ABSTRACT
Fetal intolerance of labor is a common indication for delivery by Caesarean section. Diagnosis is based on the presence of category III fetal heart rate tracing, which is an abnormal heart tracing associated with increased likelihood of fetal hypoxia and metabolic acidemia. This study analyzed data from 177 unique women who, during their prenatal visits (7-15 weeks and/or 24–32 weeks) to Atlanta area prenatal care clinics, consented to provide blood samples for DNA methylation (HumanMethylation450 BeadChip) and gene expression (Human HT-12 v4 Expression BeadChip) analyses. We focused on 57 women aged 18–36 (mean 25.4), who had DNA methylation data available from their second prenatal visit. DNA methylation patterns at CpG sites across the genome were interrogated for associations with fetal intolerance of labor. Four CpG sites (P value <8.9 × 10−9, FDR <0.05) in gene SLC9B1, a Na+/H+ exchanger, were associated with fetal intolerance of labor. DNA methylation and gene expression were negatively associated when examined longitudinally during pregnancy using a linear mixed-effects model. Positive predictive values of methylation of these four sites ranged from 0.80 to 0.89, while negative predictive values ranged from 0.91 to 0.92. The four CpG sites were also associated with fetal intolerance of labor in an independent cohort (the Johns Hopkins Prospective PPD cohort). Therefore, fetal intolerance of labor could be accurately predicted from maternal blood samples obtained between 24–32 weeks gestation. Fetal intolerance of labor may be accurately predicted from maternal blood samples obtained between 24–32 weeks gestation by assessing DNA methylation patterns of SLC9B1. The identification of pregnant women at elevated risk for fetal intolerance of labor may allow for the development of targeted treatments or management plans.
Introduction
Fetal intolerance of labor, which is also referred to as fetal distress and non-reassuring fetal status, is the most common indication for emergency Caesarean section (C-section) [Citation1,Citation2]. It is characterized by the presence of an abnormal fetal heart rate pattern, a category III tracing, detected through electronic fetal heart rate monitoring during labor [Citation3,Citation4]. Category III fetal heart rate patterns indicative of fetal intolerance of labor include absent baseline fetal heart rate variability and recurrent late and/or variable decelerations and/or bradycardia or the presence of a sinusoidal pattern, typically after the onset of contractions during the second stage of labor [Citation4,Citation5].
A category III tracing is considered abnormal as studies have demonstrated that these heart rate patterns are associated with an increased risk of fetal hypoxia and metabolic acidemia, though such tracings are not absolutely indicative of fetal hypoxia and/or acidemia [Citation6]. In the case of fetal intolerance of labor, expedited delivery, often through a C-section, is indicated to avoid fetal hypoxia, acidemia, and their subsequent consequences [Citation6–10]. Fetal hypoxia and acidemia can have drastic consequences for the fetus in the perinatal period and throughout life, including severe brain damage [Citation11], and has been previously associated with diagnosis of cerebral palsy [Citation12]. Accurate recognition and prompt management of fetal intolerance of labor is essential for decreasing the risk of fetal hypoxia and acidemia and thereby providing the best possible pregnancy outcome.
Early identification of pregnant women at elevated risk for having a pregnancy complicated by fetal intolerance of labor would provide clinical benefits, including maternal preparations to deliver at hospitals with the required resources to perform advanced monitoring and an emergency C-section, if required [Citation1]. Another potential benefit of early identification of those at risk for fetal intolerance of labor is a decrease in the time from decision to deliver until emergency C-section, which might promote adherence to the American College of Obstetricians and Gynecologists (ACOG) recommendation of a 30-minute timeframe, though previous studies have not demonstrated substantial differences in outcome due to decision to delivery time greater than 30 minutes [Citation1,Citation2,Citation13]. Beyond the immediate clinical utility, identification of biological pathways underlying the development of fetal intolerance of labor may provide insight for development of novel treatments and preventive strategies. Previous studies have identified potential biomarkers associated with fetal intolerance of labor including pregnancy-associated plasma protein-A (PAPP-A) and combinations of fetal indices, such as estimated fetal weight, serum placental growth factor, and soluble fms-like tyrosine kinase-1 [Citation14,Citation15]. These studies suggest an early pathogenesis for fetal intolerance that can be detected by screening prior to labor.
