ABSTRACT
Growth is a complex trait whose variability within a population cannot be explained solely by genetic variation. Epigenetic regulation is often suggested as an important factor shaping the phenotype, but its association with growth can be highly context- and species-dependent. Nevertheless, the mechanisms involved in epigenetic regulation of growth in fish are poorly understood. We have used reduced representation bisulphite sequencing to determine the genome-wide CpG methylation patterns in male and female Nile tilapia of different sizes but at the same early stage of domestication. The average CpG methylation level in the reduced genome representation was 63% across groups but many sites displayed group-specific methylation patterns. The number of differentially methylated (DM) CpGs was much higher when the interaction between sex and weight was included rather than when these factors were considered separately. There were 1128 DM CpGs between large and small females and 970 DM CpGs between large and small males. We have found many growth-related genes associated with DM CpGs, namely map3k5 and akt3 in females and gadd45g and ppargc1a in males. Only 5% of CpG locations associated with growth were common to both sexes. In particular, the autophagy-related gene atg14 displayed a high association of methylation with growth exclusively in males. The sexually dimorphic association between atg14 methylation and growth may uncover novel metabolic mechanisms at play during mouth brooding in Nile tilapia females. Taken together, our data suggest that epigenetic regulation of growth in Nile tilapia involves different gene networks in males and females.
Background
Growth in fish is a polygenic and complex trait, which involves a panoply of pathways and tissues [Citation1] and is influenced by the interaction with other fitness-related traits [Citation2]. The role of abiotic and biotic factors such as temperature, water current, density, diet, and social interactions in shaping fish growth or condition factor is well established, with ample experimental evidence from wild and farmed fish populations [Citation3–Citation5]. In wild populations, in particular, stochastic conditions such as food availability or disease outbreaks and additional selective pressure (e.g., predation) can lead to expression of sub-optimal growth rates or prevent accurate predictions of growth [Citation6]. In contrast, captive populations benefit from relaxed natural selection and therefore can exhibit wider phenotypic variation than their wild conspecifics [Citation7]. First, low mortality in aquaculture conditions allows for survival of phenotypes that would not persist in the wild [Citation8]. Second, specific conditions of the controlled rearing environment (food, density, temperature) are often aimed at promoting growth rate, i.e., allowing individuals with high growth potential a less restricted expression of this trait. Relaxed natural selection, when leading to increased phenotypic variation [Citation9], can help the subsequent artificial selection and/or rapidly occurring adaptation to captivity, which can be seen as one of the first steps in the domestication process [Citation10].
The initial stage of domestication represented by a parental generation (F0) produced in the wild but reared under farmed conditions is a unique model for studying growth mechanisms. It represents a particular case of reaction norms generated by a new and highly controlled environment, where individuals benefit from reduced selective pressure from the wild but are not yet affected by artificial selection. Indeed, man-assisted selection such as breeding of fast growers is often directional and stabilizing, thus domestication-related changes in the subsequent generations will be rather associated with relocation and/or flattening of reaction norms of selected and co-selected traits [Citation11].
Stable changes in phenotypic expression can be extremely fast under captivity conditions, even within a single generation of domestication [Citation12] and do not always require neo-Darwinian modulation of allelic frequencies [Citation13]. Epigenetic changes such as DNA methylation, i.e., potentially heritable modifications of chemical structure of genome without affecting its nucleotide composition, can be an important mechanism shaping phenotypic differences occurring within the same generation. Cytosine methylation is among the most studied epigenetic marks [Citation14] and it has recently been associated with hatchery rearing in coho salmon (Oncorhynchus kisutch) [Citation15]. Global changes in DNA methylation have been observed in Nile tilapia (Oreochromis niloticus) gonads after temperature treatment [Citation16] and it has been shown that differential cytosine methylation may contribute to sex-specific growth phenotypes in hybrid tilapia (O. mossambicus × O. niloticus) [Citation17]. Differences in cytosine methylation of the myogenin gene have also been linked to thermal plasticity of growth in Atlantic salmon (Salmo salar) and Senegalese sole (Solea senegalensis) [Citation18,Citation19]. Therefore, a better knowledge on phenotype-associated methylome modifications may have key implications in fish epigenetic programming in the future [Citation20].
