 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
DNA methylation is a crucial epigenetic mark associated to gene silencing, and its targeted removal is a major goal of epigenetic editing. In animal cells, DNA demethylation involves iterative 5mC oxidation by TET enzymes followed by replication-dependent dilution and/or replication-independent DNA repair of its oxidized derivatives. In contrast, plants use specific DNA glycosylases that directly excise 5mC and initiate its substitution for unmethylated C in a base excision repair process. In this work, we have fused the catalytic domain of Arabidopsis ROS1 5mC DNA glycosylase (ROS1_CD) to the DNA binding domain of yeast GAL4 (GBD). We show that the resultant GBD-ROS1_CD fusion protein binds specifically a GBD-targeted DNA sequence in vitro. We also found that transient in vivo expression of GBD-ROS1_CD in human cells specifically reactivates transcription of a methylation-silenced reporter gene, and that such reactivation requires both ROS1_CD catalytic activity and GBD binding capacity. Finally, we show that reactivation induced by GBD-ROS1_CD is accompanied by decreased methylation levels at several CpG sites of the targeted promoter. All together, these results show that plant 5mC DNA glycosylases can be used for targeted active DNA demethylation in human cells.
Introduction
DNA methylation at carbon 5 of cytosine (5-methylcytosine, 5mC) is a stable but reversible epigenetic mark associated to gene silencing, and plays essential roles in development and genome defense against transposons.Citation1 Modification of DNA methylation patterns in both normal and pathological conditions results from both methylation and demethylation processes, but in animal cells the latter are not yet well understood.Citation2 Demethylation involves either dilution of 5mC by DNA replication in the absence of methylation, designated passive DNA demethylation, or enzymatic processes for replication-independent removal of 5mC, termed active DNA demethylation.Citation3
Accumulating evidence suggests that DNA demethylation in mammalian cells involves iterative oxidation of 5mC by TET dioxygenases, followed by either passive replication-dependent dilution or active removal of oxidized 5mC derivatives through DNA repair (reviewed inCitation4,5). However, the emerging picture is still unclear because the magnitude of passive dilution compared to active DNA repair is unknownCitation2 and, moreover, oxidized forms of 5mC may behave as independent epigenetic marks on their own.Citation6
Unlike animals, plants are able to remove 5mC without prior modification. Convincing biochemical and genetic evidence indicates that proteins belonging to a plant-specific family of DNA glycosylases directly excise 5mC from DNA and initiate active DNA demethylation through a base excision repair (BER) pathway.Citation7-9 Arabidopsis thaliana REPRESSOR OF SILENCING 1 (ROS1) is a representative member of this 5mC DNA glycosylase family, whose members are uniquely characterized by a discontinuous DNA glycosylase domain,Citation10 a conserved C-terminal domain that is essential for the catalytic activity,Citation11,12 and a basic amino-terminal domain that mediates nonspecific binding to DNA but it is not required for catalysis.Citation13
The understanding of active DNA demethylation mechanisms may have broad implications for epigenetic editing, a new discipline aimed to develop molecular tools to modulate gene expression. One of its most promising approaches is to overwrite epigenetic marks at specific genome regions by targeting epigenetic effector domains (EDs) fused to a DNA binding domain (DBD) that specifically recognizes a particular DNA sequence.Citation14 These chimeric proteins can be very useful in basic researchCitation15 and might also have important applications in the study of diseases that show epigenetic alterations, such as cancer.Citation16,17 Recent studies have shown targeted DNA demethylation and gene reactivation in mammalian cells by fusing different enzymes involved in DNA demethylation to diverse DBDs.Citation18-22 However, no efforts have been reported so far to express or target plant 5mC DNA glycosylases, which directly excise 5mC from DNA.
In this work, we have explored the possibility of directing ROS1 DNA glycosylase activity to a specific target sequence in human cells. We hypothesized that it would be possible to replace Arabidopsis thaliana ROS1 N-terminal domain by a specific DBD without changing its catalytic efficiency. As a proof of concept, we have fused the catalytic domain of ROS1 (ROS1_CD) with the GAL4 DNA binding domain (GBD), a well-characterized DBD that specifically binds the GAL upstream activating sequence (UAS).Citation23 GBD has been previously used for targeting the catalytic domain of mouse DNA methyltransferases DNMT3a and DNMT3b in human cells.Citation24 The targeted binding capacity of GBD-ROS1_CD was first analyzed in vitro and then the fusion protein was transiently expressed in a human cell line. We have found that transient expression of GBD-ROS1_CD in human cells promotes targeted DNA demethylation and reactivation of a methylation-silenced reporter gene.