Previous studies have also reported associations between DNA methylation and pregnancy complications, including preeclampsia and gestational diabetes [Citation16,Citation17]. DNA methylation, the addition of a methyl group to the 5’ position of cytosine in a cytosine-guanine dinucleotide (CpG site), serves as a mechanism to regulate gene expression. This study utilizes an epigenome-wide association study to assess the relationship between individual CpG sites and fetal intolerance of labor.
Results
Of the 57 women included in this study, 12 had deliveries complicated by fetal intolerance of labor (21%). Maternal age, smoking, gestational hypertension, and gestational age at birth and gestational age at sample collection did not differ between those subjects who did and did not experience deliveries complicated by fetal intolerance of labor (0.59<P value<0.98). However, the group experiencing fetal intolerance of labor was more likely to undergo delivery by C-section (P = 2.70 × 10−5; ), which would be expected given that fetal intolerance of labor is an indication for C-section delivery.
Table 1. Cohort demographics for women with samples collected between 24–32 weeks gestation (n = 57).
Methylation of SLC9B1 predicts fetal intolerance of labor
Four CpG sites were associated with fetal intolerance of labor in maternal samples collected between 24 and 32 weeks gestation, after adjusting for maternal age and peripheral blood mononuclear cells (PBMC) cell type (FDR<0.05; , ). All four sites were annotated to the CpG island of solute carrier family 9, subfamily B, member 1 (SLC9B1) gene, alternatively known as NHEDC1, a NA+/ H+ exchanger. Methylation of these four sites was highly correlated (r = 0.86-0.98; ), and was not associated with PBMC cell type or maternal age (P>0.05). In a separate analysis of these four CpG sites in samples collected between 7–15 weeks gestation (n = 45), each site associated with fetal intolerance of labor to a lesser degree (0.001<P< 0.003), suggesting that the methylation differences associated with fetal intolerance of labor may be detectable even earlier in early pregnancy. Differential methylation region (DMR) analysis identified 18 differentially methylated regions (Supplemental Table 1), including chr4: 103940711–103941205, which contains the four CpG sites mentioned above. In the Johns Hopkins Prospective PPD cohort, these four CpG sites were also highly correlated (r = 0.91-0.97), and associated with fetal intolerance of labor (0.036<P<0.048), replicating our initial finding (, Supplemental Figures 1–3).
Figure 1. Manhattan plot showing the association of CpG sites across the genome. Each CpG site that passed quality control was assessed for associations with fetal intolerance of labor. Sites falling above the horizontal line indicate experiment-wide significance. The x-axis represents the chromosome number and the y-axis is the negative log of the P value, which is indicative of the significance level. The plot is further zoomed in to chromosome four, and the SLC9B1 gene. In the gene diagram, yellow boxes represent exons and the green box represents the location of the CpG island. The heatmap indicates the correlation between CpG sites.
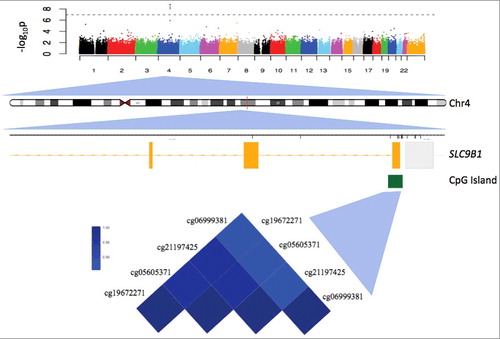
Table 2. CpG sites significantly associated with fetal intolerance of labor.
