ABSTRACT
The presence of H3K9me3 and heterochromatin protein 1 (HP1) are hallmarks of heterochromatin conserved in eukaryotes. The spreading and maintenance of H3K9me3 is effected by the functional interplay between the H3K9me3-specific histone methyltransferase Suv39h1 and HP1. This interplay is complex in mammals because the three HP1 isoforms, HP1α, β, and γ, are thought to play a redundant role in Suv39h1-dependent deposition of H3K9me3 in pericentric heterochromatin (PCH). Here, we demonstrate that despite this redundancy, HP1α and, to a lesser extent, HP1γ have a closer functional link to Suv39h1, compared to HP1β. HP1α and γ preferentially interact in vivo with Suv39h1, regulate its dynamics in heterochromatin, and increase Suv39h1 protein stability through an inhibition of MDM2-dependent Suv39h1-K87 polyubiquitination. The reverse is also observed, where Suv39h1 increases HP1α stability compared HP1β and γ. The interplay between Suv39h1 and HP1 isoforms appears to be relevant under genotoxic stress. Specifically, loss of HP1α and γ isoforms inhibits the upregulation of Suv39h1 and H3K9me3 that is observed under stress conditions. Reciprocally, Suv39h1 deficiency abrogates stress-dependent upregulation of HP1α and γ, and enhances HP1β levels. Our work defines a specific role for HP1 isoforms in regulating Suv39h1 function under stress via a feedback mechanism that likely regulates heterochromatin formation.
Introduction
The compaction of DNA inside the nucleus requires successive levels of chromatin organization that begins with the fundamental unit of chromatin packaging, the nucleosome, and ends in the highest degree of compaction found in the metaphase chromosome. The transitions between compaction states of chromatin are regulated by a number of mechanisms that include posttranslational modifications of histones, DNA methylation, non-histone structural proteins, transcription factors, and chromatin remodeling activities. Of these factors, the epigenetic histone mark trimethylation of histone H3 in lysine 9 (H3K9me3) and heterochromatin protein 1 (HP1) are highly conserved hallmarks of heterochromatin.Citation1 The primary role of H3K9me3 is to serve as a docking site of specific factors, allowing a sequence of molecular events, such as the establishment of another heterochromatin hallmark, H4K20me3, or methylation of DNA.Citation2-5 Suv39h1 was the first lysine-specific HMT activity described and is required, along with Suv39h2, for the formation and maintenance of pericentric heterochromatin (PCH).Citation6-8 Simultaneous deletion of Suv39h1/2 induces genome instability, chromosomal aberrations, decreased proliferation, cell cycle alteration, meiosis defects, and increased tumorigenesis.Citation8,9 Suv39h1 also regulates heterochromatin formation under several specific conditions including oxidative and metabolic stress.Citation10-12 Under these conditions, the stress-related NAD+-dependent deacetylase SirT1 promotes Suv39h1 stability through inhibition of Suv39h1-K87 polyubiquitination catalyzed by the E3-ubiquitin ligase MDM211. Suv39h1 and H3K9me3 have also been involved in the transient formation of heterochromatin at sites of DNA double-strand breaks.Citation13
Mammalian HP1α, HP1β, and HP1γ are small proteins of around 25 kDa that have an N-terminal chromodomain (CD) and a sequence-related C-terminal chromo shadow domain (CSD), linked by an unstructured hinge region (HR). The CD binds di-/tri-methylated form of lysine 9 on histone H3 (H3K9me2/3).Citation14 HP1 dimerization through the CSD allows binding to a penta-peptide motif, PxVxL, found in many HP1-interacting proteins.Citation15,16 HP1 proteins recognize H3K9me2/3 through their CD and interact with Suv39h1, thereby enabling spreading of H3K9me3 along a heterochromatin domain.Citation17 The interaction of Suv39h1 and HP1 proteins has largely focused on the Suv39h1-HP1β interaction. HP1β was originally found bound to the N-terminal 44 amino-acids of Suv39h1 in nuclear extracts,Citation18 and FRET analysis shows that this interaction takes place in vivo.Citation19 The Suv39h1-HP1β complex also co-sediments in a ∼20S chromatin fraction.Citation6 HP1 proteins interact with a wide range of factors from enzymes to structural proteins, allowing the direct or indirect recruitment of DNA methyltransferases, the H4K20me3 HMT Suv420h2, chromatin chaperons, chromatin remodeling factors and cohesins, among others.Citation4,5,20,21
HP1α, β, and γ exhibit a high degree of sequence conservation and are thought to be involved in heterochromatin regulation, gene expression, DNA damage signaling, DNA repair, differentiation and cell cycle regulation.Citation22-25 Several lines of evidence have shown that the mammalian HP1 proteins have isotype-specific functions. First, all three isoforms are present in PCH, but HP1β and HP1γ are also enriched in euchromatin.Citation26,27 The euchromatic localization of HP1γ has been associated with a role in gene activation.Citation28,29 Second, there are isotype-specific interactions with binding partners, as well as isotype-specific posttranslational modifications, including sumoylation, acetylation, and phosphorylation.Citation22,24,30-33 Third, all three isoforms have been implicated to varying extents in stress responses. For example, HP1α depletion impairs the binding of 53BP1 and RAD51 to double-strand breaks, thereby inhibiting DNA repair by homologous recombination.Citation34 Phosphorylation of HP1β at residue threonine 51 increases the mobility of the protein at damaged sites induced by IR.Citation35 However, these studies were performed with a specific isoform and it remains possible that all three isoforms are redundant with respect to stress responses. Finally, isotype-specific deletions in gene-targeted mice reveal unique phenotypes.Citation36-38
Here, we show that each HP1 isoform plays a specific role in regulating Suv39h1 function. Despite a strong redundancy between isoforms, under normal conditions, HP1α and γ preferentially interact with Suv39h1 and regulate its dynamics in heterochromatin in contrast to HP1β. Moreover, HP1α and, to a lesser extent, γ regulate Suv39h1 protein stability by inhibition of MDM2-dependent ubiquitination of K87 in Suv39h1. There is also reciprocity, in that Suv39h1 activity increases HP1α protein levels while decreasing both HP1β and γ, indicating the operation of a feedback regulatory network that links Suv39h1 with the HP1 isoforms. We suggest that this feedback mechanism is functionally relevant in situations of genotoxic stress that results in a specific interaction of HP1α and γ with the SirT1 protein. Importantly, loss of HP1α and HP1γ proteins abrogates stress-dependent upregulation of Suv39h1 and H3K9me3 that is likely mediated by SirT1. These data demonstrate a role for HP1 isoforms in Suv39h1 function and suggests a regulatory feedback mechanism between these factors that protect genome stability under conditions of stress.