Results
GBD-ROS1_CD binds preferentially a DNA containing a GAL4-target sequence
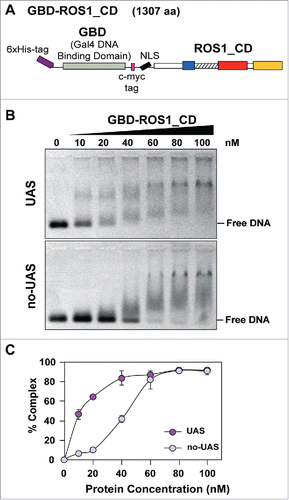
To target DNA demethylation to a specific sequence, we fused the GAL4 DNA binding domain (GBD) to a truncated ROS1 version lacking its lysine-rich N-terminal domain [ROS1_CD, previously designated as ROS1NΔ294Citation13]. We first examined the in vitro binding properties of the recombinant fusion protein, which was overexpressed in E. coli and purified as a N-terminal His-tagged protein.
We analyzed the DNA binding specificity of GBD-ROS1_CD by electrophoretic mobility shift assay (EMSA) (). Increasing concentrations of GBD-ROS1_CD were incubated with either a GBD-targeted substrate or a non-targeted substrate (). Both substrates are 60 bp-long, but the targeted DNA (UAS) contains an upstream activating sequence (5′-CGGAGGACAGTACTCCG-3′) that is absent in the non-targeted DNA (no-UAS) (Table S2). At protein concentrations lower than 60 nM, a significantly higher percentage of DNA-protein complexes were detected with the targeted DNA in comparison with the non-targeted probe (). The binding isotherm for the targeted DNA is hyperbolic (), indicative of a true equilibrium process. Using a Scatchard plot, an apparent dissociation constant (Kdapp) of 12.8 nM was estimated. A reliable estimate of Kdapp could not be obtained for the no-UAS substrate (Fig. S1). These results indicate that GBD-ROS1_CD fusion protein binds preferentially DNA molecules containing sequences targeted by the GAL4 DNA binding domain.
Figure 1. Analysis of the DNA binding capacity of GBD-ROS1_CD in vitro. (A) Schematic diagram of the His-tagged GBD-ROS1_CD recombinant fusion protein. ROS1_CD comprises residues 295–1393 of ROS1, and contains a non-contiguous DNA glycosylase domain distributed over two segments (blue and red) separated by a non-structured linker region (striped), and a C-terminal domain (yellow). (B) Increasing concentrations of purified GBD-ROS1_CD were incubated at 25°C for 30 min with 10 nM of fluorescein-labeled DNA duplex containing (UAS) or not (no-UAS) the sequence targeted by GBD. After non-denaturing gel electrophoresis, gels were scanned to detect fluorescein-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA. A representative gel for each substrate is shown. (C) Percentage of protein–DNA complexes versus protein concentration. All bands with slower mobility were used in quantitation of bound protein. Values are mean ± SE (error bars) from three independent experiments.
GBD-ROS1_CD and its mutant variants are efficiently expressed after transient transfection in HEK293 cells
We next explored the possibility to direct the DNA demethylation activity of GBD-ROS1_CD to UAS sequences in human cells by using a luciferase reporter system. The plasmid constructed for expression of GBD-ROS1_CD in human cells encodes a 1304 aa-protein with a Flag-tag at the N-terminus, and a c-myc tag and a nuclear localization signal (NLS) between GBD and ROS1_CD (). We additionally generated two control constructs encoding mutant versions of the fusion protein. GBD-ROS1_CDmut contains a mutation in the DNA glycosylase domain of ROS1 that changes to alanine an invariant aspartic acid residue (D971 in full-length ROS1) and completely abolishes the catalytic activity of the protein.Citation7 The GBDmut-ROS1_CD version contains a double mutation in the GBD domain (L32P and C38G in full-length GAL4) that abrogates its DNA binding capacity.Citation25
Figure 2. Transient expression of GBD-ROS1_CD and its mutant variants in HEK293 cells. (A-B) Schematic diagrams of effector and reporter constructs used for co-transfection of HEK293 cells. Control effector constructs contain either a mutation in the ROS1 catalytic domain (GBD-ROS1_CDmut) or two mutations in GBD that abolish binding to target UAS sequences (GBDmut-ROS1_CD). Reporter constructs contain the TK promoter fused to the firefly luciferase gene. The targeted version (5xUAS-TK-Luc) includes five copies of GBD binding sites upstream the TK promoter that are absent in the non-targeted control (TK-Luc). (C) Transient expression of GBD-ROS1_CD fusion proteins in HEK293 cells. Western-blot analysis with an anti-Flag antibody was performed in cell extracts (80 μg) prepared 48 h after co-transfection of different effector constructs with either the targeted (upper panels) or the non-targeted (lower panels) methylated reporter plasmid. Actin was used as an input control.
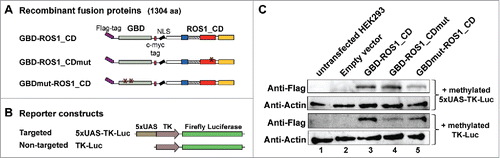
We co-transfected each expression construct in HEK293 cells with a targeted reporter plasmid expressing the firefly luciferase gene under the control of a TK promoter and five copies of the GBD-targeted sequence (5xUAS) (). A non-targeted reporter plasmid containing a TK promoter with no UAS sequence was used as control (). The firefly reporter vectors were previously methylated in vitro with M.SssI. A second reporter plasmid expressing Renilla luciferase gene (RLL) under the control of a CMV promoter (pRL-CMV, Promega) was used for normalization of transfection yield and cell number in all co-transfection experiments.
