 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Cervical screening by high-risk HPV (hrHPV) testing requires additional risk stratification (triage), as most infections are transient and only a subset of hrHPV-positive women harbours clinically relevant disease. Molecular triage markers such as microRNAs (miRNAs) and DNA methylation markers are particularly promising, as they can be objectively tested directly on hrHPV-positive scrapes and cervicovaginal self-samples. Here, we evaluated the marker potential of 10 candidate miRNAs in 209 hrHPV-positive scrapes of women with underlying precancer (cervical intraepithelial neoplasia, grade 2–3 (CIN2-3)), cancer, or without disease (CIN0/1). A predictive miRNA classifier for CIN3 detection was built using logistic regression, which was compared to and combined with DNA methylation marker FAM19A4. Markers were correlated to histology parameters and hrHPV genotype. A miRNA classifier consisting of miR-149, miR-20a, and miR-93 achieved an area under the curve (AUC) of 0.834 for CIN3 detection, which was not significantly different to that of FAM19A4 methylation (AUC: 0.862, p = 0.591). Combining miRNA and methylation analysis demonstrated complementarity between both marker types (AUC: 0.939). While the miRNA classifier seemed more predictive for CIN2, FAM19A4 methylation was particularly high in HPV16-positive and histologically advanced CIN3, i.e. CIN3 with high lesion volume. The miRNA classifier, FAM19A4 methylation, and the miRNA/methylation combination were highest in cancer-associated scrapes. In conclusion, a panel of three miRNAs is discriminatory for CIN3 in hrHPV-positive scrapes and can complement DNA methylation analysis for the efficient detection of cervical disease. Combined analysis of the two marker types warrants further evaluation as triage strategy in hrHPV-based screening.
Introduction
Testing for high-risk human papillomavirus (hrHPV) has recently replaced cytology as a primary screening tool for cervical precancer and cancer in The Netherlands and parts of Italy. Other countries are soon to follow. While primary hrHPV testing has a higher sensitivity for the detection of cervical disease than cytology, it has a suboptimal specificity as it also detects clinically irrelevant, transient infections [Citation1,Citation2]. The identification of hrHPV-positive women who are in need of treatment, i.e. women with high-grade cervical intraepithelial neoplasia (CIN2 and CIN3) or cancer, therefore requires additional triage markers. Current triage strategies for hrHPV-positive cervical scrapes include cytology and more objective, molecular alternatives such as HPV16/18 genotyping and analysis of DNA methylation markers [Citation3]. Analysis of DNA methylation marker FAM19A4, for instance, has shown convincing results for the triage of hrHPV-positive cervical scrapes in the past [Citation4].
Analysis of differential microRNA (miRNA) expression for diagnostic and prognostic purposes has gained increasing attention in recent years. Others and we have previously shown that differential miRNA expression between normal and cervical (pre)cancer cannot only be detected in cervical tissue samples but also in cervical exfoliated cells present in cervical scrapes and self-collected cervico-vaginal specimens (self-samples) [Citation5–Citation8]. Although the feasibility of miRNA analysis for the triage of hrHPV-positive women has been demonstrated, an optimal miRNA panel for the detection of CIN3 and cervical cancer still remains to be determined [Citation6]. Moreover, a direct comparison between miRNAs and other molecular markers is currently missing.
In this study, the expression levels of 10 candidate miRNAs (miR-15b, miR-125b, miR-149, miR-203a, miR-375, miR-20a, miR-31, miR-93, miR-222, let-7b) derived from an earlier feasibility study on cervical scrapes and a study on hrHPV-positive self-samples [Citation6,Citation8] were determined by qRT-PCR in 209 cervical scrapes of hrHPV-positive women (age: 30–83 years) with known underlying histology that have not been tested for miRNAs before. A predictive miRNA classifier for the detection of CIN3 was built, which was applied to the full sample series and subsequently compared to and combined with the DNA methylation marker FAM19A4 [Citation4]. Markers were correlated to hrHPV genotype and histology parameters.
Results
miRNAs discriminate between CIN3 and CIN0/1 in hrHPV-positive cervical scrapes
To assess miRNA expression in cervical scrapes a study population obtained from an outpatient cohort of hrHPV-positive women that was previously tested for DNA methylation of FAM19A4 [Citation4] was used (n = 203). The study population was supplemented with 27 cervical cancer patients. The final study population consisted of 209 hrHPV-positive cervical scrapes, as 21 samples (9%) did not pass quality control (reference gene Ct value > 32, ).