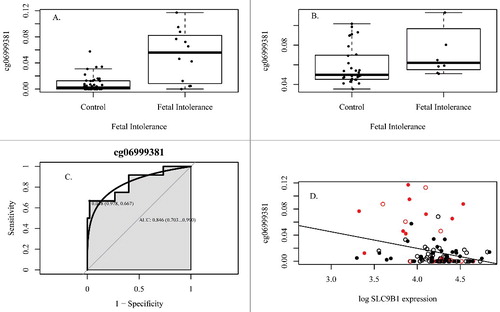
Figure 2. DNA methylation associates with fetal intolerance of labor. (A) DNA methylation of cg06999381 associates with fetal intolerance of labor between 24–32 weeks gestation in the original cohort, (B) Association with fetal intolerance of labor in the replication cohort, (C) Receiver operator characteristic curve, (D) DNA methylation associates with gene expression over pregnancy (ILMN_1724931). Red indicates fetal intolerance to labor. Open circles are samples from visit 1, closed circles are samples from visit 2. All associations are statistically significant (P<0.05).
Methylation of SLC9B1 associates with gene expression
We next sought to assess whether methylation differences of these CpG sites associate with expression of SLC9B1 and whether gene expression alone can predict fetal intolerance of labor. We therefore examined paired methylation and expression data for a subset of subjects with available expression data. Expression of SLC9B1 was represented by two probes— ILMN_1673417 and ILMN_1724931—that passed quality control. Changes in DNA methylation over time were associated with changes in gene expression for all CpG sites and SLC9B1 expression (0.003<P<0.03, , Supplemental Figures 1–3). Gene expression data was available for 65 women who also had DNA methylation data at one or more timepoints. Gene expression alone did not predict fetal intolerance of labor between 24 and 32 weeks gestation (P value non-significant) or over the whole of pregnancy (P value non-significant), suggesting that the CpG sites identified in SLC9B1 are better predictors of fetal intolerance of labor.
Predictive accuracy
Each of the four CpG sites was interrogated for its predictive accuracy of fetal intolerance of labor in maternal PBMC samples collected between 24–32 weeks gestation. The area under the receiver operating characteristic curve ranged from 0.85 to 0.87 for the four sites, representing a predictive accuracy of 85–87% (, Figure S1-S3). The thresholds for each CpG site, along with their respective sensitivity, specificity, and predictive values are shown in Supplemental Table 2.
Gene expression and cardiovascular remodeling
All 56 women with gene expression data available underwent blood pressure measurement between 24–32 weeks. Gene expression, but not DNA methylation, was associated with both high systolic (>140; P = 0.003) and diastolic (>90; P = 0.003) blood pressure (ILMN_1724931).
Discussion
We identify four CpG sites in SLC9B1 whose DNA methylation levels detected in the late second and early third trimesters are predictive of fetal intolerance of labor at delivery and was replicated in an independent sample. Each of these CpG sites was highly correlated with one another, indicating that any of these sites could serve as a proxy for the other three. As an additional line of evidence, SLC9B1 was also identified as being associated with fetal intolerance of labor in a differentially methylated region analysis. This region, and the other identified DMRs should be further investigated as potential contributors to the etiology of fetal intolerance of labor. Women enrolled in this study were medically low-risk, as they had singleton pregnancies with early initiation of prenatal care and did not have chronic health conditions. Thus, we believe these results are generalizable to other low risk populations.
Few studies have examined the role of SLC9B1, also known as NHEDC1. The SLC9 family of genes encodes Na+/H+ exchangers (NHE), which play a role in regulating pH and cell volume [Citation18,Citation19]. A recent study suggests that DNA methylation of SLC9B1 regulates its expression [Citation20], which is consistent with our finding that methylation was associated with gene expression of SLC9B1 over pregnancy. This gene was previously thought to be expressed specifically in the testes, based on a limited panel of 18 tissues, which did not include immune cells [Citation19,Citation21]. Data from the Genotype-Tissue Expression (GTEx) Project shows low-level expression of SLC9B1 in a variety of tissues, including whole blood [Citation22]. Localization to the mitochondria, as shown in the Human Protein Atlas [Citation23], suggests a role for this gene in processes associated with the electron transport chain, which produce the energy required for the cell, as well as reactive oxygen species (ROS) as byproducts.