Results
HP1α and γ interact preferentially with Suv39h1 within the PCH foci
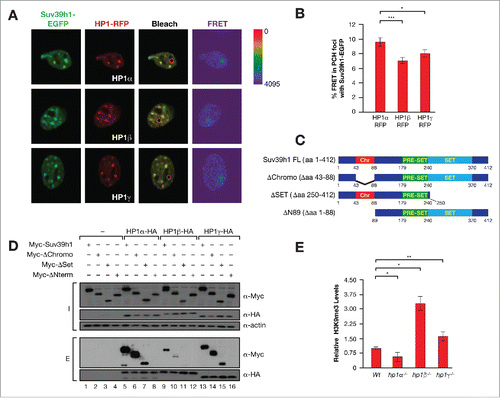
Aiming to understand the role of each HP1 isoforms in Suv39h1 function, we determined their interaction with Suv39h1 in PCH foci in vivo by fluorescence resonance energy transfer (FRET) analysis. We found that Suv39h1 bound preferentially to HP1α and, to a lesser extent, HP1γ, and less still to HP1β (). Analysis of the binding of the isoforms to Suv39h1 deletion mutants () showed that all three isoforms interacted with the N-terminal domain of Suv39h1 (). Immunoprecipitation experiments indicated that the differences in interactions of Suv39h1 and HP1 isoforms, as measured by FRET, could be directly related to differences in binding efficiency (). The preferential binding of Suv39h1 to HP1α was also indicated by the ability of HP1α to compete binding of Suv39h1 to HP1β (Fig. S1A). HP1α−deficient MEFs also contained significantly lower levels of H3K9me3 in contrast to HP1β− or HP1γ−knockout (KO) MEFs; the latter showed a mild significant increase in H3K9me3 levels compared to wild-type (WT) MEFs ().
Figure 1. (A) Representative images of FRET experiments between Suv39h1-EGFP and HP1α,β,γ-RFP in PCH foci of mouse NIH3T3 cells. Bleached foci are indicated by a red circle. (B) Quantification of the FRET assays also shown in (A). *P < 0.05; ***P < 0.0005. (C) Schematic diagram of Suv39h1 domains and constructs used in (D). (D) HA immunoprecipitation between the indicated Myc-tagged Suv39h1 constructs described in (C) and HA-tagged HP1 isoforms. Inputs (I) and Elutions (E) are shown. (E) Levels of H3K9me3 global levels in WT, HP1α, β, and γ–deficient MEFs. The levels of H3K9me3 represented were quantified from western-blots as in and normalized by histone H3 levels. *P < 0.05; **P < 0.0005.
HP1α, β, and γ have different roles in the regulation of Suv39h1 protein levels
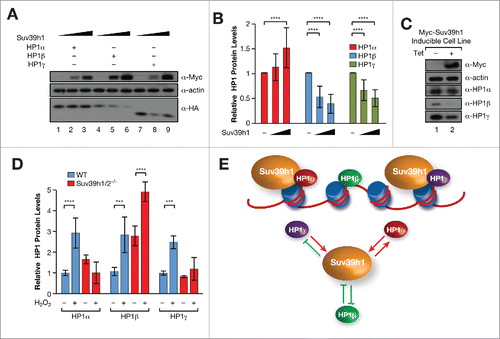
Each isoform had a different impact on Suv39h1 protein levels. Overexpression of HP1α and, to a lesser extent, γ increased Suv39h1 protein levels, while HP1β did not alter Suv39h1 levels significantly (). In agreement with these results, levels of endogenous Suv39h1 were decreased to around 50% of WT levels in HP1α and γ–deficient MEFs (). In contrast, loss of HP1β had the opposite effect, where Suv39h1 protein levels were increased almost two-fold in HP1β KO compared to WT MEFs (). HP1-mediated regulation of Suv39h1 occurred at the protein level as Suv39h1 mRNA levels in KO MEFs did not correlate with changes in protein levels (Fig. S1B). The reduced Suv39h1 protein stability in HP1β KO MEFs was intriguing given that HP1β appears to shows that weakest interaction with Suv39h1 (). HP1β effect on Suv39h1 may be indirect because loss of HP1β increases the levels of HP1γ, which in turn increases Suv39h1 protein stability ( and S1C). However, the fact that we observed increased levels of HP1β in hp1γ−/− MEFs, may indicate a direct role of HP1β in Suv39h1 protein instability ().