Figure 1. Overview of the study population.
hrHPV, high-risk human papillomavirus; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; AC, adenocarcinoma.
Candidate miRNAs were selected as the clinically most promising triage markers from previous qRT-PCR studies performed on (1) hrHPV-positive scrapes (miR-125b, miR-149, miR-203a, and miR-375) or (2) hrHPV-positive self-samples (miR-20a, miR-31, miR-93, miR-222, let-7b) obtained from screening cohorts [Citation6,Citation8]. miR-15b was derived from both studies. miRNAs selected from (1) were previously identified using miRNA microarray analysis of cervical tissue samples and shown to be either genetically (miR-15b) or epigenetically (miR-125b, miR-149, miR-203a, miR-375) regulated [Citation9,Citation10], while miRNAs from approach (2) were identified following small RNA sequencing directly performed on hrHPV-positive self-samples [Citation8].
Supplemental Figure 1 shows the expression levels of the 10 candidate miRNA markers in the different biological groups, i.e. CIN0/1, CIN2, CIN3, squamous cell carcinoma (SCC), and adenocarcinoma (AC). All miRNAs, with the exception of miR-31, were significantly differentially expressed between the histological groups (Kruskal–Wallis: p < 0.05). Since the classification of CIN2 is ambiguous [Citation11] we compared CIN0/1 to CIN3. Eight miRNAs, i.e. miR-15b, miR-125b, miR-149, miR-203a, miR-375, let-7b, miR-93, and miR-222, showed significantly differential expression between CIN0/1 and CIN3 (Wilcoxon, Benjamini-Hochberg: p < 0.05). To evaluate the discriminatory power of individual miRNAs for CIN3, we next performed univariate logistic regression on data obtained from 77 CIN0/1 and 55 CIN3. Obtained AUCs ranged from 0.571 (miR-20a) to 0.743 (miR-125b, ). Robustness of the individual miRNAs was demonstrated by comparable AUCs achieved after leave-one-out cross-validation (LOOCV).
Table 1. Performance of individual miRNAs for the detection of CIN3 in cervical scrapes. Robustness of the individual miRNAs was evaluated using LOOCV.
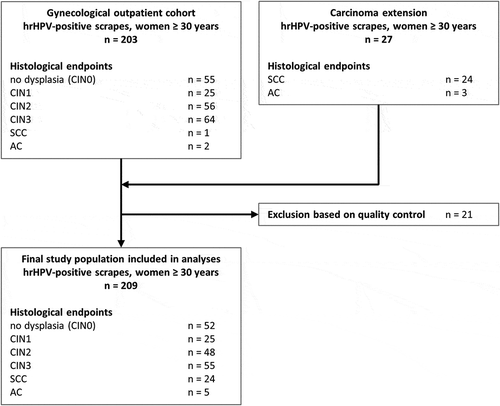
To identify the combination of miRNAs with the highest discriminatory power for CIN3, we performed multivariate logistic regression on data obtained from CIN0/1 and CIN3. The optimal miRNA classifier consisted of miR-149, miR-20a, and miR-93, and achieved an AUC of 0.834 (), which was higher than AUCs achieved by individual miRNAs. Following LOOCV, an AUC of 0.808 was obtained, indicating robustness of the miRNA classifier. Including more miRNAs in the classifier did not improve detection of CIN3 (data not shown). Applying the 3-miRNA classifier to the full sample series, predicted probabilities representing the risk of an underlying CIN3 were assigned to each sample. A clear increase in predicted probabilities with disease severity was observed (). Carcinoma samples, in particular, had very high predicted probabilities, indicating that the 3-miRNA classifier would also allow for the detection of cervical cancer even though these were not included in the building of the model.
Figure 2. Performance of the 3-miRNA classifier for the detection of cervical disease. (a) Results obtained from 77 hrHPV-positive scrapes from women without underlying disease (CIN0/1) and 55 scrapes from women with CIN3 were used to build a 3-miRNA classifier for the detection of CIN3. The diagonal line indicates an AUC of 0.5. (b) Predicted probabilities (i.e. risk of CIN3; value range 0 to 1) obtained for all samples using the 3-miRNA classifier.