The co-transfection efficiency with different combinations of constructs ranged between 70–75%, as measured by flow cytometry 48 h after co-transfection with a GFP-expressing reporter plasmid (Fig. S2). By Western-blot analysis after co-transfection with either the targeted or the non-targeted reporter vector, we confirmed the transient expression of all three GBD-ROS1_CD versions in HEK293 cells ().
Transient expression of GBD-ROS1 in HEK293 cells reactivates a methylation-silenced reporter gene
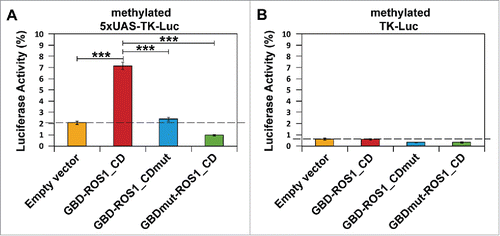
As expected, the luciferase activity of both targeted (5xUAS-TK-Luc) and non-targeted (TK-Luc) reporter plasmids in HEK293 cells was reduced by about 98% after in vitro methylation by M.SssI (). These results confirm that the TK promoter is highly sensitive to DNA methylation, as previously reported.Citation24 We determined the effect exerted by the transient expression of different GBD-ROS1_CD versions on this silenced luciferase activity (). We found that co-transfection with GBD-ROS1_CD significantly increased the luciferase activity of the targeted (5xUAS-TK-Luc) reporter gene by 3.4-fold (P < 0.001) () compared to that observed with the empty expression vector. In contrast, no significant increase was detected in the untargeted (TK-Luc) gene (). When either the GBD-ROS1_CDmut or the GBDmut-ROS1_CD mutant variants were transiently expressed, no significant increase in luciferase activity was detected, either with the targeted or the non-targeted reporter gene (). All together, these results suggest that both ROS1_CD catalytic activity and specific DNA binding by GBD are required for targeted gene reactivation by the fusion protein GBD-ROS1_CD in vivo.
Figure 3. Targeted reactivation of luciferase activity by transient expression of GBD-ROS1_CD. Cells were co-transfected with the indicated effector construct and either the targeted (A) or non-targeted (B) version of the in vitro-methylated reporter plasmid. Luciferase activity, determined 48 h after co-transfection, is shown relative to that detected after co-transfection with empty vector and unmethylated reporter. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences (P < 0.001; Student's unpaired t-test).
Targeted transcriptional activation of the silenced reporter gene by GBD-ROS1_CD
We next examined whether changes in luciferase activity correlate with mRNA levels of the luciferase reporter gene. We isolated total RNA 48 h after co-transfection of either the targeted or the non-targeted reporter plasmid with constructs expressing different GBD-ROS1_CD versions, and levels of the firefly luciferase transcript were analyzed by quantitative real-time PCR (qRT-PCR). As expected, in vitro methylation of the reporter plasmids strongly decreased transcript levels of both the targeted (5xUAS-TK-Luc) and the non-targeted (TK-Luc) genes ().
Figure 4. Targeted transcriptional activation of luciferase reporter gene by transient expression of GBD-ROS1. Cells were co-transfected with the indicated effector construct and either the targeted (A) or non-targeted (B) version of the in vitro methylated reporter plasmid. Levels of firefly luciferase transcript, assessed 48 h after transfection by qRT-PCR and normalized to GAPDH, are shown relative to those detected after co-transfection with empty vector and unmethylated reporter. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences (P < 0.05; Student's unpaired t-test).
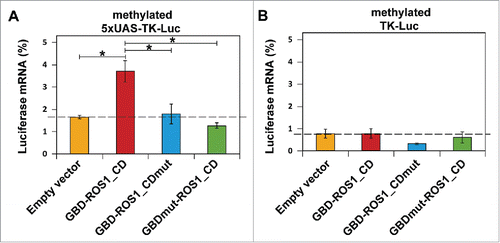
We found that the transient expression of GBD-ROS1_CD significantly increased mRNA levels of the silenced targeted (5xUAS-TK-Luc) gene by 2.2-fold (P < 0.05), compared to those observed with the empty expression vector (). In contrast, we did not observe any noticeable effect of GBD-ROS1_CD on the non-targeted (TK-Luc) gene mRNA levels (). As previously observed with luciferase activity, neither the GBD-ROS1_CDmut nor the GBDmut-ROS1_CD mutant variant significantly increased luciferase transcript levels. All together, these results indicate that transient expression of GBD-ROS1_CD in human cells specifically activates transcription of a methylated GBD-targeted gene, and that such activation requires both the catalytic activity of ROS1_CD and the DNA binding capacity of GBD.