Figure 2. (A) Western-blot of Myc-Suv39h1 levels in presence or absence of FLAG-tagged HP1 isoforms in 293F cells. Right, quantification of n = 3 experiments. *P < 0.05; **P < 0.005. (B) Quantification of endogenous Suv39h1 protein levels in WT, HP1α−, β−, and γ–deficient MEFs from western-blots as in . The relative levels were normalized compared to Suv39h1 levels in WT cells. *P < 0.05. (C) Levels of all three HP1 isoforms in the same cells as in (B). (D) Schematic diagram of the Suv39h1 chromodomain (Chr) or N-terminal domain (Nt) GFP fusions used in (E). (E) Levels of the indicated GFP constructs in presence or absence of HP1α overexpression in 293F cells. Quantification of n = 3 experiments as in (E). The relative levels of GFP fussions were normalized compared to GFP levels. ****P < 0.0001. (F) Schematic diagram of the HP1α, β domains and the constructs used including the WT isoforms, HP1α containing the hinge region (HR) of HP1β [HP1α(β)] and HP1β containing the hinge region (HR) of HP1α [HP1β(α)]. Suv39h1 protein levels upon overexpression of the indicated HP1α, β constructs shown in (F). Levels are represented relative to empty vector overexpression (−).*P < 0.05; **P > 0.005; ***P < 0.0005.
![Figure 2. (A) Western-blot of Myc-Suv39h1 levels in presence or absence of FLAG-tagged HP1 isoforms in 293F cells. Right, quantification of n = 3 experiments. *P < 0.05; **P < 0.005. (B) Quantification of endogenous Suv39h1 protein levels in WT, HP1α−, β−, and γ–deficient MEFs from western-blots as in Fig. 5D. The relative levels were normalized compared to Suv39h1 levels in WT cells. *P < 0.05. (C) Levels of all three HP1 isoforms in the same cells as in (B). (D) Schematic diagram of the Suv39h1 chromodomain (Chr) or N-terminal domain (Nt) GFP fusions used in (E). (E) Levels of the indicated GFP constructs in presence or absence of HP1α overexpression in 293F cells. Quantification of n = 3 experiments as in (E). The relative levels of GFP fussions were normalized compared to GFP levels. ****P < 0.0001. (F) Schematic diagram of the HP1α, β domains and the constructs used including the WT isoforms, HP1α containing the hinge region (HR) of HP1β [HP1α(β)] and HP1β containing the hinge region (HR) of HP1α [HP1β(α)]. Suv39h1 protein levels upon overexpression of the indicated HP1α, β constructs shown in (F). Levels are represented relative to empty vector overexpression (−).*P < 0.05; **P > 0.005; ***P < 0.0005.](/cms/asset/b5eaa2e1-ad53-476a-8378-5c9df1e07d07/kepi_a_1278096_f0002_oc.gif)
HP1α directly regulates Suv39h1 protein levels through a mechanism that requires the N-terminal domain of Suv39h1 and involves the hinge region of HP1α
Our previous studies demonstrated that the N-terminal Suv39h1 CD, at residues 44–88 of Suv39h1 is required for protein stability [11]. We next investigated whether the HP1 interaction domain that encompasses residues 1–44 of Suv39h1 is required for the CD-dependent regulation of Suv39h1 protein stability. To that end, we fused the N-terminal region of Suv39h1 containing both domains (NT; residues 1–118) to GFP and tested the effect of HP1α expression on the protein levels of the GFP fusion in 239F cells (). We observed a seven-fold increase NT-GFP protein levels in cells expressing HP1α. HP1α expression did not upregulate GFP fused to the CD (GFP-CHR) or the first 44 N-terminal residues to Suv39h1 (GFP-N44) ( and S2A–B). Considering that the first 44 residues of Suv49h1 are required for interaction with HP1α, these results indicated that the mechanism by which HP1α increases Suv39h1 protein stability is direct and also requires the Suv39h1 CD. Given the close sequence identity between all three isoforms, the different effects of HP1 isoforms on Suv39h1 activity was intriguing. Particularly interesting was the case of the two closest HP1 isoforms HP1α and HP1β, which showed the most divergent effects on Suv39h1 protein levels (). Since the main differences between these isoforms are localized in the hinge region (HR) of the protein (), we next investigated whether the HR was responsible for the differences between them. Accordingly, we swapped the HR between HP1α and HP1β and tested the effect of these mutants in Suv39h1 stability. We found that both mutants HP1α(β-HR) and HP1β(α-HR) increased Suv39h1 protein stability to levels midway between those found for each isoform (). These observations indicate that the HP1 HR may participate in the isoform-specific stability of Suv39h1.
HP1α and HP1γ increase Suv39h1 stability by inhibiting polyubiquitination of K87 in Suv39h1 chromodomain
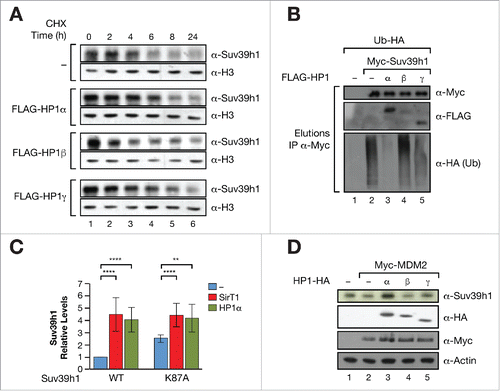
In order to further explore the effect of HP1 proteins on Suv39h1 protein stability we undertook cycloheximide (CHX) chase experiments that measured the effect of each HP1 isoform on the half-life of endogenous Suv39h1. We found that both HP1α and γ increased Suv39h1 protein half-life, while HP1β did not (). As explained, we have previously shown that the stability of Suv39h1 is regulated by polyubiquitination. To test the hypothesis that HP1 regulation of Suv39h1 stability is mediated by polyubiquitination we immunoprecipitated Suv39h1 from cells expressing each HP1 isoform together with HA-tagged ubiquitin, and studied the rate of HA incorporation in Suv39h1. We found that HP1α, and to a lesser extent HP1γ, induced a clear decrease in Suv39h1 polyubiquitination while HP1β did not have any effect (). Given we had previously identified the lysine K87 in Suv39h1 CD as the main polyubiquitination site of Suv39h1,Citation11 we were next prompted to investigate whether the K87 residue was involved in the mechanism by which HP1α regulates Suv39h1 activity. As observed for SirT1, HP1α-induced upregulation of Suv39h1 was significantly decreased in K87 mutant (). Consistently, HP1α overexpression was able to protect against the Suv39h1 degradation induced by MDM2, indicating that both SirT1 and HP1 regulate Suv39h1 stability through the same mechanism (). HP1γ and HP1β, either had a very mild protective effect (HP1γ) or had no effect (HP1β) on MDM2-induced downregulation of Suv39h1, underscoring the higher binding affinity of HP1α for Suv39h1 compared to other isoforms.