AUC, area under the curve; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; AC, adenocarcinoma.
Combined analysis of miRNA expression and DNA methylation improves CIN3 detection
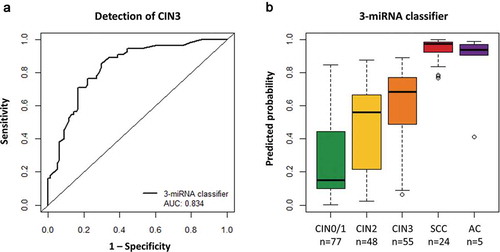
To allow for a direct comparison between the miRNA classifier and the DNA methylation marker FAM19A4, predicted probabilities were calculated for the previously obtained FAM19A4 methylation data [Citation4] using univariate logistic regression analysis. In the same set of samples, the AUC of FAM19A4 methylation analysis (0.862, LOOCV: 0.849) was not significantly different to that of the 3-miRNA classifier (DeLong: p = 0.591, ). We next evaluated the potential complementarity between miRNA and methylation analysis for the detection of CIN3. To this end, a classifier was built based on the three miRNAs from the 3-miRNA classifier and FAM19A4 methylation. Multivariate logistic regression was performed on data obtained from CIN0/1 and CIN3. In the combined model all three miRNAs and FAM19A4 methylation were retained. The model achieved an AUC of 0.939 (LOOCV: 0.918; ) and had a significantly better performance than the 3-miRNA classifier (DeLong: p = 0.0003) or FAM19A4 methylation alone (DeLong: p = 0.007). This indicates that miRNA and methylation markers are to a certain extent complementary and identify a slightly different subset of CIN3 lesions. When applying the combined miRNA/methylation classifier to the full sample series, including CIN2, SCC, and AC samples, the predicted probabilities were found to increase with cervical disease severity (). Compared to the 3-miRNA classifier (), the combined classifier generally assigned lower predicted probabilities to CIN0/1, while the majority of CIN3 lesions obtained very high predicted probabilities. This led to a better separation between the two groups, in line with the higher AUC. Cervical SCC and AC were characterized by predicted probabilities close to 1, indicating that a combined miRNA/methylation classifier is highly discriminatory for cervical carcinomas.
Figure 3. Performance of the combined miRNA/methylation classifier for the detection of cervical disease. (a) Results obtained from 77 hrHPV-positive scrapes from women without underlying disease (CIN0/1) and 55 scrapes from women with CIN3 were used to build a FAM19A4 methylation classifier [Citation4] and a combined miRNA/methylation classifier for the detection of CIN3. The diagonal line indicates an AUC of 0.5. There was no significant difference between the 3-miRNA classifier and FAM19A4 methylation analysis (DeLong: p = 0.591). The miRNA/methylation classifier performed significantly better than the 3-miRNA classifier (DeLong: p = 0.0003) and FAM19A4 methylation analysis (DeLong: p = 0.007). (b) Predicted probabilities (i.e. risk of CIN3; value range 0 to 1) obtained for all samples using the combined miRNA/methylation classifier.
AUC, area under the curve; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; AC, adenocarcinoma.