In this study, we assessed the size-related differences in genome-wide methylation status between F0 generation male and female Nile tilapia (O. niloticus), which were born in the wild and reared under hatchery conditions. Growth of this important aquaculture species is a highly plastic [Citation21] and heritable trait [Citation22]. Thus, detecting methylation changes related to extreme growth levels can undercover new regulatory mechanisms of myogenesis and provide potential epigenetic biomarkers of such trait. Moreover, due to the magnitude of sexual size dimorphism in Nile tilapia (with males being considerably larger than females) [Citation23], we hypothesized that there would be sex-specific differences between methylation profiles in both large and small individuals. By using the wild-born but hatchery-reared individuals, we are simulating the first steps of domestication, i.e., acclimation to farmed conditions and expression of reaction norms that are not necessarily attained in wild conditions. Such a less restricted expression of growth potential allows us to get insight into potential epigenetically regulated functions specific to growth and to sex in fish.
Results
Experimental groups and library characterization
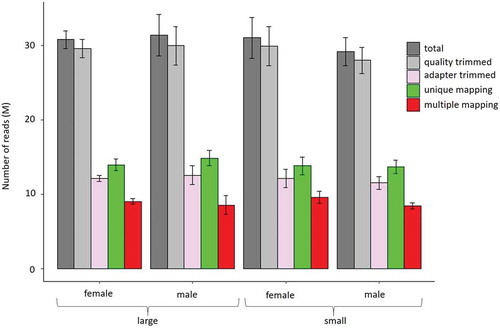
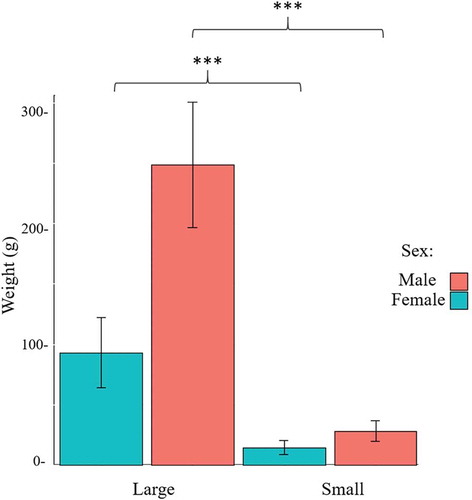
Within full-sib groups reared in common garden, the large individuals were 8.7- and 5.7-fold heavier than their small counterparts, in the case of males and females, respectively (). A mean of 30 M raw reads was obtained per library, of which 15 M were uniquely mapped (i.e., 50% mapping efficiency) and an additional 8 M reads could be multiple mapped (, Supplementary Table 1). Neither individual samples nor compared groups differ significantly with respect to their raw, trimmed, or mapped reads. The mean CpG coverage of reads mapping uniquely to the fraction of the genome that was successfully aligned with reads is 31-fold or higher (Supplementary Figure 2). Absence of a peak at the right side of the histogram indicates that the data did not contain polymerase chain reaction (PCR) duplicates.
Figure 1. Average weight of Nile tilapia, according to sex and weight (n = 6). Large females were statistically heavier than small females (t-test, p-value <0.001) and lighter than large males (t-test, p-value <0.001), while small males were statistically lighter than large males (t-test, p-value <0.001) and heavier than small females (t-test, p-value <0.001).
The fast muscle methylome in Nile tilapia
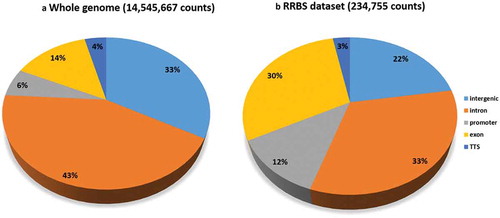
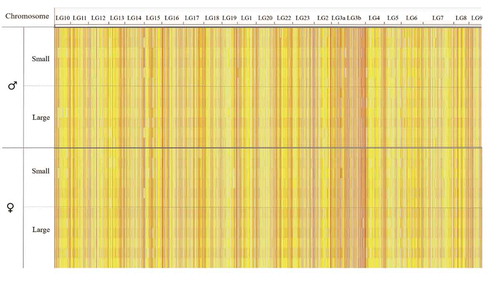
The global cytosine count corresponded to 244 M reads with a fold coverage was 3.2. Average methylation corresponded to 63% (), mean methylation levels per chromosome were homogeneous (Supplementary Table 2), and the distribution of methylated cytosines was similar across all samples (). However, methylation levels differed between chromosomes, namely between chromosome LG3b and LG4 (). In addition, the genomic context of the RRBS dataset differed from that in the whole Nile tilapia genome (). Indeed, promoter-annotated cytosines were enriched twofold in the reduced representation genome compared to whole genome. A twofold increase in CpG sites found in the RRBS dataset compared to the whole genome was also observed for exon regions.