Transient expression of GBD-ROS1_CD induces partial demethylation of the 5xUAS region
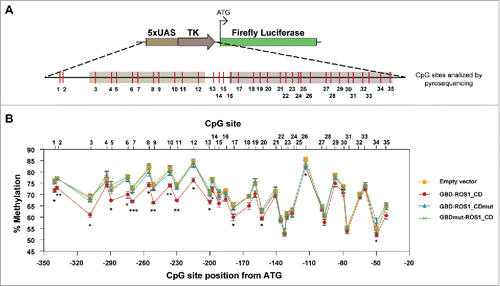
The results reported above strongly suggest that GBD-ROS1_CD can induce the transcriptional reactivation of a methylation-silenced gene. To examine whether such reactivation involves DNA methylation changes, we performed bisulfite DNA pyrosequencing of the 5xUAS region after transient expression of either GBD-ROS1_CD or its mutant variants. A total of 35 CpG sites distributed over a 302-bp region were analyzed (). We found a modest, but statistically significant, decrease of methylation levels in a region covering about 200 bp, when compared to those detected in cells transfected with the empty vector. Importantly, DNA demethylation declined as distance from the 5xUAS region increased. This result suggests that the DNA demethylation range is determined by the distance to the site of GBD-targeting. Very similar results were observed when comparing cells expressing wild type GBD-ROS1_CD with control cells expressing either the GBD-ROS1_CDmut or the GBDmut-ROS1 mutant variant. These results strongly suggest that the 5mC DNA glycosylase activity of ROS1 can be targeted to specific sequences to induce active DNA demethylation.
Figure 5. DNA methylation analysis of the targeted 5xUAS-TK region after transient expression of GBD-ROS1_CD. (A) Schematic diagram of the analyzed region. CpG sites are shown in red. (B) Quantitative methylation analysis. Cells were co-transfected with the indicated effector construct and the targeted methylated reporter plasmid. Plasmid DNA was re-isolated 48 h after co-transfection, bisulfite-treated, PCR-amplified, and pyrosequenced. The graph shows the percentage of methylation at different positions. Values are means ± SE (error bars) from three independent transfection experiments. Asterisks indicate statistically significant differences between GBD-ROS1 and the empty vector (*: P < 0.05; **: P < 0.01; ***: P < 0.001; Student's unpaired t-test).
Discussion
The GBD-ROS1_CD fusion protein comprises a version of ROS1 lacking the N-terminal 294 amino acids, a region that binds non-specifically both unmethylated and methylated DNA, but it is not essential for enzymatic activity.Citation13 We hypothesized that replacing the N-terminal domain with a well-characterized DBD would render a protein able to target specific DNA sequences. By using in vitro DNA binding assays, we have found that GBD-ROS1_CD fusion protein binds preferentially DNA molecules containing the GAL4 DNA binding domain target sequence. We also detected some non-specific binding to DNA lacking the target sequence. Although it cannot be ruled out that the ROS1_CD portion of the protein may contribute to such non-specific binding, there might be alternative factors involved. GBD contains a Cys6-zinc cluster motifCitation26 and, as other zinc-coordinating DNA-binding proteins is expected to bind non-specifically to DNA while searching for its target site among an enormous amount of non-specific sites.Citation27 Furthermore, it has been reported that GBD is able to recognizes a variety of UAS sequences that adjust to the general formula (A/C)GGN10–12CCG and that DNA binding specificity is more stringent in vivo than in vitro.Citation28
We have shown that transient expression of GBD-ROS1_CD in HEK293 cells significantly increases transcription of a methylated 5xUAS-TK-Luc reporter gene. Importantly, reactivation is not observed if the GBD region contains two mutations abrogating its DNA binding capacity. This result suggests that, even if the non-GBD portion might retain some non-specific DNA binding capacity in vivo, it does not lead to efficient reactivation. Furthermore, the fact that reactivation was not observed when the reporter plasmid did not include the targeted UAS sequence strongly suggests that GBD-ROS1_CD has a low off-target rate in vivo.
Another important observation is that reactivation depends upon the 5mC DNA glycosylase activity of ROS1_CD. This result rules out any indirect effect arising from targeting a voluminous protein to the methylated promoter, for example by preventing recruitment of methyl-CpG binding proteins (MBP) and other repressor factors. In fact, we have found that HEK293 cells that transiently express GBD-ROS1_CD display reduced methylation levels at several CpG sites in the 5xUAS region, but that such decrease is not observed in cells expressing a catalytically inactive mutant version or a binding deficient mutant variant, this strongly suggests that the significant increase in luciferase activity induced by GBD-ROS1_CD was directly caused by a targeted DNA demethylation activity.
