol 2012; 34(3):383-99; PMID:22461019; http://dx.doi.org/10.1007/s00281-012-0304-1
- Barthel SR, Wiese GK, Cho J, Opperman MJ, Hays DL, Siddiqui J, Pienta KJ, Furie B, Dimitroff CJ. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc Natl Acad Sci U S A 2009; 106(46):19491-6; PMID:19889975; http://dx.doi.org/10.1073/pnas.0906074106
- Desiderio V, Papagerakis P, Tirino V, Zheng L, Matossian M, Prince ME, Paino F, Mele L, Papaccio F, Montella R, et al. Increased fucosylation has a pivotal role in invasive and metastatic properties of head and neck cancer stem cells. Oncotarget 2015; 6(1):71-84; PMID:25428916
- Martín-Satué M, Castellarnau CD, Blanco J. Overexpression of α(1,3)-fucosyltransferase VII is sufficient for the acquisition of lung colonization phenotype in human lung adenocarcinoma HAL-24Luc cells. Br J Cancer 1999; 80(8):1169-74; PMID:10376968; http://dx.doi.org/10.1038/sj.bjc.6690482
- Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem 2006; 281(5):2572-7; PMID:16316986; http://dx.doi.org/10.1074/jbc.M510893200
- Zhao Y, Itoh S, Wang X, Isaji T, Miyoshi E, Kariya Y, Miyazaki K, Kawasaki N, Taniguchi N, Gu J. Deletion of core fucosylation on alpha3beta1 integrin down-regulates its functions. J Biol Chem 2006; 281(50):38343-50; PMID:17043354; http://dx.doi.org/10.1074/jbc.M608764200
- Sato Y, Nakata K, Kato Y, Shima M, Ishii N, Koji T, Taketa K, Endo Y, Nagataki S. Early recognition of Hepatocellular carcinoma based on altered profiles of Alpha-Fetoprotein. N Eng J Med 1993; 328(25):1802-6; PMID:7684823; http://dx.doi.org/10.1056/NEJM199306243282502
- Kurebayashi J, Nomura T, Hirono M, Okubo S, Udagawa K, Shiiki S, Ikeda M, Nakashima K, Tanaka K, Sonoo H. Combined measurement of serum sialyl Lewis X with serum CA15-3 in breast cancer patients. Jpn J Clin Oncol 2006; 36(3):150-3; PMID:16520359; http://dx.doi.org/10.1093/jjco/hyi235
- Merry CR, Forrest ME, Sabers JN, Beard L, Gao X-H, Hatzoglou M, Jackson MW, Wang Z, Markowitz SD, Khalil AM. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genetics 2015; 24(21):6240-53; PMID:26307088; http://dx.doi.org/10.1093/hmg/ddv343
- Weake V. Histone ubiquitylation control of gene expression. In: Workman JL, Abmayr SM, editors. Fundamentals of Chromatin: Springer New York; 2014:p. 257-307.