Table 1. Global cytosine methylation level in CpG, CHG, and CHH contexts; number of cytosines analysed and cytosine coverage with respect to the whole Nile tilapia genome.
Figure 3. Heat map of methylation levels of 234,755 CpG sites distributed across the Nile tilapia linkage groups (columns) and separated in sex- and weight-specific groups (rows, n = 6). Methylation levels below 40%, between 40% and 60%, and above 60% are indicated in blue, yellow, and red, respectively.
Sex- and weight-specific differentially methylated (DM) cytosines
Unequal distribution of DM CpG sites between sexes
A total of 234,755 CpG positions that were covered in every sample at least once were used for analysis of differential methylation between sexes. Interestingly, neither genomic context () nor chromosomal distribution (Supplementary Table 3, Supplementary Figure 3) were similar between the sets of hyper- and hypomethylated CpG sites. For example, transcription termination sites were represented by 10% of hypomethylated CpG sites in males but absent in hypermethylated CpG sites.
Figure 5. Genomic context of sex-specific differentially methylated cytosines (hypo- and hypermethylated). Twelve fish per sex and 234,755 cytosine positions in total were compared.
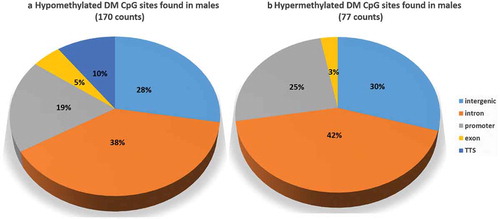
Moreover, compared to the CpG sites available in the RRBS dataset () there was an approximately twofold increase in the proportions of all sex-related DM CpG sites () found in promoters and a sixfold decrease in those found in exons. In general, the distribution of DM CpG sites across the genome was very heterogeneous (Supplementary Table 3). shows the genes that are associated with DM CpGs and that are functionally relevant to muscle-related or epigenetics-related processes. Out of a set of selected genes connected to DM CpG between males and females, the gene encoding for atg14 (autophagy-related protein 14) is linked with four exonic CpG sites, all hypomethylated in males. Other hypomethylated DM CpG sites in males were associated with hdac11 (histone deacetylase 11), tgfb2 (transforming growth factor beta 2), and lepr (leptin receptor) and one hypermethylated CpG site is connected with itga4 (integrin subunit alpha 4).
Table 2. List of functionally relevant genes associated with CpG sites that are differentially methylated between sexes in Nile tilapia.
Substantial differences in CpG methylation between large and small individuals of the same sex
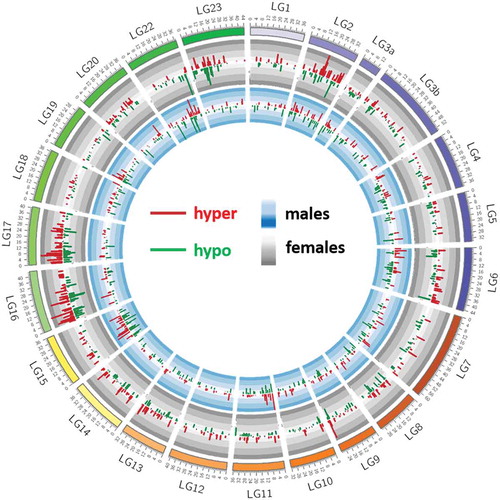
Out of approximately 300k CpG locations, a total of 1128 and 970 DM CpG positions (false discovery rate (FDR) <1%, difference >25%) were found in large individuals when compared to small ones within females and males, respectively (Supplementary Table 4a). The overall genomic context in both sets of DM CpG positions was similar (Supplementary Figure 4). However, the distribution of DM CpG sites across chromosomes differed between male- and female-specific comparisons (). Chromosomes 2, 15, and 16 are the most enriched in DM CpGs in females, and chromosomes 2, 3a, 15, and 22 in males (). In males, chromosomes with the highest number of DM CpG sites also contained the sites with the highest values of differential methylation, particularly hypermethylated sites. The shortest linkage group (LG3a) contains one of the highest proportions of DM CpG per nucleotide, whereas LG3b shows one of the lowest proportions of DM CpG per nucleotide.