By using bisulfite-based methods, previous studies have reported targeted DNA demethylation in mammalian cells with TET proteins fused to ZFPs, TALEs, or dCas9 protein.Citation18,19,21,22 TET enzymes mediate DNA demethylation indirectly through oxidation of 5mC to 5hmC, and further oxidation of 5hmC to 5fC and 5caC.Citation29,30 However, bisulfite sequencing does not distinguish between unmethylated C and either 5fC or 5caC, since these two derivatives are not resistant to deamination.Citation31 Therefore, distinguishing bona fide DNA demethylation from 5mC oxidation in the context of TET-targeting is problematic. A further complicating factor arises when distinguishing passive from active DNA demethylation. So far, studies reporting TET-mediated demethylation used potentially replicating DNA targets, which makes difficult to rule out passive dilution. In the present study, however, passive DNA demethylation is excluded because the reporter plasmid lacks an origin of replication.
Active DNA demethylation induced by the transient expression of GBD-ROS1 leads to a modest decrease in DNA methylation levels. The possibility that at least part of the 5mC loss is being counteracted by de novo and maintenance DNA methylation cannot be ruled out. In any case, such a limited demethylation exerts a significant effect on transcription activity. Several studies have previously suggested that partial changes in methylation might be sufficient to induce gene expression reactivation, emphasizing the potential of DNA demethylation for modulation of gene expression. For example, TDG fused to the Nuclear Factor kB (NF-kB) DNA binding domain caused a 5–10% reduction in methylation levels at the target CpG sites, leading to a 3-fold increase induction of gene expression in 3T3 cells.Citation20 Furthermore, TET2 fused to a ZFP (CD54) that binds ICAM-1 promoter, partially demethylates the CpG sites in the surrounding region of the ZF binding site in A2780 cells, leading to a 2-fold increase in gene expression.Citation18 Conversely, there is also evidence showing that increasing methylation of a single CpG site can be sufficient to repress gene expression.Citation32
In conclusion, this work demonstrates that targeted expression of ROS1 catalytic domain in human cells reactivates a methylation-silenced gene in a replication-independent manner. Further work with different types of engineered DBDs and endogenous targets will be required to establish the utility of plant 5mC DNA glycosylases in epigenetic editing.
Materials and methods
GBD-ROS1_CD expression and purification in E. coli
The pET28c-ROS1NΔ294 plasmidCitation13 was used as a backbone to obtain a plasmid expressing GBD-ROS1_CD protein with an N-terminal His6-tag. A fragment containing the GAL4 DNA Binding Domain (GBD, 1–147 amino acids), a c-myc tag and a nuclear localization signal (NLS, PKKKKRK), was amplified by PCR from pcDNA3.1-GBD,Citation24 using primers with NheI and EcoRI restriction sites (Table S1). The PCR product was digested, subcloned between the His6-tag and ROS1_CD into pET28c-ROS1NΔ294, and verified by DNA sequencing. The recombinant His-GBD-ROS1_CD protein was overexpressed and purified as previously described.Citation13
Plasmid construction for GBD-ROS1_CD expression in human cells
A DNA fragment containing the ROS1_CD coding sequence was released from pET28c-ROS1NΔ294 by EcoRI-HindIII digestion and subcloned into pCMVTag2B vector (Stratagene) to obtain pCMVTag2B-ROS1_CD. Then, a fragment containing GAL4 DNA Binding Domain (GBD, 1–147 amino acids), a c-myc tag, and a nuclear localization signal (NLS, PKKKKRK), was amplified from pcDNA3.1-GBD using primers with a PstI restriction site (Table S1). The amplification product was digested with PstI and inserted into pCMVTag2B-ROS1_CD, downstream the Flag-tag and upstream ROS1_CD, to obtain pCMVTag2B-GBD-ROS1_CD. Constructs expressing the catalytically inactive and the binding-deficient GBD-ROS1_CD mutant versions (GBD-ROS1_CDmut and GBDmut-ROS1_CD, respectively) were obtained by site-directed mutagenesis of pCMVTag2B-GBD-ROS1_CD using the Quick-Change II XL kit (Stratagene) and specific oligonucleotides (Table S1). Mutational changes were confirmed by DNA sequencing.
Luciferase reporter plasmids
The reporter plasmid 5xUAS-TK-LucCitation24 is derived from the pGL3-basic vector (Promega) and contains five repeats of the upstream activating sequence (5xUAS) and the minimal human herpesvirus 1 thymidylate kinase (TK) promoter (156 bp) upstream the firefly luciferase reporter gene. The non-targeted (TK-Luc) reporter plasmid was obtained by replacing the 5xUAS-TK sequence with a TK-PCR fragment flanked by XhoI and HindIII sites (Table S1). A reporter plasmid containing the Renilla luciferase reporter gene under the control of the cytomegalovirus (CMV) promoter (pRL-CMV; Promega) was used for normalization of transfection yield and cell number in all co-transfection experiments involving luciferase expression analysis.