Early in normal pregnancy, a relative hypoxic state is essential for proper placentation and embryogenesis, but oxygen requirements increase around 11–12 weeks gestation [Citation24]. Oxygen tension is regulated by transcription factors, including the hypoxia-inducible factor 1 (HIF-1). In normal pregnancy, HIF-1 is rapidly degraded [Citation24]. However, increased ROS production by the mitochondria can inhibit the degradation of HIF-1, which may impair placental function and lead to pregnancy complications [Citation25,Citation26]. Additionally, ROS production may result in insufficient energy generation and sodium ion influx into the cell, which has been associated with both cardiac pathology and mitochondrial function [Citation25,Citation27]. We hypothesize that alterations in the methylation and expression of sodium-hydrogen transporters that contribute to proper mitochondrial function, including SLC9B1, may contribute to cardiac pathology and changes in pH, which are both risk factors for fetal intolerance of labor [Citation28,Citation29]. This hypothesis is further supported by our findings that gene expression of SLC9B1 is associated with high maternal systolic and diastolic blood pressure, and with a report from the literature that showed preeclampsia, which is characterized by high blood pressure, is associated with abnormal fetal heart rate [Citation30]. Previous studies have also shown increased ROS production in both the mother and fetus is associated with fetal intolerance of labor [Citation31]. Future studies should further examine the role of SLC9B1 in human fertility and pregnancy outcomes, especially in the context of cardiovascular dysfunction.
This study has several limitations. First, the difference in methylation between women with pregnancies complicated by fetal intolerance of labor and those not complicated by fetal intolerance of labor is relatively small, making the development of a targeted assay for methylation of these CpG sites difficult due to the limited discriminatory power available for common techniques. Additionally, although methylation of these CpG sites was associated with fetal intolerance of labor in the PPD cohort, the range of methylation at these sites was higher in this cohort, potentially due to differences in processing or quality control, or inherent differences in methylation between PBMCs in the original cohort and whole blood in the Johns Hopkins Prospective PPD cohort. Finally, type III fetal heart rate tracings are not absolutely indicative of fetal hypoxia or metabolic acidemia, but there are not currently standards for further evaluation of fetuses intra-labor. Other methods to diagnose fetal intolerance of labor are not routinely performed and lack sufficient clinical evidence for routine implementation. Despite these limitations, receiver operator characteristic curves of the 4 CpG sites in SLC9B1 suggested that the positive and negative predictive values, sensitivity, and specificity, are all well within the range for potential development into a clinically useful diagnostic test (C).
DNA methylation patterns in maternal blood at four CpG sites in SLC9B1 are predictive of fetal intolerance of labor during the late second and early third trimester. These sites have highly positive and negative predictive values, indicating that they may be clinically relevant for the detection and management of fetal intolerance of labor. Future studies should work to develop a robust targeted assay to measure DNA methylation at one or more of these CpG sites so that the clinical utility of DNA methylation at these sites can be further evaluated for its predictive power in other studies throughout pregnancy.
Methods
Study subjects
Subjects included in this study (n = 69) are being enrolled into an ongoing pregnancy cohort study investigating the microbiome and epigenome and the outcome of preterm birth (R01NR014800, R01MD009064) for which pregnant African American women are being recruited from outpatient prenatal care clinics affiliated with two Atlanta metro area hospitals: Emory University Midtown Hospital and Grady Memorial Hospital. A flow chart describing subjects in each analysis is shown in Supplemental Figure 4. These two hospitals represent private and public hospitals, respectively, that serve women of a wide range of socioeconomic status. Eligible women for this study were African American by self-report, between 18–35 years of age, having a singleton pregnancy, having fewer than four previous births, are able to understand written and spoken English, without chronic medical conditions, enrolled between March 2014 and August 2015 and experiencing labor (spontaneous or induced). Not experiencing labor, fetal death before labor and congenital abnormalities of the fetus were criteria for post-enrollment exclusion. Demographic and clinical obstetrical data (including estimated date of confinement, gestational age at delivery, pregnancy complications, labor and delivery course, and blood pressure) were collected through self-report questionnaires and prenatal, labor, and delivery medical chart abstraction under the supervision of a qualified physician (ALD). The diagnosis of fetal intolerance of labor was based upon the medical record documentation by the attending obstetrician or midwife determining the presence of a category III fetal heart rate tracing during labor, and after delivery each chart was reviewed by a qualified physician to confirm the diagnosis. All subjects provided written informed consent. This study was approved by the Emory Institutional Review Board.