Figure 6. Chromosomal distribution of weight-specific differentially methylated CpG sites in females (1128 positions) and males (970 positions). Small individuals are set as reference for both sexes separately. Methylation differences in males are represented in the inner circle (blue background) and those of females in outer circle (grey background). Histograms pointing inwards (dark green) and outwards (dark red) represent hypomethylated and hypermethylated sites, respectively.
A list of genes associated with growth-related DM CpG loci in females (Supplementary Table 4a) and males (Supplementary Table 4b) was created based on their functional relevance to muscle- or epigenetics-related processes. Among the selected genes associated with DM CpG in females, mettl9 (methyltransferase-like protein precursor 9) is associated with the highest number of DM CpG (five in total for variant X4 and X8 of mettl9), all hypomethylated in large females and close to the transcription start site (TSS) of that gene. Their frequency ratio (number of DM CpG linked with the specific gene/number of all CpG sites related to the gene and available in the RRBS data) was higher than 50%. A gene encoding for ppp1r3c (protein phosphatase 1 regulatory subunit 3C) was represented by 2 DM CpG sites, one hypomethylated and situated in exon and another one hypermethylated and situated in intergenic region. Similarly, histone deacetylase gene (hdac11) was also represented by two DM CpG, one hypo- and one hypermethylated. We observed hypermethylation in exonic regions of gata6 and xpo5 (exportin 5), in intergenic regions close to dmap1 (DNA methyltransferase 1-associated protein) and intronic region of ranbp2, encoding for a SUMO E3 ligase (RAN binding protein 2). Two genes, map3k5 (mitogen-activated kinase kinase kinase 5, known also as ask1) and esrrg (encoding for oestrogen-related receptor gamma), were affected by hypermethylation of multiple CpG sites (five and four, respectively) in their promoter regions, very close to their TSS. In contrast, CpG hypomethylation was detected in introns of igf2bp2 (insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2)), dner (delta/notch like EGF repeat containing gene), and itgbl2 (integrin beta-like 2), as well as in the intergenic region close to prlr (prolactin receptor).
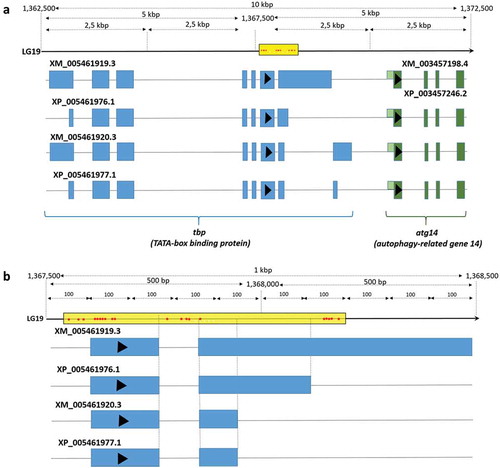
In males, the highest number of DM CpG was associated with atg14 (autophagy-related gene 14). This gene was represented by 20 DM CpG (87% of all CpG sites associated with atg14 and available in the RRBS dataset), all hypomethylated in large males and situated in introns or exons of tbp2 (TATA-box binding protein 2). Other hypomethylated CpG sites were associated with ppargc1a (peroxisome proliferator-activated receptor-γ coactivator-1α, also called PGC-1alpha), in the promoter region of gdf3 (growth differentiation factor 3) and in the third exon of gadd45a (growth arrest and DNA damage-inducible protein GADD45 gamma-like). We also observed hypomethylation of five CpG sites situated in the promoter region of hadh (hydroxyacyl-CoA dehydrogenase) and two CpGs located in the exon region of phf20 gene (PHD finger protein 20), as well as hypermethylation of CpG sites associated with gata6 and xpo5.