Figure 3. (A) Cycloheximide (CHX) chase experiment to determine endogenous Suv39h1 protein stability in absence or presence of empty vector (−) or FLAG-tagged HP1α, β or γ after 0, 2, 5, 10, and 24 h of CHX treatment. (B) Ubiquitination of Suv39h1 in presence or absence of Suv39h1 isoforms. Myc-Suv39h1 was purified from 293F cells expressing Ubiquitin-HA and the indicated HP1 constructs. After immunoprecipitation, equal levels of immunopurified Suv39h1 were loaded in a gel and the levels of ubiquitination (HA-tag incorporation) were tested. The inputs of the initial immunoprecipitation experiment are shown in Fig. S2D. (C) Levels of Suv39h1 in presence or absence of SirT1 or HP1α. The relative levels were normalized compared to Suv39h1 levels in WT cells upon overexpression of empty vector (−). **P < 0.005; ****P < 0.0001. (D) Effect of HP1α, β, and γ in MDM2-dependent degradation of Suv39h1. Western blot of the levels of Suv39h1 in H1299 cells transfected with MDM2−/+ HP1 indicated isoforms.
HP1α and HP1γ promote Suv39h1 stability in PCH foci
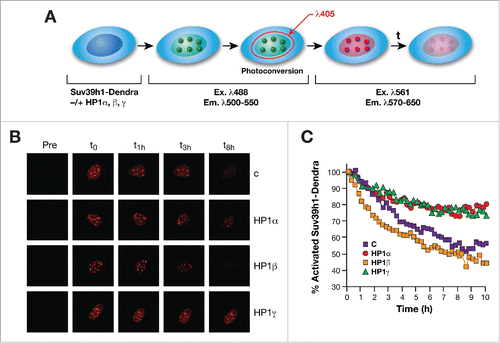
To further investigate the role played by HP1 isoforms in regulating Suv39h1 protein stability we performed in vivo “pulse-chase” experiments using the Dendra fluorophore (see Materials and Methods for details). Dendra fluorophore can be activated by irradiation at 405 nm wavelength. Activation results in photoconversion that changes the fluorophore's properties so that it is now excited and emits at different wavelengths. This enables specific detection of the activated Dendra. In our experiment, we expressed Dendra-fused Suv39h1 in WT MEFs together with the HP1 isoforms, induced photo-conversion, and followed the levels of Suv39h1 in PCH foci (). We found that upregulation of HP1α or γ increased significantly the stability of Suv39h1 in vivo in PCH foci. In contrast, HP1β did not affect Suv39h1 levels in PCH foci ().
Figure 4. (A) Schematic design of Dendra2-Suv39H1 photoconversion experiments. NIH3T3 cells co-expressing the indicated HP1 isoforms or empty vector (C) together with Dendra2-Suv39H1 were photoconverted using 405 nm laser diode and imaged every 15 min during 10 h. (B) Time-lapse series of confocal images of photoconverted Dendra2-Suv39H1 red fluorescence in foci of indicated cells. A 488 nm laser and 561 nm laser were used to excite non-photoconverted Dendra2-Suv39h1 (green) (data not shown) and photoconverted Dendra2-Suv39H1(red). (C) Normalized mean fluorescence values overtime obtained from individual photoconverted foci of the induced cells using ImageJ/fiji open source analysis software.
HP1α and HP1γ regulate Suv39h1 dynamics under normal and stress conditions
We next investigated whether HP1 isotypes affect the dynamics of Suv39h1 in PCH under normal conditions or genotoxic stress. We performed FRAP experiments to measure the dynamics of Suv39h1 in PCH foci of MEFs derived from mice WT or deficient for HP1α, HP1β, or HP1γ and found that loss of HP1α and HP1γ decreased both the turnover rate [t1/2] and the mobile fraction of Suv39h1-EGFP ( and ) confirming a direct link between HP1α and HP1γ with Suv39h1. Overexpression of HP1α and HP1γ had the opposite effect and increased both turnover of Suv39h1 and the mobile fraction in the PCH foci (). Neither HP1β loss nor overexpression had any significant effect on Suv39h1 turnover in the PCH foci (). However, overexpression of HP1β did increase Suv39h1 mobile fraction ( and ), indicating that HP1β may indirectly affect Suv39h1 dynamics in PCH foci. We next addressed the effect of genotoxic stress. FRAP analysis of HP1 KO MEFs under oxidative stress conditions showed that the turnover rate of Suv39h1 in PCH foci increased by approximately 20% in hp1α−/− or hp1γ−/− and WT MEFs compared to the same genotypes under normal conditions; Suv39h1 turnover did not change in stressed hp1β−/− cells. In contrast to WT cells, the mobile fraction of Suv39h1 did not change in any of the KO MEFs upon oxidative stress. Several other lines of evidence support a direct role of HP1α and γ in Suv39h1 dynamics under stress. First, in agreement with a role of both isoforms in Suv39h1 and H3K9me3 upregulation under stress, loss of HP1α, and, to a significant lesser extent gamma, abrogated the global increase of both Suv39h1 and H3K9me3 observed upon different types of genotoxic stress, such as IR or oxidative stress ( and data not shown). Interestingly, and in agreement with our previous results (), Suv39h1 and H3K9me3 levels that were not significantly increased by stress conditions in HP1β-deficient cells compared to WT cells. Second, oxidative stress also induced specific binding of SirT1 to HP1α, but not to HP1β (). HP1γ also behaved like HP1α, although the interaction was significantly weaker (). These data indicate that HP1α and SirT1 may cooperate to increase Suv39h1 protein stability under oxidative stress raising the possibility of a role for HP1α in SirT1-dependent facultative heterochromatin formation.