![Figure 3. Performance of the combined miRNA/methylation classifier for the detection of cervical disease. (a) Results obtained from 77 hrHPV-positive scrapes from women without underlying disease (CIN0/1) and 55 scrapes from women with CIN3 were used to build a FAM19A4 methylation classifier [Citation4] and a combined miRNA/methylation classifier for the detection of CIN3. The diagonal line indicates an AUC of 0.5. There was no significant difference between the 3-miRNA classifier and FAM19A4 methylation analysis (DeLong: p = 0.591). The miRNA/methylation classifier performed significantly better than the 3-miRNA classifier (DeLong: p = 0.0003) and FAM19A4 methylation analysis (DeLong: p = 0.007). (b) Predicted probabilities (i.e. risk of CIN3; value range 0 to 1) obtained for all samples using the combined miRNA/methylation classifier.AUC, area under the curve; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; AC, adenocarcinoma.](/cms/asset/222d7121-60a0-406b-8e2c-f0a2dcb1b68c/kepi_a_1600390_f0003_oc.jpg)
miRNA and methylation results in relation to clinical and viral parameters
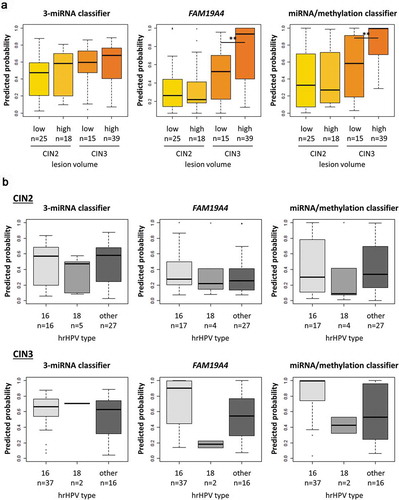
In an attempt to further compare the different classifiers, underlying CIN2 and CIN3 lesions were stratified (1) by lesion volume into low- and high-volume lesions [Citation12] () and () by hrHPV type into HPV16, HPV18, and other hrHPV types (). The predicted probabilities of the miRNA classifier were slightly increased in high-volume lesions compared to low-volume lesions in both CIN2 and CIN3 (). Predicted probabilities obtained with FAM19A4 methylation and the miRNA/methylation classifier, on the other hand, were significantly increased in the high-volume CIN3 lesions and no relation to lesion size was seen within CIN2 lesions. Stratification of CIN2 and CIN3 lesions by hrHPV type indicated a trend towards higher predicted probabilities for FAM19A4 and the combined miRNA/methylation classifier in cervical scrapes with HPV16-positive underlying CIN3 lesions compared to non-16 hrHPV type-associated CIN3 (). There were no clear differences between hrHPV types with the 3-miRNA classifier, FAM19A4, or the combined miRNA/methylation classifier in other histological groups (data not shown).
Figure 4. 3-miRNA classifier and FAM19A4 methylation in relation to lesion volume of CIN2 and CIN3 lesions and hrHPV genotype. Predicted probabilities obtained by the 3-miRNA classifier, FAM19A4 methylation analysis, or the combined miRNA/methylation classifier in hrHPV-positive cervical scrapes of women with CIN2/3, stratified by A lesion size (lesion size was not available for 5 CIN2 and 1 CIN3) and B hrHPV type. (Partial) hrHPV genotyping was performed on the same cervical scrape used for miRNA expression and FAM19A4 methylation analysis. ** p < 0.005.
CIN, cervical intraepithelial neoplasia.
Discussion
The use of molecular markers for the triage of hrHPV-positive women is objective and may allow for high-throughput analysis of large numbers of cervical samples. In this study, we evaluated 10 candidate miRNA triage markers and compared miRNA expression analysis to the DNA methylation marker FAM19A4. Logistic regression was used to build a 3-miRNA classifier with high discriminatory power for CIN3. We demonstrate that miRNA expression analysis is complementary to FAM19A4 methylation and that combining miRNA expression and DNA methylation analysis improves detection of CIN3.
In general, higher predicted probabilities were obtained in CIN2 using the 3-miRNA classifier compared to the combined miRNA/methylation classifier, whereas in CIN3 the reverse was seen. Further stratification of CIN2 and CIN3 lesions by lesion volume showed that the highest predicted probabilities were obtained in CIN3 lesions with a high volume when using FAM19A4 methylation. This is in line with previous data demonstrating the highest methylation levels in advanced CIN3 [Citation13]. Importantly, when combining miRNA and methylation analysis, both CIN2 and CIN3 lesions were captured. Stratification of CIN2 and CIN3 lesions by hrHPV type, moreover, indicated that methylation of FAM19A4 is particularly high in HPV16-associated CIN3. Interestingly, increased methylation levels of ASCL1, LHX8, and ST6GALNAC5 in HPV16-compared to other hrHPV type-associated CIN3 lesions and cervical carcinomas have recently been reported in a South African study cohort of women living with HIV [Citation14]. Hypermethylation of host cell genes in cervical carcinogenesis has been linked to HPV16 E6 and E7 mediated upregulation of the DNA methyltransferase DNMT1 [Citation15,Citation16]. The high carcinogenic potential of HPV16 may, therefore, relate to its capacity to activate DNMT1. Because studies on the effect of other hrHPV types on DNMT1 and other methyl transferases are currently missing, this notion warrants further investigation.