- Johnson C, Warmoes MO, Shen X, Locasale JW. Epigenetics and cancer metabolism. Cancer Letters 2015; 356(2):309-14; PMID:24125862; http://dx.doi.org/10.1016/j.canlet.2013.09.043
- Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genetics Dev 2012; 22(1):50-5; PMID:22402447; http://dx.doi.org/10.1016/j.gde.2012.02.008
- Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM, Ausseil F, Vispé S, Arimondo PB. DNA methylation inhibitors in cancer: Recent and future approaches. Biochimie 2012; 94(11):2280-96; PMID:22967704; http://dx.doi.org/10.1016/j.biochi.2012.07.025
- ClinicalTrials 2015. NIH [accessed 2015 mar 9]. Available from: https://clinicaltrials.gov/ct2/results?term=decitabine+breast&Search=Search
- Ari F, Napieralski R, Ulukaya E, Dere E, Colling C, Honert K, Krüger A, Kiechle M, Schmitt M. Modulation of protein expression levels and DNA methylation status of breast cancer metastasis genes by anthracycline‐based chemotherapy and the demethylating agent decitabine. Cell Biochem Function 2011; 29(8):651-9; PMID:21887697; http://dx.doi.org/10.1002/cbf.1801
- Horvat T, Deželjin M, Redžić I, Barišić D, Herak Bosnar M, Lauc G, Zoldos V. Reversibility of membrane N-Glycome of HeLa cells upon treatment with epigenetic inhibitors. PLoS One 2013; 8(1):e54672; PMID:23336012; http://dx.doi.org/10.1371/journal.pone.0054672
- Lauc G, Krištić J, Zoldoš V. Glycans – the third revolution in evolution. Frontiers Genetics 2014; 5:145; PMID:24904645; http://dx.doi.org/10.3389/fgene.2014.00145
- Chakraborty AK, de Frietas Sousa J, Chakraborty D, Funasaka Y, Bhattacharya M, Chatterjee A, Pawelek J. GnT-V expression and metastatic phenotypes in macrophage–melanoma fusion hybrids is down-regulated by 5-Aza-dC: Evidence for methylation sensitive, extragenic regulation of GnT-V transcription. Gene 2006; 374(0):166-73; PMID:16556489; http://dx.doi.org/10.1016/j.gene.2006.01.031
- Chachadi VB, Cheng H, Klinkebiel D, Christman JK, Cheng PW. 5-Aza-2′-deoxycytidine increases sialyl Lewis X on MUC1 by stimulating β-galactoside:α2,3-sialyltransferase 6 gene. Int J Biochem Cell Biol 2011; 43(4):586-93; PMID:21168523; http://dx.doi.org/10.1016/j.biocel.2010.12.015
- Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med 2015; 4(5):745-58; PMID:25684390; http://dx.doi.org/10.1002/cam4.409
- Timsah Z, Ahmed Z, Ivan C, Berrout J, Gagea M, Zhou Y, Pena GN, Hu X, Vallien C, Kingsley CV, et al. Grb2 depletion under non-stimulated conditions inhibits PTEN, promotes Akt-induced tumor formation and contributes to poor prognosis in ovarian cancer. Oncogene 2016; 35(17):2186-96; PMID:26212011; http://dx.doi.org/10.1038/onc.2015.279
- Lesniak D, Sabri S, Xu Y, Graham K, Bhatnagar P, Suresh M, Abdulkarim B. Spontaneous epithelial-mesenchymal transition and resistance to HER-2-targeted therapies in HER-2-positive luminal breast cancer. 2013; 8(8):e71987; PMID:23991019, http://dx.doi.org/10.1371/journal.pone.0071987
- Huang C, Park CC, Hilsenbeck SG, Ward R, Rimawi MF, Wang YC, Shou J, Bissell MJ, Osborne CK, Schiff R. b1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast Cancer Res 2011; 13:R84; PMID:21884573; http://dx.doi.org/10.1186/bcr2936
- Schultz MJ, Swindall AF, Wright JW, Sztul ES, Landen CN, Bellis SL. ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J Ovarian Res 2013; 6(1):25; PMID:23578204; http://dx.doi.org/10.1186/1757-2215-6-25
- Le QT, Chen E, Salim A, Cao H, Kong CS, Whyte R, Donington J, Cannon W, Wakelee H, Tibshirani R, et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res 2006; 12(5):1507-14; PMID:16533775; http://dx.doi.org/10.1158/1078-0432.CCR-05-2049
- Meixensberger J, Dings J, Kuhnigk H, Roosen K. Studies of tissue PO2 in normal and pathological human brain cortex. Acta Neurochir Suppl 1993; 59:58-63; PMID:7906079