Figure 5. (A) FRAP assays of Suv39h1-EGFP in PCH foci of NIH3T3 upon expression of empty vector, or the indicated HP1 isoforms. (B) Similar experiment as in (A) performed in WT, HP1α, β, and γ–deficient MEFs under normal (left) or oxidative stress conditions (right). (C) Quantification of Suv39h1-EGFP half time [t1/2] and mobile fraction in the FRAPs shown in (A) and (B). (D) Levels of Suv39h1, H3K9me3, and histone H3 in WT and KO HP1 MEFs upon normal and stress conditions (IR). (E) Interaction between SirT1 and the HP1 isoforms under untreated or oxidative stress conditions. FLAG immunoprecipitation of extracts from 293F cells expressing FLAG-SirT1 −/+ HA-tagged HP1 isoforms in the indicated conditions.
![Figure 5. (A) FRAP assays of Suv39h1-EGFP in PCH foci of NIH3T3 upon expression of empty vector, or the indicated HP1 isoforms. (B) Similar experiment as in (A) performed in WT, HP1α, β, and γ–deficient MEFs under normal (left) or oxidative stress conditions (right). (C) Quantification of Suv39h1-EGFP half time [t1/2] and mobile fraction in the FRAPs shown in (A) and (B). (D) Levels of Suv39h1, H3K9me3, and histone H3 in WT and KO HP1 MEFs upon normal and stress conditions (IR). (E) Interaction between SirT1 and the HP1 isoforms under untreated or oxidative stress conditions. FLAG immunoprecipitation of extracts from 293F cells expressing FLAG-SirT1 −/+ HA-tagged HP1 isoforms in the indicated conditions.](/cms/asset/94b7924d-6e09-4368-8fff-01cf0286b65d/kepi_a_1278096_f0005_oc.gif)
Feedback regulation of HP1 isoforms by Suv39h1
Having shown that Suv39h1 protein stability is regulated by HP1 isoforms (), we tested whether the reverse was also true, that is, whether Suv39h1 can regulate the stability of HP1 isoforms. We observed that Suv39h1 overexpression resulted in higher levels of HA-tagged HP1α, while the levels of both HA-HP1β and HA-HP1γ decreased ( and ). These results indicate the existence of isotype-specific feedback regulation of HP1 proteins by Suv39h1. To confirm that this regulatory feedback mechanism also operates on endogenous HP1 proteins, we setup a Myc-Suv39h1 tetracycline-inducible (Tet-ON) cell line in 293F cells. Surprisingly, endogenous HP1α levels were not altered by Tet-induction of Myc-Suv39h1 (), although we observed a decrease in endogenous HP1β and HP1γ protein levels () consistent with our results obtained using HA-tagged-HP1β and γ ( and ). We also tested the effect of Suv39h1/2 depletion on HP1 levels. We found that Suv39h1/2 depletion did not affect HP1α protein levels (Fig. S3A), which was also the case for HP1γ, although HP1β protein levels were elevated (Fig. S3A). We were puzzled by the apparent discrepancies we obtained, but were mindful of our observation that it is under conditions of cellular stress that Suv39h1 is upregulated ( to ). We hypothesized that it is in the context of cellular stress that we should test the role of Suv39h1 upregulation on the stability of HP1 isoforms. To that end, we measured the levels of endogenous HP1α, β, and γ under normal conditions and oxidative stress. We found that under conditions of oxidative stress Suv39h1/2 depleted cells do not have elevated levels of HP1α and γ ( and S3A) by contrast to WT cells treated in the same way (). Strikingly, the levels of HP1β were further increased by oxidative stress in Suv39h1/2−/− MEFs. These results strongly indicate a direct and isoform-specific role for Suv39h1 in stress-dependent regulation of HP1 protein levels.
Figure 6. (A) Levels of HA-tagged HP1 isoforms upon increased expression of Myc-Suv39h1 in NIH3T3 cells. (B) Quantification of n = 3 experiments as in (A). Levels of each HP1 isoform were normalized relative to empty vector. ****P < 0.0001 (C) Western-blot of endogenous HP1 isoforms in a Suv39h1-inducible 293-TREX (Invitrogen) cell line. The levels of the Suv39h1 were induced with tetracycline. (D) Quantification of n = 3 experiments of HP1 protein levels in WT or Suv39h1 KO cells under stress. ***P < 0.0005; ****P < 0.0001.(E) Proposed model. Above, in normal conditions, HP1α and γ interact in vivo with Suv39h1 and regulate its dynamics, function, and stability in heterochromatin. Below, we have unveiled a complex interplay between Suv39h1 and each of the three HP1 isoforms, modulating upregulation of each other upon stress conditions.