The 10 candidate miRNAs analysed in this study have previously been investigated as triage markers in either hrHPV-positive scrapes, hrHPV-positive self-samples, or both, and current data on 8 out of the 10 analysed miRNAs showed good concordance with previous results [Citation6,Citation8]. The optimal miRNA panel for detection of CIN3 consisted of miR-149, miR-20a, and miR-93. The most discriminatory individual miRNA marker (miR-125b) was not selected by our statistical analysis in the final miRNA classifier, which illustrates that combining a few complementary miRNAs has benefits for the detection of CIN3 over selecting the seemingly best miRNA markers.
In cervical cancer cell lines SiHa and HeLa, miR-149 has been shown to act as tumour suppressor by inhibiting proliferation and promoting apoptosis [Citation17]. Both miR-20a and miR-93 belong to the miR-17–92 family, a highly conserved, polycistronic cluster of miRNAs. MiRNAs belonging to the mir-17–92 have been associated to carcinogenesis, in particular by inhibition of apoptosis via E2F transcription factors, and are therefore referred to as oncomiR-1 cluster [Citation18,Citation19]. In cervical cancer, both miR-20a and miR-93 have been shown contribute to disease progression [Citation20–Citation24]. This indicates that the miRNAs in our 3-miRNA classifier are biologically relevant in cervical carcinogenesis.
Due to their short length, miRNAs are considered to be very stable molecules that can be successfully detected even in degraded RNA preparations [Citation25]. Analysis of differentially expressed miRNAs might, therefore, be especially promising in low-resource settings, where sample quality is compromised as a result of storage and transportation under suboptimal conditions. In our study, miRNA expression analysis of 10 targets and two reference genes was performed from low amounts of clinical material, i.e. 20 ng small RNA, and the invalid rate due to high reference gene Ct values was low (3%), highlighting the value of miRNA expression analysis in cervical scrapes. Considering that miRNA expression and DNA methylation analysis currently require two individual workflows, i.e. RNA isolation, reverse transcription, and qPCR or DNA isolation, bisulfite-conversion, and qMSP, respectively, we acknowledge that combined analysis of miRNAs and methylation markers in its current format is not practical. While simultaneous analysis of the transcriptome (mRNA) and DNA methylome is feasible at rather high costs for limited numbers of samples at present [Citation26,Citation27], advancing (targeted) sequencing techniques might allow for simultaneous analysis of miRNA expression and DNA methylation at moderate prices in the future.
A limitation of the study is that for the purpose of a direct comparison between miRNA and DNA methylation markers we used a selected series of cervical samples from women referred to a gynecological outpatient clinic, and only included CIN0/1 lesions with normal cytology as controls. Therefore, the results cannot be directly translated to a screening setting and further studies are warranted. Moreover, FAM19A4 was the only DNA methylation marker investigated. Recent studies, however, have described other promising DNA methylation markers for hrHPV-based screening that remain to be tested in combination with differentially expressed miRNAs [Citation28,Citation29].
In conclusion and to the best of our knowledge, this is the first study on combining miRNA and methylation markers for the detection of cervical disease. This study shows that miRNA expression can complement DNA methylation analysis for the efficient detection of cervical disease on hrHPV-positive cervical scrapes, and that combined analysis improves detection of CIN3 lesions. Our data suggest that molecular testing of cervical scrapes offers a promising triage strategy in hrHPV-based screening.
Materials & methods
Study population and HPV testing
The sample series included cervical scrapes of hrHPV-positive women between 30 and 70 years of age who participated in a gynecological outpatient study between December 2010 and December 2013. Detailed characteristics, inclusion criteria, and follow-up procedures have been described previously [Citation4]. FAM19A4 methylation status and HPV genotyping have previously been reported [Citation4]. The study cohort comprised 56 CIN2, 64 CIN3 (including 2 adenocarcinomas in situ (AIS)), 1 SCC, and 2 AC (including 1 adenosquamous carcinoma (ASC)), of which the corresponding hrHPV-positive cervical scrapes were included in this study (). Moreover, hrHPV-positive cervical scrapes of women without cervical disease (CIN0, n = 55) or with underlying CIN1 (n = 25), which had normal cytology (Pap1) and who were not referred to the outpatient clinic based on abnormal cytology in a previous scrape were included as controls. Absence or presence of underlying disease was histologically confirmed for all samples. Histological outcomes were based on colposcopy-directed biopsies, or, if classified worse, on the histology result of the specimen excised by LLETZ, conisation, or hysterectomy.