Biological sample collection and DNA extraction
Venous blood samples were collected during each of two prenatal visits (between 7–15 weeks and 24–32 weeks), when an additional 12 mL of peripheral blood was drawn into a tube containing EDTA using the same needle stick as for the routine blood draws. Blood was transferred into SepMate tubes with a Ficoll density gradient to isolate peripheral blood mononuclear cells (PBMCs) from whole blood. PBMCs were stored in AllProtect Buffer (Qiagen, Catalog # 76405) at −80°C until a simultaneous DNA and RNA extraction using the AllPrep RNA/DNA Mini Kit (Qiagen, Catalog # 80204, Venlo, Netherlands) was performed according to manufacturer's instructions. DNA quantification and quality was assessed using the Quant-it Pico Green kit (Invitrogen, Catalog# P11496, Carlsbad, California). RNA quantification was assessed using the Agilent 2100 Bioanalyzer. The average RIN score was 8.6, with a standard deviation of 1.4.
DNA methylation
DNA methylation from samples of maternal PBMCs was interrogated for each subject using the HumanMethylation450 BeadChip, which evaluates >450,000 CpG sites across the genome. Briefly, 1 μg of DNA was processed and hybridized to the HumanMethylation450 BeadChip (Illumina) according to manufacturer's instructions. Initial data quality control was performed using the R package CpGassoc [Citation32]. Any CpG site with low signal or missing data for greater than 10% of samples was removed, and any sample with missing data for greater than 5% of CpG sites was removed. Cross-reactive probes were removed [Citation33]. Following quality control, 449,094 probes were included in subsequent analyses. One sample collected between 7–15 weeks gestation failed quality control. Beta values (β) were calculated for each CpG site as the ratio of methylated (M) to methylated and unmethylated (U) signal: β = M/M+U. β-mixture quantile normalization was performed as previously described [Citation34]. Briefly, BMIQ involves fitting a three-state β-mixture model, transforming the distribution of type 2 probes to match the type 1 distribution, followed by a dilation transformation [Citation34]. DNA methylation data can be accessed through NCBI's Gene Expression Omnibus, GSE107459.
RNA expression
RNA expression from maternal PBMCs was interrogated for a subset of subjects for which RNA was available. Briefly, 750 ng of RNA was directly hybridized to the HumanHT-12 v4 BeadChip (Illumina, San Diego, CA) according to manufacturer's instructions. The BeadChips were scanned using the iScan scanner, and the raw data was analyzed using the Expression Module of GenomeStudio Software (Illumina, San Diego, CA). Samples with probe detection rates <90% or with an average intensity of <50% of the experiment-wide sample mean were excluded, resulting in one sample collected between 24–32 weeks being dropped due to quality control. Probes with detection P values >0.01 in >90% of the samples were excluded. Data was then quantile-normalized and log2 transformed. Following quality control, 18,634 expression probes were included in subsequent analyses. RNA expression data can be accessed through NCBI's Gene Expression Omnibus, GSE107437.