Discussion
Our work suggests that Suv39h1, a key enzymatic activity responsible for generating the H3K9me3 determinant of the “histone code” at PCH,Citation7 is regulated by HP1 proteins in an isotype-specific manner. HP1α and, to a lesser extent, HP1γ directly regulate Suv39h1 activity both under normal conditions and under conditions of cellular stress. Under normal conditions, both HP1α and HP1γ bind preferentially to Suv39h1 in vitro and in vivo in the PCH foci, and their deficiency alters the in vivo dynamics and stability of Suv39h1 and the global levels of H3K9me3. The relatively weak interaction of HP1β and Suv39h1 was intriguing, especially given that the Suv39h1-HP1β was the first interaction of Su39h1 and HP1 characterized, at the time of the isolation of Suv39h1.Citation6,18 We observed a clear antagonism between Suv39h1 and HP1β, compared to the interaction of Suv39h1 and HP1α. This finding is very surprising given that HP1α and β show the highest degree of identity and mainly diverge only in the HR.Citation46 Swapping the hinge region between HP1α and β resulted in a halfway effect, between both isoforms, on Suv39h1 stability indicating that the HP1 HR is involved in regulating Suv39h1 stability. Our observation that the loss of HP1β increases Suv39h1 protein levels may be, at least partially, an indirect effect for several reasons. First, HP1β overexpression did not alter Suv39h1 stability or Suv39h1 polyubiquitination levels. Second, neither HP1β overexpression nor its deficiency had a significant impact in Suv39h1 turnover rate and mobile fraction in PCH. Third, we have observed a compensatory effect between HP1β and γ protein levels (), by which HP1β loss induces HP1γ increased levels and vice versa. Based on this latter observation we suggest that the effect of HP1β-deficiency on Suv39h1 levels may be largely due to the upregulation of HP1γ, and not by a direct effect of HP1β on Suv39h1.
Our work also reports, for the first time, that the HP1α and γ isoforms regulate Suv39h1 protein stability. Dissection of the molecular mechanism by which both isotypes regulate Suv39h1 protein stability show that they enhance Suv39h1 protein stability by inhibiting MDM2-dependent polyubiquitination of K87 in Suv39h1. This results in a significant lengthening of Suv39h1 protein half-life. Regulation of Suv39h1 levels is likely to be relevant in physiological situations where genome integrity may have become compromised in a Suv39h1-dependent manner, such as in aging,Citation39 cancer,Citation40 diseaseCitation41-43 and cellular (oxidative and metabolic) stress.Citation11 Exploring the role of HP1 isotypes in regulating Suv39h1 protein stability under conditions of cellular stress revealed that loss of HP1α, and to a lesser extent loss of HP1γ, abolished the upregulation of Suv39h1 protein levels and H3K9me3 that results from cellular stress (). The lack of upregulation of Suv39h1 protein levels and H3K9me3 under stress () was similar to that observed using SirT1-deficient cells,Citation10,11 indicating that SirT1 may cooperate with both HP1α and γ to increase the levels of active Suv39h1 protein in response to stress, perhaps as part of a SirT1-HP1-Suv39h1 complex.
The complex interplay between HP1 isotypes and Suv39h1 is reinforced by our observation that Suv39h1 also regulates HP1α and γ stability under both normal and stress conditions. Interestingly, recent studies have suggested that proteins that control Suv39h1 stability, such as Lamin A,Citation44,45 also regulate HP1 protein stability, further supporting the notion that the stability of both Suv39h1 and HP1 is likely coordinated. The fact that the interaction of Suv39h1 with HP1α and γ isotypes affects their stability, particularly under conditions of cellular stress, strongly indicates that the Suv39h1/HP1α,γ feedback mechanism is relevant to the stress response. The complexity of the interplay between Suv39h1 and the two HP1 isotypes is highlighted by our observation that Suv39h1 upregulation increased HP1α stability but decreased HP1γ stability. The functional link between Suv39h1 and these two isoforms is not simple and further studies will be aimed at defining the interactions, especially during the response to stress.
Taken together our results indicate that there is a complex, isotype-specific interplay between Suv39h1 and each of the HP1 isoforms, which results in coordinated regulation of Suv39h1/HP1 dynamics and protein stability under normal conditions and particularly under conditions of cellular stress. These data provide a novel paradigm for understanding the role of each of the HP1 isoforms in genome stability and heterochromatin regulation under stress.
Materials and methods
Plasmids and antibodies
Suv39h1, GFP fusions and SirT1 expression constructs were previously described.Citation10,11,47 Expression vectors of FLAG or HA-tagged HP1 isoforms (FLAG-HP1α/β/γ and HP1-HA α/β/γ, respectively) were generated from HP1-GFP vectors previously describedCitation48 using pcDNA4T0 (Invitrogen) by standard PCR-based cloning strategy. HP1 α(β) and HP1 β(α) expression constructs, were generated by exchanging hinge regions of HP1 α and HP1β (aa78-117 and aa79-113, respectively). The following primary antibodies were: α-Myc (Cell Signaling); α-GFP (Millipore); α-FLAG M2, α-HA, and α-Actin, (Sigma-Aldrich); and α-HP1 α, β, and γ (Euromedex).