In vitro methylation of firefly-luciferase reporter plasmid DNA (50 μg) was performed by incubation with M.SssI CpG methyltransferase (160 U, New England Biolabs) in a reaction mixture containing 50 mM NaCl, 10 mM Tris–HCl pH 7.9, 10 mM EDTA and 0.8 mM S-adenosylmethionine (SAM), at 37°C overnight. After incubation, plasmid DNA was purified by ethanol precipitation. Methylation was verified by confirming full resistance to HpaII digestion.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotides used to generate DNA duplexes for electrophoretic mobility shift assays (EMSA) (Table S2) were synthesized by Operon or Integrated DNA Technologies (IDT) and purified by polyacrylamide gel electrophoresis (PAGE) or dual HPLC before use. Double-stranded DNA substrates were prepared by mixing a 5 μM solution of a 5′-fluorescein-labeled oligonucleotide (upper-strand) with a 10 μM solution of an unlabeled oligomer (lower-strand), heating to 95°C for 5 min and slowly cooling to room temperature.
For EMSA reactions, increasing amounts of GBD-ROS1_CD were incubated with fluorescein-labeled duplex DNA (10 nM). DNA binding reactions were carried out at 25°C for 30 min, in 10 mM Tris–HCl, pH 8.0, 1 mM DTT, 10 μg/ml BSA, 20 μM ZnSO4, in a final volume of 10 μl. Complexes were electrophoresed through 0.2% agarose gels in 1X TAE (40 mM Tris–HCl, pH 8.0, 20 mM acetic acid, 1 mM EDTA). Electrophoresis was carried out in 1X TAE for 40 min at 80 V at room temperature. Fluorescein-labeled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
The dissociation constant was calculated as described in.Citation33 Data were analyzed using linear regression with Sigmaplot software according to Scatchard equation:(1)
(1)
where r is the ratio of bound DNA to total DNA ([GBDROS1-DNA] / [total DNA]), [P] is GBD-ROS1_CD concentration ([GBDROS1]), Kdapp is the apparent dissociation constant and n is the number of binding sites for GBD-ROS1_CD protein. The Kdapp value corresponds to the negative reciprocal of the slope of the line, determined by linear regression with Sigmaplot software.
Cell culture and co-transfection experiments
Human embryonic kidney HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) High Glucose (4.5 g/L, Biowest) supplemented with 10% fetal bovine serum (FBS, Biowest, previously inactivated at 56°C for 1 h), and stabilized 1% penicillin-streptomycin solution (Sigma). Cells were cultured at 37°C in a humidified 5% CO2-containing atmosphere and seeded in 6-well plates, at a density of 6×105 cells/well, one day before transfection. The co-transfection mix included the indicated GBD-ROS1_CD expression plasmid (250 ng/well), the firefly luciferase reporter plasmid (250 ng/well) and the Renilla luciferase control plasmid (25 ng/well), all three diluted in 500 μl Opti-MEM® I Reduced Serum Media (Invitrogen) and 5 μl Lipofectamine LTX™ Reagent (Invitrogen). After incubating for 30 min at room temperature, the co-transfection mix was added drop by drop to cells, previously changed to a fresh culture medium (1.5 ml/well). Forty-eight hours after co-transfection, cells were trypsinized and harvested by centrifugation at 1800 rpm for 5 min for further analysis.
Luciferase activity assay
Luciferase activity assay was performed using the Dual-Luciferase® Reporter Assay System (Promega). Cells were washed once with PBS and lysed with 1X Passive Lysis Buffer (1X PLB). After incubating for 15 min at 25°C and gently shaking, lysate samples were clarified by centrifuging for 30 sec at 17000 g and clear lysates were transferred to new tubes. Luciferase Assay Reagent II (LAR II) (50 μl/well) was added to Greiner Bio-One μClear White Plates, and mixed with 20 μl of clear lysate to determine firefly luciferase activity. The firefly luciferase reaction was stopped and the Renilla luciferase reaction was simultaneously initiated by adding 50 μl of Stop & Glo® Reagent. Measurements were carried out in a TECAN infinite F200 PRO microplate reader and using the i-Control 1.7 software. Renilla luciferase activity was used to normalize transfection yield and cell number in all co-transfection experiments.
Detection of protein expression by western blot analysis
Transient expression of GBD fusion proteins in HEK293 cells after co-transfection was monitored by western-blot using an anti-Flag antibody (Sigma). Briefly, cells were lysed in 1X RIPA buffer (Sigma), containing 1% protease inhibitor cocktail set III, EDTA-free (Calbiochem), and protein concentration was determined by the Bradford assay.Citation34 Total cell lysates (80 μg) were subjected to SDS-PAGE (7% acrylamide/bisacrylamide, 37.5:1), and transferred to nitrocellulose membranes at 30 V at 4°C overnight. After blotting, the membranes were blocked with blocking solution [3% skim milk in TBS (10 mM Tris-HCl pH 8 and 150 mM NaCl)] for 1 h at room temperature and washed three times with TBS. Then, membranes were incubated with Monoclonal Anti-Flag® M2, Clone M2 (F1804) antibody (Sigma) at a 1:1000 dilution in blocking solution at 4°C overnight, washed three times with TBS supplemented with 0.5% Tween-20 (TBS-T), and incubated with horseradish peroxidase-conjugated anti-mouse (Cell Signaling) at a 1:3000 dilution in blocking solution for 1 h at room temperature. Finally, after three washes with TBS-T, membranes were incubated with ECL Prime Western Blotting Detection Reagent (GE Healthcare) and images were captured using the LAS-3000 analyzer (Fujifilm). To verify equal loading of total cell lysates, the Monoclonal Anti-Actin Antibody Clone AC-40 (Sigma) was used at 1:2000 dilution in blocking solution at 4°C overnight, following the protocol described above.