- Hancock RL, Dunne K, Walport LJ, Flashman E, Kawamura A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics 2015; 7(5):791-811; PMID:25832587; http://dx.doi.org/10.2217/epi.51.24
- Kieda C, El Hafny-Rahbi B, Collet G, Lamerant-Fayel N, Grillon C, Guichard A, Dulak J, Jozkowicz A, Kotlinowski J, Fylaktakidou KC, et al. Stable tumor vessel normalization with pO2 increase and endothelial PTEN activation by inositol trispyrophosphate brings novel tumor treatment. J Mol Med 2013; 91(7):883-99; PMID:23471434; http://dx.doi.org/10.1007/s00109-013-0992-6
- Comerford KM, Wallace TJ, Karhausen J, Louis NA, Montalto MC, Colgan SP. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res 2002; 62(12):3387-94; PMID:12067980
- Collet G, Lamerant-Fayel N, Tertil M, El Hafny-Rahbi B, Stepniewski J, Guichard A, Foucault-Collet A, Klimkiewicz K, Petoud S, Matejuk A, et al. Hypoxia-regulated overexpression of soluble VEGFR2 controls angiogenesis and inhibits tumor growth. Mol Cancer Ther 2014; 13(1):165-78; PMID:24170768; http://dx.doi.org/10.1158/1535-7163.MCT-13-0637
- Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics: The epicenter of the hypoxic response. Epigenetics 2010; 5(4):293-6; PMID:20418669; http://dx.doi.org/10.4161/epi.5.4.11684
- Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics 2007; 2(2):119-25; PMID:17965619; http://dx.doi.org/10.4161/epi.2.2.4613
- Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a Berberine-Induced miRNA, Directly Down-Regulates Human Methionine Adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One 2013; 8(9):e75628; PMID:24098708; http://dx.doi.org/10.1371/journal.pone.0075628
- Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology 2007; 45(5):1306-12; PMID:17464973; http://dx.doi.org/10.1002/hep.21650
- Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther 2011; 10(6):1113-23; PMID:21460102; http://dx.doi.org/10.1158/1535-7163.MCT-10-1010
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol 2007; 27(5):1859-67; PMID:17194750; http://dx.doi.org/10.1128/MCB.01395-06
- Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C, et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology 2011; 54(6):2064-75; PMID:22144109; http://dx.doi.org/10.1002/hep.24614
- Sauermann M, Sahin O, Sultmann H, Hahne F, Blaszkiewicz S, Majety M, Zatloukal K, Füzesi L, Poustka A, Wiemann S, et al. Reduced expression of vacuole membrane protein 1 affects the invasion capacity of tumor cells. Oncogene 2008; 27(9):1320-6; PMID:17724469; http://dx.doi.org/10.1038/sj.onc.1210743
- Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest 2010; 120(11):4141-54; PMID:20972335; http://dx.doi.org/10.1172/JCI42980
- Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, Jiang Y, Kung HF, Lai L, Jiang BH. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One 2011; 6(4):0019139; PMID:21544242; http://dx.doi.org/10.1371/journal.pone.0019139
- Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem 2011; 286(3):2047-56; PMID:21081489; http://dx.doi.org/10.1074/jbc.M110.158790
- Koike T, Kimura N, Miyazaki K, Yabuta T, Kumamoto K, Takenoshita S, Chen J, Kobayashi M, Hosokawa M, Taniguchi A, et al. Hypoxia induces adhesion molecules on cancer cells: A missing link between Warburg effect and induction of selectin-ligand carbohydrates. Proc Natl Acad Sci U S A 2004; 101(21):8132-7; PMID:15141079; http://dx.doi.org/10.1073/pnas.0402088101
- Kannagi R, Sakuma K, Miyazaki K, Lim KT, Yusa A, Yin J, Izawa M. Altered expression of glycan genes in cancers induced by epigenetic silencing and tumor hypoxia: Clues in the ongoing search for new tumor markers. Cancer Sci 2010; 101(3):586-93; PMID:20085584; http://dx.doi.org/10.1111/j.1349-7006.2009.01455.x