Immunofluorescence, FRET, FRAP and Dendra assays
Immunofluorescence and FRAP assays of Suv39h1-EGFP were performed as previously described.Citation11,49 For Dendra experiments, cells were seeded in a 35 mm glass bottom culture dishes (Mattek Corporation). Cells were transfected with expression vectors of Suv39h1-Dendra2 −/+ FLAG-HP1α/β/γ and transferred 24 h later to phenol-red free media as Minimum Essential Medium Eagle (Sigma-Aldrich) supplemented with penicillin-G and sodium bicarbonate at 37°C in a humidified 5% CO2 atmosphere. Cells were live-imaged on a Leica TCS SP5 confocal microscope (63x plan-apochromat, NA 1.4) for 10 h. Z-stack volumes (0.5 um) were captured at 15 min intervals until 10 h. For photoconversion experiments, a 405 nm laser light was used in a region of interest (ROI) for 1s bursts at 2% of the total laser power. Post-acquisition image analyses were performed using ImageJ/Fiji opensource sortware (http://imagej.nih.gov/ij/).
Immunoprecipitations, Suv39h1-stability and ubiquitination studies
Cell extracts were prepared according to the Dignam protocol.Citation50 The Western Blot was realized using the appropriated antibodies and densitometric analysis was performed with Quantity One software (Biorad). Immunoprecipitations were performed with FLAG-agarose (Sigma), HA-agarose (Sigma), or agarose crosslinked with Myc-tag antibody, as previously described,Citation51 and bound proteins were eluted with 0.2 M of glycine pH 2.3. For Suv39h1 stability assays, NIH3T3 cells expressing empty vector, FLAG-HP1α, β or γ were incubated with 200 μg/ml cyclohexamide (FLUKA) for 0, 2, 4, 6, 8, 24, 30 h and the levels of endogenous Suv39h1 were analyzed by western-blot (Millipore). Ubiquitination assays using Myc-Suv39h1 and FLAG-HP1α, β or γ were performed as previously described.Citation11
Statistical analysis
Statistical analysis of different experiments (n ≥ 3) was performed using t-student test. The graph show the average and the bars indicate the standard deviation except for FRET experiments in that represent standard error.
Cells and treatments
Suv39h1/2−/−, hp1α−/−, hp1β−/− and hp1γ−/− were previously described.Citation36,37,43 Suv39h1-inducible 293 cells were generated in 293TREX cells (Invitrogen) by blasticidin and zeocin selection. Myc-Suv39h1 expression was induced with tetracycline as previously described.Citation47 Transfection experiments were performed with PEI (Polysciences Inc.), and cells were harvested after 48–72 h for analysis. In oxidative stress treatments, cells were incubated with 2–5 mM of H2O2 for 1 h at 37°C before analysis. For IR experiments, indicated MEFs were irradiated with 7 Gy and grown for 30 min or 2 h before analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
AV, LB-P and HR-V conceived the study and designed the experiments. AV supervised the work and wrote the manuscript with the help of PBS. LS and PBS collaborated in the experimental design and discussion of the work. LB-P, HR-V performed most of the experiments with the support of JG and AM-D. LS and NK-G performed the FRAP assays. LBP and CC carried out Dendra experiments. JPB and PS generated the WT and HP1 KO MEFs.
KEPI_A_1278096_s02.pdf
Download PDF (301 KB)Acknowledgments
The authors want to thank the members of the Vaquero lab for fruitful discussion. We thank Olivia Belbin and l'Alberto Lleó (IIB Sant Pau, Barcelona), Peter Hemmerich (Leibniz Institute for Age Research, Jena, Germany), and Pierre Chambon (IGBMC, Strasbourg, France) for kindly sharing reagents. We also thank Thomas Fuhrmann and Thomas Jenuwein (MPI, Freiburg, Germany) for Suv39h1/2 MEFs.
Funding
This work was supported by Spanish Ministry of Economy and competitiveness-MINECO [SAF2011-25860, SAF2014-55964R to A.V.] confunded by FEDER funds/ European Regional Development Fund (ERDF)-a way to Build Europe, and AGAUR [2009SGR-914, 2014SGR-400). HR-V was a fellow of the Spanish Ministry of Economy and competitiveness-MINECO.
References
- Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol 2003; 13:R895-8; PMID:14654010
- Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 2004; 117:2491-501; PMID:15128874; http://dx.doi.org/10.1242/jcs.01238
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18:1251-62; PMID:15145825; http://dx.doi.org/10.1101/gad.300704
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 2003; 31:2305-12; PMID:12711675
- Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 2007; 21:1169-78; PMID:17470536; http://dx.doi.org/10.1101/gad.1536807
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. Embo J 1999; 18:1923-38; PMID:10202156; http://dx.doi.org/10.1093/emboj/18.7.1923
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406:593-9; PMID:10949293; http://dx.doi.org/10.1038/35020506
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107:323-37; PMID:11701123
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol 2001; 11:266-73; PMID:11356363; http://dx.doi.org/10.1016/S0962-8924(01)02001-3
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 2007; 450:440-4; PMID:18004385; http://dx.doi.org/10.1038/nature06268
- Bosch-Presegue L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, Serrano L, Vaquero A. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell 2011; 42:210-23; PMID:21504832; http://dx.doi.org/10.1016/j.molcel.2011.02.034
- Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 2008; 133:627-39; PMID:18485871; http://dx.doi.org/10.1016/j.cell.2008.03.030
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A 2014; 111:9169-74; PMID:24927542; http://dx.doi.org/10.1073/pnas.1403565111
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 2002; 416:103-7; PMID:11882902; http://dx.doi.org/10.1038/nature722
- Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol 2000; 10:27-30; PMID:10660299; http://dx.doi.org/10.1016/S0960-9822(99)00260-2
- Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. Embo J 2004; 23:489-99; PMID:14765118; http://dx.doi.org/10.1038/sj.emboj.7600088
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074-80; PMID:11498575; http://dx.doi.org/10.1126/science.1063127
- Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol 2000; 20:3728-41; PMID:10779362; http://dx.doi.org/10.1128/MCB.20.10.3728-3741.2000
- Krouwels IM, Wiesmeijer K, Abraham TE, Molenaar C, Verwoerd NP, Tanke HJ, Dirks RW. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol 2005; 170:537-49; PMID:16103223; http://dx.doi.org/10.1083/jcb.200502154
- Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Worz S, Sadic D, Schulte M, Mallm JP, Maiser A, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev 2013; 27:859-72; PMID:23599346; http://dx.doi.org/10.1101/gad.210377.112
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 1999; 4:529-40; PMID:10549285; http://dx.doi.org/10.1016/S1097-2765(00)80204-X
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 2004; 5:296-304; PMID:15071554; http://dx.doi.org/10.1038/nrm1355
- Hediger F, Gasser SM. Heterochromatin protein 1: don't judge the book by its cover! Curr Opin Genet Dev 2006; 16:143-50; PMID:16503133; http://dx.doi.org/10.1038/nrclinonc.2010.9-c1
- Kwon SH, Workman JL. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 2011; 33:280-9; PMID:21271610; http://dx.doi.org/10.1002/bies.201000138
- Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol 2009; 29:6335-40; PMID:19805510; http://dx.doi.org/10.1128/MCB.01048-09
- Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet 2000; 90:279-84; PMID:11124534; http://dx.doi.org/10/56789
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 1999; 108:220-34; PMID:10460410; http://dx.doi.org/10.1007/s004120050372
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 2005; 19:381-91; PMID:16061184; http://dx.doi.org/10.1016/j.molcel.2005.06.011
- Font-Burgada J, Rossell D, Auer H, Azorin F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev 2008; 22:3007-23; PMID:18981478; http://dx.doi.org/10.1101/gad.481408
- Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama J. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol 2011; 31:1186-200; PMID:21245376; http://dx.doi.org/10.1128/MCB.01012-10
- Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol 2006; 8:407-15; PMID:16531993; http://dx.doi.org/10.1038/ncb1383
- Munari F, Gajda MJ, Hiragami-Hamada K, Fischle W, Zweckstetter M. Characterization of the effects of phosphorylation by CK2 on the structure and binding properties of human HP1beta. FEBS Lett 2014; 588:1094-9; PMID:24561199; http://dx.doi.org/10.1016/j.febslet.2014.02.019
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 2011; 43:220-7; PMID:21317888; http://dx.doi.org/10.1038/ng.765
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol 2011; 193:81-95; PMID:21464229; http://dx.doi.org/10.1083/jcb.201101030
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. Paving the way for H2AX phosphorylation: chromatin changes in the DNA damage response. Cell Cycle 2009; 8:1494-500; PMID:19377276; http://dx.doi.org/10.4161/cc.8.10.8501
- Aucott R, Bullwinkel J, Yu Y, Shi W, Billur M, Brown JP, Menzel U, Kioussis D, Wang G, Reisert I, et al. HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J Cell Biol 2008; 183:597-606; PMID:19015315; http://dx.doi.org/10.1083/jcb.200804041
- Brown JP, Bullwinkel J, Baron-Luhr B, Billur M, Schneider P, Winking H, Singh PB. HP1gamma function is required for male germ cell survival and spermatogenesis. Epigenetics Chromatin 2010; 3:9; PMID:20423503; http://dx.doi.org/10.1186/1756-8935-3-9
- Singh PB. HP1 proteins–what is the essential interaction? Genetika 2010; 46:1424-9; PMID:21250545; http://dx.doi.org/10.1134/S1022795410100297
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015; 348:1160-3; PMID:25931448; http://dx.doi.org/10.1126/science.aaa1356
- Khanal P, Kim G, Lim SC, Yun HJ, Lee KY, Choi HK, Choi HS. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. Faseb J 2013; 27:4606-18; PMID:23934277; http://dx.doi.org/10.1096/fj.13-236851
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med 2005; 11:440-5; PMID:15750600; http://dx.doi.org/10.1038/nm1204
- Liu B, Wang Z, Zhang L, Ghosh S, Zheng H, Zhou Z. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat Commun 2013; 4:1868; PMID:23695662; http://dx.doi.org/10.1038/ncomms2885
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A 2006; 103:8703-8; PMID:16738054; http://dx.doi.org/10.1073/pnas.0602569103
- Mungamuri SK, Qiao RF, Yao S, Manfredi JJ, Gu W, Aaronson SA. USP7 Enforces Heterochromatinization of p53 Target Promoters by Protecting SUV39H1 from MDM2-Mediated Degradation. Cell Rep 2016; 14:2528-37; PMID:26971997; http://dx.doi.org/10.1016/j.celrep.2016.02.049
- Chaturvedi P, Parnaik VK. Lamin A rod domain mutants target heterochromatin protein 1alpha and beta for proteasomal degradation by activation of F-box protein, FBXW10. PLoS One 2010; 5:e10620; PMID:20498703; http://dx.doi.org/10.1371/journal.pone.0010620
- Singh PB, Georgatos SD. HP1: facts, open questions, and speculation. J Struct Biol 2002; 140:10-6; PMID:12490149; http://dx.doi.org/10.1016/S1047-8477(02)00536-1
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 2004; 16:93-105; PMID:15469825; http://dx.doi.org/10.1016/j.molcel.2004.08.031
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell 2004; 15:2819-33; PMID:15064352; http://dx.doi.org/10.1091/mbc.E03-11-0827
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 2007; 26:5505-20; PMID:17694090; http://dx.doi.org/10.1038/sj.onc.1210617
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol 1983; 101:582-98; PMID:6888276; http://dx.doi.org/10.1016/0076-6879(83)01039-3
- Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol 2004; 375:393-414; PMID:14870680; http://dx.doi.org/10.1016/S0076-6879(03)75025-3