Additionally, we included hrHPV-positive scrapes of women with underlying SCC (n = 24) and AC (n = 3) who underwent treatment for cervical cancer at the Center for Gynecological Oncology Amsterdam (Amsterdam University Medical Center, location AMC and VUmc, and Antoni van Leeuwenhoek Hospital), Amsterdam, The Netherlands, between October 2016 and December 2017. These samples were tested for hrHPV using the HPV-Risk Assay (Self-screen B.V.), providing partial genotyping results [Citation30,Citation31].
Of the 230 hrHPV-positive cervical scrapes, 21 were excluded based on quality control for either miRNA expression analysis, DNA methylation analysis, or both (). The final study cohort therefore consisted of 209 scrapes comprised of 52 underlying CIN0 (median age: 39.5 years, range: 31–63), 25 CIN1 (median age: 35.0 years, range: 30–66), 48 CIN2 (median age: 34.5 years, range: 30–56), 55 CIN3 (including 2 AIS, median age: 35.0 years, range: 30–60), 24 SCC (median age: 48.5 years, range: 30–83), and five AC (including 1 ASC, median age: 50.0 years, range: 36–63).
RNA isolation and quantitative RT-PCR
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The Qubit® microRNA Assay kit was used to quantify small RNA concentrations on a Qubit® 2.0 Fluorometer (both Thermo Fisher Scientific).
Expression of hsa-let-7b-5p, hsa-miR-15b-5p, hsa-miR-125b-5p, hsa-miR-149-5p, hsa-miR-20a-5p, hsa-miR-203a-3p, hsa-miR-222-3p, hsa-miR-31-5p, hsa-miR-375, and hsa-miR-93-5p was measured using TaqMan microRNA assays (002619, 000390, 000449, 002255, 000580, 000507, 002276, 002279, 000564, 001090; Thermo Fisher Scientific). RNU24 and hsa-miR-423-3p were included as reference genes as these were previously found to be most stable in this sample type (001001, 002626; Thermo Fisher Scientific) [Citation6,Citation32].
Reverse transcription (RT) was multiplexed by combining specific RT primers for all targets as a primer pool. cDNA was synthesized from 20 ng small RNA template if available. For 37 samples the maximum possible amount (<20 ng) of RNA was used, of which 7 (19%) did not pass quality control. Each 16 µl reaction contained 6 µl primer pool, 0.3 µl dNTPs (100mM), 1.5 µl RT buffer, 0.19 µl RNase inhibitor (20 U/µl) and 3 µl MultiScribe Reverse Transcriptase (TaqMan microRNA Reverse Transcription kit, Thermo Fisher Scientific). Quantitative PCR reactions were performed on the ViiA7TM Real-Time PCR System (Thermo Fisher Scientific) in a 384-well format. Each 10 µl reaction consisted of 5 µl TaqMan® Universal Master Mix II, 0.5 µl miRNA specific TaqMan assays (both Thermo Fisher Scientific), 3.5 µl H2O and 1 µl cDNA. Cycle conditions for cDNA synthesis and PCR were used according to the manufacturer’s protocols.
Data were normalized to the geometric mean Ct of RNU24 and hsa-miR-423-3p applying the method [Citation33]. Samples with a geometric mean Ct of RNU24 and hsa-miR-423 ≥ 32 did not pass quality control.
DNA isolation and FAM19A4 methylation analysis
Cervical scrapes originating from an outpatient cohort had previously been tested for FAM19A4 methylation [Citation4]. Data were normalized to ACTB using the method [Citation33]. To ensure sample quality, samples with an ACTB Ct value ≥32 were excluded from the final study cohort. For the additional cervical scrapes from cervical cancer patients, DNA was extracted using the NucleoMag 96 Tissue kit (Macherey-Nagel) and a Microlab Star robotic system (Hamilton) according to the manufacturer’s protocol. Extracted DNA was subjected to bisulphite-conversion using the EZ DNA Methylation Kit (Zymo Research). For FAM19A4 methylation analysis, the QIAsure Methylation Test (Self-screen B.V.) was used.