Only a few growth-related DM CpG sites are shared between males and females
Out of approximately 1000 possible DM CpG locations, only 57 (5.7%) were common to both sexes. Among them, 28 showed the same methylation changes in both sexes (i.e., if hypermethylated in big females, they were also hypermethylated in big males) (Supplementary Figure 5, Supplementary Table 4b). Surprisingly, the remaining (29) common DM CpG locations showed opposite patterns of methylation associated with growth (Supplementary Table 4b). Several genes common for female- and male-specific analysis were selected based on their functional relevance to muscle- or epigenetics-related processes () and showing similar or opposite patterns of methylation related to growth. Among them, the aforementioned gata6 was related to hypermethylated DM CpGs in both large males and large females. In contrast, xpo5, a gene encoding for Exportin 5, was associated with a CpG site significantly hypermethylated in large males, but hypomethylated in large females.
Table 3. List of functionally relevant genes associated with weight-related DM CpG sites that are found separately in both sexes in Nile tilapia.
The comparison between large and small fish yields few DM CpGs when sex information is omitted
A total of 117 DM CpG sites were found when the 12 largest individuals were compared to the 12 smallest individuals from both sexes (Supplementary Table 4a). Their distribution was uneven and different from sex-specific DM CpG distribution (Supplementary Figure 6).
shows a subset of weight-specific CpG sites associated with genes involved in growth in fish. Gata6 contained the highest number of DM CpG sites (4), all exonic and hypermethylated. A gene encoding for igf2bp2 (IGF2BP2) was associated with one intronic hypermethylated DM CpG, and itgbl2 was associated with one intergenic hypomethylated CpG. The first gene was also identified in the size-related comparison within females, whereas the second was found both within females (Supplementary Table 5a) and males (Supplementary Table 5b). Only a few unique genes were associated with DM CpG sites in this group, namely cdh2 (cadherin 2) and hadh, a gene that was previously found to be size-related in males.
Table 4. List of functionally relevant genes associated with DM CpG sites related to growth in Nile tilapia.
A large number of DM CpG-associated genes are related to growth or involved in epigenetic regulation
Although Functional Annotation analysis did not yield any significant enrichment (Supplementary Table 6), many manually annotated genes were functionally related to growth and epigenetic processes (). In the comparison between males and females, we highlighted hdac11, a gene coding for a key epigenetic histone modification factor (histone deacetylase) that is capable of decreasing myoD-dependent transcription [Citation24]. It is important to notice that histone deacetylases were also found in other comparisons. Among other sex-related and DM CpG-associated genes, lepr can have a role in proliferation and differentiation of skeletal myoblasts [Citation25] whereas itga4 is involved in myogenesis [Citation26]. Muscle atrophy was a common function of tgfb2, a transforming growth factor [Citation27], and of atg14, from a family of autophagy-related genes involved in muscle degradation [Citation28,Citation29]. Interestingly, the last gene was also associated with many DM CpGs between large and small males, but not between large and small females.
Figure 7. Schematic representation of genes associated with differentially methylated CpGs in the comparison between males (a), females (b), growth (c), or sex (d) and that play a role in muscle growth or epigenetic regulation processes. Genes in black are specific to one comparison, whereas genes in red are associated with differentially methylated CpGs in more than one comparison. In the male-specific comparison, gadd45g is connected to several other differentially methylated genes. Gadd45g activates the p38MAPK pathway, which is important in muscle growth and regeneration.