Statistical analysis
Demographic characteristics were compared between women with and without pregnancies complicated by fetal intolerance of labor using Student's t tests. The R package CpGassoc was used to perform an epigenome-wide association study (EWAS) to assess the associations between maternal DNA methylation at each CpG site on the array and fetal intolerance of labor for samples collected between 24–32 weeks gestation. For each CpG site, the methylation proportion was regressed on an indicator for fetal intolerance and covariates, which included chip, maternal age, and cell type proportions (CD8+T, monocytes, B cells, natural killer), estimated using the referenced dataset developed by Reinius at al. and implemented used the approach described by Houseman et al [Citation35–37]. CpG sites that were significantly associated with fetal intolerance for labor in samples collected between 24–32 weeks were assessed for associations earlier in pregnancy, between 7–15 weeks gestation, using a linear regression that controlled for maternal age, chip, and cellular heterogeneity as above. The false discovery rate (FDR) was controlled at 0.05. Gene symbols and probe locations were assigned using the HumanMethylation450 BeadChip annotation file distributed by Illumina. To determine whether methylation of these CpG sites was influenced by cellular heterogeneity, linear regressions were performed that modeled methylation at each CpG site as a function of estimated cell type proportions. Pearson's correlation coefficient was calculated for each pair of CpG sites, and between each CpG site at the two timepoints. Predictive accuracy was assessed using the area under the curve (AUC), sensitivity, and specificity calculations, as determined by the R package pROC. Longitudinal associations between each CpG site and gene expression were assessed using a linear mixed-effects model that regressed the log2 expression signal for each gene on methylation of a single CpG site with subject as a random effect and adjustment for maternal age and cellular heterogeneity. Similarly, a linear mixed-effects model was used to interrogate associations between fetal intolerance and gene expression over pregnancy. Subject samples were included in this analysis if participants contributed blood at 24–32 weeks, a subset of which also contributed blood at 7–15 weeks (n = 54). As an additional confirmation of the associations between fetal intolerance and DNA methylation, we performed DMR analysis using the R package ChAMP [Citation38].
To assess the associations between fetal intolerance and gene expression, 65 women, recruited from the same study, were used. The log2 of the gene expression signal was regressed on fetal intolerance of labor after adjusting for cellular heterogeneity and maternal age. Subjects with available measures for blood pressure values were used to assess associations between gene expression and these cardiovascular measures. Blood pressure was dichotomized as being above or below 90 for diastolic blood pressure at least once during pregnancy and above or below 140 for systolic blood pressure at least once during pregnancy. Each outcome was regressed on the log2 gene expression signal, after adjusting for maternal age and cellular heterogeneity. Gene expression was interrogated by two probes that differed by only 2 base pairs; therefore, data from the probe with the highest number of samples that met the detection P value threshold (ILMN_1724931) is presented.
Replication cohort and analysis
A replication cohort of 43 women was recruited as part of the Johns Hopkins Prospective PPD cohort recruited at the Women's Mood Disorders Center at Johns Hopkins. Subjects were prospectively followed during pregnancy and after delivery in order to identify genetic and clinical characteristics that precede the development of a postpartum depressive episode. The average age of the participants was 31.3 years. This cohort was ethnically diverse: 65% of participants were Caucasian, 23% of participants were African American, and the remainder participants identified as Hispanic (2%), Asian or Pacific Islander (5%), or other (5%). In this cohort, 8 women experienced pregnancies complicated by fetal intolerance to labor (21%), and 4 had missing data. Epigenetic data were generated on the HumanMethylation450 BeadChip as described previously [Citation39]. The presence of fetal intolerance of labor was determined through medical record abstraction. Briefly, sample quality assessment and microarray analysis were conducted at The Sidney Kimmel Cancer Center Microarray Core Facility at Johns Hopkins University. Images were processed in Illumina's iScan scanner and data were extracted using Methylation Module of GenomeStudio v1.0 Software. Background and Illumina probe type correction and normalization were performed by the Dasen function in the wateRmelon package in R [Citation40]. The association between methylation at each of the CpG sites to be replicated and fetal intolerance was assessed using a linear regression. Pearson's correlation coefficients were calculated for each pair of CpG sites.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
supp_data_1411444.zip
Download Zip (414.6 KB)Acknowledgements
We thank the Emory Integrated Genomics Core for processing DNA and RNA samples. We thank Samantha Resin for technical assistance. We also are grateful to the participants enrolled in this cohort who provided samples and medical records for this study.
Additional information
Funding
References
- Chauhan SP, Magann EF, Scott JR, et al. Cesarean delivery for fetal distress: rate and risk factors. Obstetrical Gynecol Sur. 2003;58:337–350. doi:10.1097/01.OGX.0000066802.19138.AE.