Statistical analysis
To compare miRNA expression levels between CIN0/1, CIN2, CIN3, SCC, and AC, we performed an omnibus Kruskal–Wallis test followed by a Wilcoxon rank test with a significance level of 0.05 (two-sided) when the omnibus test showed a significant result (p < 0.05). P-values from the post-hoc test were corrected with the Benjamini-Hochberg correction method. Because screening programs are designed to identify and treat high-grade CIN to prevent cervical cancer, logistic regression was performed on square root transformed delta Ct ratios obtained from CIN0/1 (controls) and CIN3 lesions (cases). CIN2 lesions were not included in logistic regression analysis, as their classification is ambiguous [Citation11]. Univariate logistic regression was performed to evaluate the discriminatory power of individual miRNAs and FAM19A4 methylation analysis. Multivariate logistic regression followed by backward elimination was used to identify the optimal multi-miRNA classifier and to build a combined miRNA/methylation classifier for the detection of CIN3. As a result of univariate and multivariate logistic regression, predicted probabilities, i.e. values between 0 and 1 representing the risk of an underlying CIN3, were assigned to each sample. Receiver-operated characteristic (ROC) curve analysis was carried out to evaluate the performance of the miRNA classifiers in detecting CIN3. Robustness of the classifiers was evaluated using leave-one-out cross-validation (LOOCV). For comparison of the obtained AUCs DeLong’s test for two correlated ROC curves was used [Citation34]. The resulting multi-marker classifiers were subsequently applied on data obtained from CIN2 and carcinoma samples.
Stratification of CIN2 and CIN3 lesions
In an attempt to stratify CIN2 and CIN3 lesions in relation to lesion size, a rough estimation of the lesion volume (low/high) was used [Citation12]. In short, a biopsy diagnosed as a CIN2 or CIN3 lesion was classified as high-volume if the histological outcome of the corresponding LLETZ (or conisation/hysterectomy) specimen was ≥CIN2 or ≥CIN3, respectively. A biopsy diagnosed as a CIN2 or CIN3 lesion was considered low-volume if the paired LLETZ had a lower histological outcome, i.e. ≤CIN1 or ≤CIN2, respectively. Biopsy specimens of high-grade CIN for which no LLETZ was performed (according to physician’s advice) were classified as low-volume. Moreover, CIN2 and CIN3 lesions were also stratified according to hrHPV genotypes. For this purpose, multiple infections including HPV16 or HPV18 were categorized as HPV16-positive or HPV18-positive, respectively, and multiple infections containing both HPV16 and HPV18 were assigned to the HPV16-positive group [Citation35].
Ethical approval and consent to participate
The outpatient clinic study was approved by the Medical Ethical Committee of all participating hospitals (METc-VUmc-2009/178) and registered in the Dutch National Trial Registry (NTR2447). Collection of additional carcinoma samples was approved by the Medical Ethical Committee of all participating hospitals (METc-VUmc- 2016/213). All participants gave informed consent.
Supplemental Material
Download PDF (199 KB)Acknowledgments
This work is dedicated to our colleague Prof.dr. P.J.F. Snijders, who passed away on 27 May 2018. The authors are indebted to A.P. van Splunter for excellent technical assistance and our colleagues from the Center for Gynecological Oncology Amsterdam for sample collection.
Disclosure statement
RDMS, CJLMM, and DAMH are minority stakeholders of Self-screen B.V., a spin-off company of VU University medical center; and since September 2017 CJLMM is director of Self-screen B.V., which holds patents related to the work. CJLMM owns a small number of shares of Qiagen, has occasionally been consultant for Qiagen and until April 2016 was a minority shareholder of Diassay B.V. DAMH occasionally serves on the scientific advisory board of Pfizer and Bristol-Meyer Squibb and has been on the speakers’ bureau of Qiagen. RDMS and CJLMM are inventors on a patent application on the use of miRNAs for cervical cancer detection. No potential conflicts of interest were disclosed by the other authors.
Supplementary material
Supplementary data can be accessed here.
Additional information
Funding
References
- Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30(Suppl 5):F88–F99.
- Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383:524–532.
- Luttmer R, De Strooper LMA, Steenbergen RDM, et al. Management of high-risk HPV-positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn. 2016;16:961–974.
- Luttmer R, De Strooper LMA, Berkhof J, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high-risk HPV-positive women of a gynecologic outpatient population (COMETH study). Int J Cancer. 2016;138:992–1002.