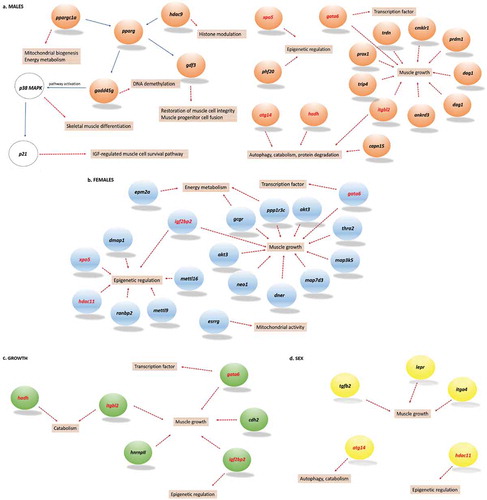
In females, a comparison between large and small fish showed DM CpG-associated genes whose function was related to growth, myogenesis, or mitochondrial activity (Supplementary Table 4a). For example, ppp1r3c is involved in glycogen metabolism [Citation30], itgbl2 (Integrin subunit beta-like 2) might contribute to muscle hypertrophy [Citation31], dner (Delta and Notch-like epidermal growth factor-related receptor) plays a role in signalling in muscle stem cells [Citation32], neo1 (encoding Neogenin) promotes myotube formation and regulates myofiber size [Citation33], map3k5 (Mitogen Activated Protein Kinase Kinase Kinase 5) promotes myogenic differentiation [Citation34], esrrg regulates mitochondrial activity [Citation35], and igf2bp2 (Insulin-like growth factor 2 binding protein 2) is involved in myoblast proliferation and myogenesis [Citation36]. The latter can also be involved in epigenetic regulation [Citation37]. Epigenetic processes were also highlighted by other genes, such as xpo5 involved in transfer of miRNA from nucleus to cytoplasm [Citation38] and dmap1 involved in global maintenance methylation [Citation39]. In addition, ranbp2 can modify histone deacetylase HDAC4 [Citation40].
A male-specific comparison between large and small fish showed similar genes related to epigenetic regulation (Supplementary Table 4b), such as hdac9, xpo5, and phf20, a gene encoding for PHD finger protein 20 and involved in histone acetylation [Citation41]. Many DM CpG sites were associated with atg14, but situated in the last introns and exons of the vertebrate-specific tbp2 gene [Citation42] () encoding for TATA-box binding protein 2, required for transcription initiation of promoters that contain a TATA-box [Citation42]. Other growth-related and DM CpG-associated genes found in the male-specific comparison were hadh involved in catabolism, gadd45g (growth arrest and DNA damage-inducible protein gamma-like) causing muscle atrophy [Citation43], and gdf3 and ppargc1a (encoding for peroxisome proliferator-activated receptor gamma) both involved in muscle metabolism [Citation44,Citation45].
Figure 8. Location at 10k (a) and 1k (b) base pair resolution of 20 DM CpG sites situated within introns and exons of tbp2 (TATA-box binding protein 2) and associated with the transcription starts site (TSS) of atg14 (autophagy-related gene 14).
The comparison between large and small fish regardless of sex () revealed a growth-related gene cdh2 (encoding cadherin 2) that promotes skeletal muscle differentiation [Citation46] and was only identified in this comparison. In addition, the transcription factor gata6 was found both in size- and sex-specific comparisons between large and small fish (), and is involved in regulation of growth in smooth muscle [Citation47].
Discussion
In this study, we report the reduced representation profiling of the fast muscle methylome in Nile tilapia groups with different growth rates. All fish were first generation of captive-reared descendants of wild parents; therefore, their phenotypic expression is on the one hand unrestricted by selective pressures present in wild but absent in hatchery conditions, and on the other hand not yet affected by artificial selection due to selective breeding. To the best of our knowledge, this is the first single-cytosine resolution study of methylation in fast muscle of Nile tilapia in the context of sex, growth, and the early domestication process in fish. Previous studies on genome-wide Nile tilapia methylation were performed on gonadal tissue and at a lower resolution using MeDIP technique, then bisulphite treatment of a restricted number of genes [Citation48]. To date, there is only one report on genome-wide methylation patterns in tilapia [Citation17], suggesting that DNA methylation in skeletal muscle contributes to sex-specific phenotypes. However, this whole genome bisulphite sequencing study focused on a hybrid tilapia species (O. niloticus × O. mossambicus) and the analysis was performed at the level of DM regions, which fails to detect unique but highly DM cytosines and to annotate them separately to specific genes. Importantly, it focused only on sex-specific differences, without taking into account the growth/size of the fish, which is known to be sexually dimorphic in most tilapia species. Therefore, some differences in methylation specifically due to size might have been mistakenly highlighted as sex-specific. In contrast, our study established a multiple factor analysis of methylation data at single nucleotide resolution for sex and growth in combination and independently. Some genes or gene families found to be associated with differential methylation between sexes in hybrid tilapia [Citation17] were also found in Nile tilapia, but not necessarily with the same patterns of methylation. For example, a member of solute carrier family 22 (Supplementary Table 4a) previously found to be hypermethylated in males compared to females was linked in the present study to two hypermethylated CpGs only in large females compared to smaller ones. A gene from the family