- Yin J, Hashimoto A, Izawa M, Miyazaki K, Chen GY, Takematsu H, Kozutsumi Y, Suzuki A, Furuhata K, Cheng FL, et al. Hypoxic culture induces expression of sialin, a sialic acid transporter, and cancer-associated gangliosides containing non-human sialic acid on human cancer cells. Cancer Res 2006; 66(6):2937-45; PMID:16540641; http://dx.doi.org/10.1158/0008-5472.CAN-05-2615
- Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem 2002; 277(26):23111-5; PMID:11943784; http://dx.doi.org/10.1074/jbc.M202487200
- Gornik O, Wagner J, Pucic M, Knezevic A, Redzic I, Lauc G. Stability of N-glycan profiles in human plasma. Glycobiology 2009; 19(12):1547-53; PMID:19726492; http://dx.doi.org/10.1093/glycob/cwp134
- Lauc G, Krištić J, Zoldoš V. Glycans–the third revolution in evolution. Frontiers Genetics 2014; 5:145; PMID:24904645; http://dx.doi.org/10.3389/fgene.2014.00145
- Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer 2013; 13(12):858-70; PMID:24263189; http://dx.doi.org/10.1038/nrc3628
- Zoldoš V, Horvat T, Lauc G. Glycomics meets genomics, epigenomics and other high throughput omics for system biology studies. Curr Opin Chem Biol 2013; 17(1):34-40; PMID:23287290; http://dx.doi.org/10.1016/j.cbpa.2012.12.007
- Lauc G, Huffman JE, Pučić M, Zgaga L, Adamczyk B, Mužinić A, Novokmet M, Polašek O, Gornik O, Krištić J, et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. 2013; 9(1):e1003225; PMID:23382691; http://dx.doi.org/10.1371/journal.pgen.1003225
- Haakensen VD, Steinfeld I, Saldova R, Shehni AA, Kifer I, Naume B, Rudd PM, Børresen-Dale AL, Yakhini Z. Serum N-glycan analysis in breast cancer patients – Relation to tumour biology and clinical outcome. Mol Oncol 2016; 10(1):59-72; PMID:26321095; http://dx.doi.org/10.1016/j.molonc.2015.08.002.
- ClinicalTrials.gov Ovarian 2015. NIH [accessed 2015 Sep 26] Available from: https://clinicaltrials.gov/ct2/results?term=decitabine+ovarian&Search=Search
- Breast cancer clinicals Trials with Decitabine 2015. NIH. [accessed 2015 Sep 26] Available from: https://clinicaltrials.gov/ct2/results?term=decitabine+breast&Search=Search
- Vasconcelos-Dos-Santos A, Oliveira IA, Lucena MC, Mantuano NR, Whelan SA, Dias WB, Todeschini AR. Biosynthetic machinery involved in aberrant glycosylation: Promising targets for developing of drugs against cancer. Front Oncol 2015; 5:138; PMID:26161361; http://dx.doi.org/10.3389/fonc.2015.00138
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. A Functional genetic approach identifies the PI3K pathway as a major determinant of Trastuzumab resistance in breast cancer. Cancer Cell 2007; 12(4):395-402; PMID:17936563; http://dx.doi.org/10.1016/j.ccr.2007.08.030
- Abdullah LN, Chow EK-H. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med 2013; 2:3; PMID:23369605; http://dx.doi.org/10.1186/2001-1326-2-3
- Ween MP, Armstrong MA, Oehler MK, Ricciardelli C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Critical Rev Oncology/Hematol 2015; 96(2):220-56; PMID:26100653; http://dx.doi.org/10.1016/j.critrevonc.2015.05.012
- Borley J, Brown R. Epigenetic mechanisms and therapeutic targets of chemotherapy resistance in epithelial ovarian cancer. Ann Med 2015; 47(5):359-69; PMID:26158617; http://dx.doi.org/10.3109/07853890.2015.1043140
- Ren J, Chen Y, Song H, Chen L, Wang R. Inhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell line. J Cell Biochem 2013; 114(6):1395-403; PMID:23255418; http://dx.doi.org/10.1002/jcb.24481
- Houghton JA, Houghton PJ. Elucidation of pathways of 5-fluorouracil metabolism in xenografts of human colorectal adenocarcinoma. Eur J Cancer Clin Oncol 1983; 19(6):807-15; PMID:6191988; http://dx.doi.org/10.1016/0277-5379(83)90013-5