- Tian Q, Li Y, Wang F, et al. MicroRNA detection in cervical exfoliated cells as a triage for human papillomavirus – positive women. J Natl Cancer Inst. 2014;106:dju241.
- Babion I, Snoek BC, Novianti PW, et al. Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study. Clin Epigenetics. 2018;10:76.
- Ivanov M, Titov S, Glushkov S, et al. Detection of high-grade neoplasia in air-dried cervical PAP smears by a microRNA-based classifier. Oncol Rep. 2018;39:1099–1111.
- Snoek BC, Verlaat W, Babion I, et al. Genome-wide microRNA analysis of HPV-positive self-samples yields novel triage markers for early detection of cervical cancer. Int J Cancer. 2019;144:372–379.
- Wilting SM, Snijders PJF, Verlaat W, et al. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2013;32:106–116.
- Wilting SM, Verlaat W, Jaspers A, et al. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics. 2013;8:220–228.
- Herrington CS. The terminology of pre-invasive cervical lesions in the UK cervical screening programme. Cytopathology. 2015;26:346–350.
- Luttmer R, De Strooper LMA, Dijkstra MG, et al. FAM19A4 methylation analysis in self-samples compared with cervical scrapes for detecting cervical (pre)cancer in HPV-positive women. Br J Cancer. 2016;115:579–587.
- De Strooper LMA, Meijer CJLM, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila). 2014;7:1251–1257.
- Kremer W, van Zummeren M, Heideman D, et al. HPV16-related cervical cancers and precancers have increased levels of host cell DNA methylation in women living with HIV. Int J Mol Sci. 2018;19:3297.
- Burgers WA, Blanchon L, Pradhan S, et al. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655.
- Au Yeung CL, Tsang WP, Tsang TY, et al. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–1604.
- Qian B, Zhao L, Wang X, et al. miR-149 regulates the proliferation and apoptosis of cervical cancer cells by targeting GIT1. Biomed Pharmacother. 2018;105:1106–1116.
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134.
- Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18:262–267.
- Wu S, Huang S, Ding J, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308.
- Honegger A, Schilling D, Bastian S, et al. Dependence of intracellular and exosomal microRNAs on Viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11:e1004712..
- Zhang J, Wang F, Xu J, et al. Micro ribonucleic acid-93 promotes oncogenesis of cervical cancer by targeting RAB11 family interacting protein 1. J Obstet Gynaecol Res. 2016;42:1168–1179.
- Liu X. Up-regulation of miR-20a by HPV16 E6 exerts growth-promoting effects by targeting PDCD6 in cervical carcinoma cells. Biomed Pharmacother. 2018;102:996–1002.
- Zhang X, Li F, Zhu L. Clinical significance and functions of microRNA-93/CDKN1A axis in human cervical cancer. Life Sci. 2018;209:242–248.
- Jung M, Schaefer A, Steiner I, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006.
- Hu Y, Huang K, An Q, et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17:88.
- Macaulay IC, Ponting CP, Voet T. Single-Cell Multiomics: multiple Measurements from Single Cells. Trends Genet. 2017;33:155–168.
- Verlaat W, Snoek BC, Heideman DAM, et al. Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin Cancer Res. 2018;24:3456–3464.
- van Leeuwen RW, Oštrbenk A, Poljak M, et al. DNA methylation markers as a triage test for identification of cervical lesions in a high risk human papillomavirus positive screening cohort. Int J Cancer. 2019;144:746–754.
- Hesselink AT, Berkhof J, van der Salm ML, et al. Clinical validation of the HPV-risk assay, a novel real-time PCR assay for detection of high-risk human papillomavirus DNA by targeting the E7 region. J Clin Microbiol. 2014;52:890–896.
- Polman NJ, Oštrbenk A, Xu L, et al. Evaluation of the clinical performance of the HPV-risk assay using the VALGENT-3 panel. J Clin Microbiol. 2017;55:3544–3551.
- Babion I, Snoek BC, van de Wiel MA, et al. A strategy to find suitable reference genes for miRNA quantitative PCR analysis and its application to cervical specimens. J Mol Diagnostics. 2017;19:625–637.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408.
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845.
- IARC Working. Group on the evaluation of carcinogenic risks to humans. biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1–441.