of histone deacetylases, hdac4, was associated with a hypermethylated region in female hybrid tilapia compared to males [Citation17]. In our study, we detected two histone deacetylases, hdac9 (with hypermethylated CpG sites in large males compared to small males) and hdac11. The latter was associated with hypomethylated CpG in males compared to females, both in hybrid tilapia and in Nile tilapia, but it was also linked to two hypermethylated CpG sites in large females compared to small ones in the present study. It is important to keep in mind that lack of detection of a specific gene by RRBS does not mean that differences in methylation do not exist, since the data represent only a fraction of the whole genome. Nevertheless, our study represents a complementary and more accurate insight into sex- and growth-specific methylation patterns and provides a list of DM CpG-associated genes relevant to muscle growth and epigenetic processes, found specifically or commonly to growth, sex, or interaction between both (). As the fish were reared in the same tank, we cannot exclude the possibility that differences in growth rate may be attributed to nutrient condition and feeding behaviour, as well as epigenetic changes.
Our results indicate that global methylation levels in Nile tilapia fast muscle are above 60%, similar to those found in brain of zebrafish [Citation49] of skeletal muscle of tilapia [Citation17]. Moreover, the alignment efficiency of raw reads was on average 50% (, Supplementary Table 1), which is in accordance with another study using the same protocol and alignment method in zebrafish [Citation49]. The optimized double-digestion step of our protocol allowed us to study a larger portion of the genome and the choice of Taqα1 in addition to MspI extended the coverage of high CpG density regions as shown in mammalian genomes [Citation50] and in our twofold enrichment of promoter-annotated cytosines. The methylation levels are highly heterogeneous between the chromosomes, and these non-random methylation patterns can be placed in the genomic context (inter- or intragenomic region, promoter, intron exon, or transcription termination site) and assigned to a specific gene. However, it is difficult to predict how methylated cytosines could affect the gene only based on its genomic context. For example, in contrast to generally repressive role of higher methylation near the TSS, gene body methylation can increase or at least not preclude its expression [Citation51]. The order of exons affected by methylation influences gene expression differently [Citation52] and intragenic DNA methylation can also provide splicing signals [Citation53] and suppress cryptic promoters found within transcription units [Citation54]. Finally, CpG methylation can impact distal genes in 5ʹ and 3ʹ direction or act as distal enhancer of one gene while being promoter of another one [Citation55], meaning that transcriptional information would be needed to predict a regulatory outcome of DNA methylation. Our study lacks the experimentally validated connection between changes in methylation of specific DM CpG sites and their consequence in terms of gene expression or phenotype. However, the known functions of DM genes give us an insight into the potential consequences on growth or growth-related processes. As automated functional annotation analysis did not yield any significantly enriched biological processes associated with genes containing DM CpG sites (Supplementary Table 6), we manually selected the genes with growth-related or epigenetics-related functions based on available literature. To our knowledge, none of the available tools for systematic analysis of functional annotation are adapted to RRBS datasets, since they do not consider the number of DM CpG sites associated with a single gene nor the level of methylation differences between compared groups.
Global methylation levels are similar between males and females
The sex-dependent comparison is interesting from an aquaculture point of view, since in Nile tilapia, as in most tilapiine species, males naturally grow faster and attain larger size than females [Citation56]. Factors related to spawning (nutrient loss during spawning, fasting, and post-hatching) have been proposed to explain such dimorphism [Citation57], but epigenetic mechanisms that could affect muscle growth in a sex-dependent manner were never reported. Only 0.01% of all compared CpG sites were DM between males and females and mostly hypomethylated in males. The highest number of DM CpG sites was found in chromosome LG2, which is not the chromosome bearing the highest number of sex-linked markers [Citation56]. Interestingly, while the reduced representation dataset is twofold enriched in promoter- and exon-related CpGs compared to those representing the whole genome, sex-related DM CpGs are even more frequent in promoters but extremely low in exon regions when compared to the raw RRBS dataset. Therefore, methylation upstream of the TSS might be a more frequent sex-dependent mechanism of transcriptional regulation than the methylation within the gene body.