- Schauberger CW, Chauhan SP. Emergency cesarean section and the 30-minute rule: definitions. Am J Perinatol. 2009;26:221–226. doi:10.1055/s-0028-1103033. PMID:19031352
- Bergmans MG. How to diagnose intrapartum fetal distress? Eur J Obstet Gynecol,Reproduct Biol.1998;79:119–121. doi:10.1016/S0301-2115(97)00271-6. PMID:9720826
- Penning S, Garite TJ. Management of fetal distress. Obstet Gynecol Clin North Am. 1999;26:259–74. doi:10.1016/S0889-8545(05)70073-5. PMID:10399760
- Liston R, Sawchuck D, Young D. Society of O, Gynaecologists of C, British Columbia Perinatal Health P. Fetal health surveillance: antepartum and intrapartum consensus guideline. J Obstet Gynaecol Can. 2007;29:S3–S56. doi:10.1016/S1701-2163(16)32615-9. PMID:17845745
- Maso G, Piccoli M, De Seta F, Parolin S, et al. Intrapartum fetal heart rate monitoring interpretation in labour: a critical appraisal. Minerva Ginecol. 2015;67:65–79. PMID:25411863
- Wood S, Ross S, Sauve R. Cesarean section and subsequent stillbirth, Is confounding by indication responsible for the apparent association? An updated cohort analysis of a large perinatal database. PloS one. 2015;10:e0136272. doi:10.1371/journal.pone.0136272. PMID:26331274
- Sperling LS, Henriksen TB, Ulrichsen H, et al. Indications for cesarean section in singleton pregnancies in two Danish counties with different cesarean section rates. Acta obstetricia et gynecologica Scandinavica. 1994;73:129–135. doi:10.3109/00016349409013415. PMID:8116351
- Benzouina S, Boubkraoui Mel M, Mrabet M, et al. Fetal outcome in emergency versus elective cesarean sections at Souissi Maternity Hospital, Rabat, Morocco. Pan Afr Med J. 2016;23:197. doi:10.11604/pamj.2016.23.197.7401. PMID:27347286
- Silberstein T, Sheiner E, Salem SY, et al. Fetal heart rate monitoring category 3 during the 2nd stage of labor is an independent predictor of fetal acidosis. The Journal of Maternal-fetal & Neonatal Medicine: the Official Journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2017;30(3):257–260.
- Jensen A, Garnier Y, Berger R. Dynamics of fetal circulatory responses to hypoxia and asphyxia. Eur J Obstet, Gynecol Reproduct Biol. 1999;84:155–172. doi:10.1016/S0301-2115(98)00325-X. PMID:10428339
- MacLennan AH, Thompson SC, Gecz J. Cerebral palsy: causes, pathways, and the role of genetic variants. Am J Obstet Gynecol. 2015;213:779–788. doi:10.1016/j.ajog.2015.05.034. PMID:26003063
- Roy KK, Baruah J, Kumar S, et al. Cesarean section for suspected fetal distress, continuous fetal heart monitoring and decision to delivery time. Indian J Pediatr. 2008;75:1249–1252. doi:10.1007/s12098-008-0245-9. PMID:19190880
- Uccella S, Colombo GF, Bulgheroni CM, et al. First-trimester maternal serum screening and the risk for fetal distress during labor. American journal of obstetrics and gynecology. 2009;201(166): e1–e6.
- Valino N, Giunta G, Gallo DM, et al. Biophysical and biochemical markers at 35–37 weeks' gestation in the prediction of adverse perinatal outcome. Ultrasound Obstet Gynecol the Official J Inter Soc Ultrasound Obstet Gynecol. 2016;47:203–209. doi:10.1002/uog.15663.
- Anderson CM, Ralph JL, Wright ML, et al. DNA methylation as a biomarker for preeclampsia. Biol Res Nurs. 2014;16:409–20. doi:10.1177/1099800413508645. PMID:24165327
- Wu P, Farrell WE, Haworth KE, et al. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics: Official J DNA Methylation Soc. 2016. Epub ahead of print. doi:10.1080/15592294.2016.1166321.