Detection of co-transfection efficiency by flow cytometry
Co-transfection efficiency of HEK293 cells was monitored by green fluorescent protein (GFP) signal count by flow cytometry. Cells were co-transfected with Renilla luciferase reporter plasmid (25 ng/well), GBD-ROS1_CD or GBD-ROS1_CDmut expressing constructs (250 ng/well), the methylated reporter plasmid 5xUAS-TK-Luc (250 ng/well) and pACGFP-C1 plasmid (250 ng/well, Clontech), as described above. Cells were harvested 48 h after co-transfection, washed once with PBS and resuspended in PBS. GFP expression was analyzed using a BD FACSCalibur flow cytometer (Becton Dickinson Biosciences) and CellQuest software. Data were analyzed with FlowJo software.
Analysis of firefly luciferase reporter gene expression by quantitative Real-Time PCR
Total RNA was extracted from HEK293 cells 48 h after co-transfection using the RNeasy Mini Kit 74016 (Qiagen). 1 µg RNA was treated with 1 U DNase I, RNase-free (Thermo Scientific), and subsequently used for cDNA synthesis using the qSCRIPT™ cDNA synthesis Kit (Quanta Biosciences), following the manufacturer's instructions. Then, 1 µl cDNA synthesis reaction was used as template in a 10 µl PCR reaction with specific primers (Table S3) and iQ SYBR® Green Supermix (Bio-Rad). All reactions were carried out on the CFX Connect Real-Time System (Bio-Rad) and data were analyzed using the CFX Manager Software (Bio-Rad). Expression levels relative to GAPDH were determined using the 2−ΔΔCt method.Citation35
DNA methylation analysis
The firefly luciferase reporter plasmid was re-isolated 48 h after transfection by resuspending cells in 500 μl Cytoplasmic Extraction Buffer [CPEB pH 7.9: 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 1% protease inhibitor cocktail (535140, Calbiochem), 1% phosphatase inhibitor cocktails 2 and 3 (p5726 and p0044, Sigma), 0.075% igepal CA-630 (Sigma)]. After incubating 30 min on ice and centrifuging 10 min 1800 rpm at 4°C, pellet was resuspended in 250 μl CPEB without igepal. After incubating 10 min on ice and centrifuging 10 min 1800 rpm at 4°C, the pellet was resuspended in 500 μl Nuclear Extraction Buffer (NEB: 10 mM HEPES pH 7.9, 420 mM NaCl, 1.2 mM MgCl2, 0.2 mM EDTA, 25% glycerol). The suspension was incubated for 30 min on ice with repeated vortexing, and then centrifuged for 20 min at 13400 rpm and room temperature. The pellet was resuspended in 250 μl NEB, and 250 μl Buffer 2 and 350 μl Buffer 3 of NZYMiniprep (NZYTech Genes & Enzymes) kit. Henceforth, NZYMiniprep (NZYTech Genes & Enzymes) manufacturer's protocol was followed, eluting plasmid DNA in 25 μl sterile distilled water.
Re-isolated plasmid DNA (20 μl) was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo Research) following the manufacturer's instructions and eluted in 12 μl Elution Buffer. A 2-μl aliquot of bisulfite-treated plasmid DNA was used for PCR amplification with Immolase DNA Polymerase (Bioline) using specific primers (Table S4). DNA pyrosequencing was performed with 15 μl of PCR products and specific sequencing primers (Table S4) on a PyroMark Q24 instrument (Qiagen), according to the manufacturer's guidelines. Analysis of the percentage of methylation at each CpG was determined using PyroMark Q24 Software (Qiagen).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Spanish Ministry of Science and Innovation and the European Regional Development Fund under grant BFU2013-43269-P; and by the Junta de Andalucía and the European Regional Development Fund under grant P11-CVI-7576. J.T.P.D. was the recipient of a PhD FPU Fellowship from the Spanish Ministry of Education. We thank Dr. A. Jeltsch (Stuttgart University, Germany) for kindly providing plasmids 5xUAS-TK-Luc and pcDNA3.1-GBD. We also thank members of our laboratory for helpful discussions and advice.
Author contributions
J.T.P.D., R.R.A and T.R.A conceived and designed the study. R.R.A and T.R.A supervised the study. J.T.P.D. acquired data. J.T.P.D., R.R.A and T.R.A interpreted data. J.T.P.D., R.R.A and T.R.A wrote the manuscript. All authors read and approved the manuscript.
KEPI_A_1294306_supplemental_data.zip
Download Zip (399.6 KB)References
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11:204-20; PMID:20142834; http://dx.doi.org/10.1038/nrg2719
- Schuermann D, Weber AR, Schar P. Active DNA demethylation by DNA repair: Facts and uncertainties. DNA Repair (Amst) 2016; 44:92-102; PMID:27247237; http://dx.doi.org/10.1016/j.dnarep.2016.05.013
- Kress C, Thomassin H, Grange T. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Lett 2001; 494:135-40; PMID:11311228; http://dx.doi.org/10.1016/S0014-5793(01)02328-6
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014; 156:45-68; PMID:24439369; http://dx.doi.org/10.1016/j.cell.2013.12.019
- Bochtler M, Kolano A, Xu GL. DNA demethylation pathways: Additional players and regulators. Bioessays 2017; 39:1-13; http://dx.doi.org/10.1002/bies.201600178
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013; 152:1146-59; PMID:23434322; http://dx.doi.org/10.1016/j.cell.2013.02.004
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA 2006; 103:6853-8; PMID:16624880; http://dx.doi.org/10.1073/pnas.0601109103
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006; 124:495-506; PMID:16469697; http://dx.doi.org/10.1016/j.cell.2005.12.034
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA 2006; 103:11796-801; PMID:16864782; http://dx.doi.org/10.1073/pnas.0603563103
- Ponferrada-Marín MI, Parrilla-Doblas JT, Roldán-Arjona T, Ariza RR. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res 2011; 39:1473-84; PMID:21036872; http://dx.doi.org/10.1093/nar/gkq982
- Hong S, Hashimoto H, Kow YW, Zhang X, Cheng X. The carboxy-terminal domain of ROS1 is essential for 5-methylcytosine DNA glycosylase activity. J Mol Biol 2014; 426:3703-12; PMID:25240767; http://dx.doi.org/10.1016/j.jmb.2014.09.010
- Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, Huh JH. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci U S A 2010; 107:19225-30; PMID:20974931; http://dx.doi.org/10.1073/pnas.1014348107
- Ponferrada-Marín MI, Martínez-Macías MI, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J Biol Chem 2010; 285:23032-9; PMID:20489198; http://dx.doi.org/10.1074/jbc.M110.124578
- de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res 2012; 40:10596-613; PMID:23002135; http://dx.doi.org/10.1093/nar/gks863
- Kungulovski G, Jeltsch A. Epigenome editing: State of the art, concepts, and perspectives. Trends Genet 2016; 32:101-13; PMID:26732754; http://dx.doi.org/10.1016/j.tig.2015.12.001
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597-610; PMID:16136652; http://dx.doi.org/10.1038/nrg1655
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007; 8:286-98; PMID:17339880; http://dx.doi.org/10.1038/nrg2005
- Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. Induced DNA demethylation by targeting ten-eleven translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 2014; 42:1563-74; PMID:24194590; http://dx.doi.org/10.1093/nar/gkt1019
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 2013; 31:1137-42; PMID:24108092; http://dx.doi.org/10.1038/nbt.2726
- Gregory DJ, Mikhaylova L, Fedulov AV. Selective DNA demethylation by fusion of TDG with a sequence-specific DNA-binding domain. Epigenetics 2012; 7:344-9; PMID:22419066; http://dx.doi.org/10.4161/epi.19509
- Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu G-l, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery 2016; 2:16009; PMID:27462456; http://dx.doi.org/10.1038/celldisc.2016.9
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell 2016; 167:233-47 e17; PMID:27662091; http://dx.doi.org/10.1016/j.cell.2016.08.056
- Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 1985; 40:767-74; PMID:3886158; http://dx.doi.org/10.1016/0092-8674(85)90336-8
- Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res 2007; 35:100-12; PMID:17151075; http://dx.doi.org/10.1093/nar/gkl1035
- Johnston M, Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A 1987; 84:2401-5; PMID:3550810; http://dx.doi.org/10.1073/pnas.84.8.2401
- Pan T, Coleman JE. GAL4 transcription factor is not a “zinc finger” but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A 1990; 87:2077-81; PMID:2107541; http://dx.doi.org/10.1073/pnas.87.6.2077
- Iwahara J, Levy Y. Speed-stability paradox in DNA-scanning by zinc-finger proteins. Transcription 2013; 4:58-61; PMID:23412360; http://dx.doi.org/10.4161/trns.23584
- Vashee S, Xu H, Johnston SA, Kodadek T. How do “Zn2 cys6” proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J Biol Chem 1993; 268:24699-706; PMID:8227030
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333:1300-3; PMID:21778364; http://dx.doi.org/10.1126/science.1210597
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930-5; PMID:19372391; http://dx.doi.org/10.1126/science.1170116
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14:341-56; PMID:23698584; http://dx.doi.org/10.1038/nrm3589
- Pogribny IP, Pogribna M, Christman JK, James SJ. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: possible in vivo relevance during tumorigenesis. Cancer Res 2000; 60:588-94; PMID:10676641
- Miao F, Bouziane M, O'Connor TR. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res 1998; 26:4034-41; PMID:9705516; http://dx.doi.org/10.1093/nar/26.17.4034
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/10.1016/0003-2697(76)90527-3
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