- Fuster DG, Alexander RT. Traditional and emerging roles for the SLC9 Na+/H+ exchangers. Pflugers Arch. 2014;466:61–76. doi:10.1007/s00424-013-1408-8. PMID:24337822
- Donowitz M, Ming Tse C, Fuster D. SLC9/NHE gene family, a plasma membrane and organellar family of Na(+)/H(+) exchangers. Mol Aspects Med. 2013;34:236–251. doi:10.1016/j.mam.2012.05.001. PMID:23506868
- Kumar PL, James PF. Identification and characterization of methylation-dependent/independent DNA regulatory elements in the human SLC9B1 gene. Gene. 2015;561:235–248. doi:10.1016/j.gene.2015.02.050. PMID:25701605
- Ye G, Chen C, Han D, et al. Cloning of a novel human NHEDC1 (Na+/H+ exchanger like domain containing 1) gene expressed specifically in testis. Mol Biol Rep. 2006;33:175–180. doi:10.1007/s11033-006-0010-y. PMID:16850186
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi:10.1038/ng.2653. PMID:23715323
- Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based Map Human Proteome Sci. 2015;347:1260419.
- Patel J, Landers K, Mortimer RH, et al. Regulation of hypoxia inducible factors (HIF) in hypoxia and normoxia during placental development. Placenta. 2010;31:951–7. doi:10.1016/j.placenta.2010.08.008. PMID:20869770
- Maltepe E, Saugstad OD. Oxygen in health and disease: regulation of oxygen homeostasis–clinical implications. Pediatric Res. 2009;65:261–268. doi:10.1203/PDR.0b013e31818fc83f. PMID:18852690
- Herrera EA, Krause B, Ebensperger G, et al. The placental pursuit for an adequate oxidant balance between the mother and the fetus. Front Pharmacol. 2014;5:149. doi:10.3389/fphar.2014.00149. PMID:25009498
- Murphy E, Eisner DA. Regulation of intracellular and mitochondrial sodium in health and disease. Circ Res. 2009;104:292–303. doi:10.1161/CIRCRESAHA.108.189050. PMID:19213964
- Omo-Aghoja L. Maternal and fetal Acid-base chemistry: a major determinant of perinatal outcome. Ann Med Health Sci Res. 2014;4:8–17. doi:10.4103/2141-9248.126602. PMID:24669324
- Gravett C, Eckert LO, Gravett MG, et al. Brighton Collaboration Non-reassuring fetal status Working G. Non-reassuring fetal status: Case definition & guidelines for data collection, analysis, and presentation of immunization safety data. Vaccine. 2016;34:6084–6092. doi:10.1016/j.vaccine.2016.03.043. PMID:27461459
- Sharma KJ, Esakoff TF, Guillet A, et al. Pregnancies complicated by both preeclampsia and growth restriction between 34 and 37 weeks' gestation are associated with adverse perinatal outcomes. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2016:1–4.
- Raicevic S, Cubrilo D, Arsenijevic S, et al. Oxidative stress in fetal distress: potential prospects for diagnosis. Oxidative Med Cell Longevity. 2010;3:214–218. doi:10.4161/oxim.3.3.12070. PMID:20716946
- Barfield RT, Kilaru V, Smith AK, et al. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28:1280–1281. doi:10.1093/bioinformatics/bts124. PMID:22451269
- Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics: Official J DNA Methylat Soc. 2013;8:203–209. doi:10.4161/epi.23470.
- Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. doi:10.1093/bioinformatics/bts680. PMID:23175756
- Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14:R115. doi:10.1186/gb-2013-14-10-r115. PMID:24138928
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformat. 2012;13:86. doi:10.1186/1471-2105-13-86. PMID:22568884
- Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS one. 2012;7:e41361. doi:10.1371/journal.pone.0041361. PMID:22848472
- Morris TJ, Butcher LM, Feber A, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–430. doi:10.1093/bioinformatics/btt684. PMID:24336642
- Guintivano J, Arad M, Gould TD, et al. Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Mol Psychiatr. 2014;19:560–567. doi:10.1038/mp.2013.62. PMID:23689534
- Pidsley R, CC YW, Volta M, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi:10.1186/1471-2164-14-293. PMID:23631413