Although functional analysis did not show any significant enrichment of specific functions (Supplementary Table 6), several sex-dependent DM CpGs were associated with genes that have a known function in muscle-related processes (). Hdac11 (histone deacetylase) is an important epigenetic histone modification factor that can decrease myoD-dependent transcription and thus inhibit myoblast differentiation [Citation24]. Indeed, elevated expression of hdac11 decreased the levels of histone acetylation at promoter regions of mef2c and myoG, which prevented myo-D-dependent myoblast differentiation. Hdac11 is also involved in many immune functions [Citation58]. Lepr can have a role in proliferation and differentiation of skeletal myoblasts [Citation25] and is a potential candidate for growth rate and fat depositions traits in Durocs pigs [Citation59] and is DM between normal and heat-stressed pigs which can affect muscle development and meat quality [Citation60]. Sexual dimorphism of leptin receptors expression in skeletal muscle was also found in human [Citation61]. Itga4 plays a role in myogenesis [Citation26] and is highly downregulated in Myo-deleted mice [Citation62]. Tgfb2 is a transforming growth factor that plays an important role in skeletal muscle atrophy [Citation27]. Four CpG hypomethylated in males were found in exon of atg14, a key component in autophagy, a process of cellular self-renewal [Citation28]. Atg14 also plays an important role in hepatic lipid metabolism [Citation63] and can, by similarity to other autophagy-related genes, have a role in regulating muscle mass due to its potential role in muscle degradation as shown in case of fasting rainbow trout [Citation29].
The importance of sex in the growth-related muscle methylome
Although body weight differences between small and large groups were smaller in females than in males, females displayed more growth-related DM CpG than males. However, their genomic context was very similar between both sexes and close to the genomic context of all CpG sites found in RRBS dataset. In addition, the chromosomal distribution of DM CpGs is strikingly different between sexes, suggesting that different growth-related gene networks are epigenetically modulated in female and male of Nile tilapia. Indeed, out of approximately 1000 possible common positions of sex-related DM CpG sites, only 57 were common to both sexes. For example, the promoters of mettl9 and mettl16 methyltransferase-like genes absent in the DM CpG male dataset were highly represented by hypomethylated CpG in large females. The function of proteins encoded by these methyltransferase-like genes is not well known but mettl16 is suggested to catalyse the methyl marks on RNA, another epigenetic regulator [Citation64] and the mettl3, another gene from methyltransferase-like group can promote muscle cell differentiation by affecting myoD transcription [Citation65]. Large females were also represented by two CpGs associated with ppp1r3c, which is associated with energy metabolism in skeletal muscle [Citation30]. Indeed, ppp1r3c is also called protein targeting to glycogen, since it has an important role in glycogen metabolism [Citation66]. Interestingly, the two DM CpGs showed distinct features. One CpG was hypermethylated and found in an intergenic region whereas the other one, hypomethylated, was found in one exon. In addition, myogenesis, growth, and mitochondrial activity were the main functions of many more genes associated with DM CpG between large and small females, as shown in .
Another important gene with two hypermethylated intronic CpG sites in large females encodes IGF2BP2. Its methylation is inversely associated with its mRNA abundance [Citation67]. In addition, IGF2BP2 is involved in satellite-cell-mediated skeletal muscle differentiation, proliferation, and regeneration [Citation36]. Moreover, IGF2BP2 is reported to enhance mRNA stability through recognition of RNA N 6-methyladenosine [Citation37]. Several genes coding for proteins involved in epigenetic regulation were also DM in females (). Among them, HDAC11 was discussed above, since it was also associated with sex-related DM CpG sites. Exportin 5 is involved in export of miRNA from nucleus to cytoplasm [Citation38] and DMAP1 is involved in global maintenance methylation [Citation39]. In addition, ranbp2 encodes for a SUMO E3 ligase that is known to modify histone deacetylase HDAC4 [Citation40]. Cytosine methylation could interact with histone modifiers or regulate miRNA to control the growth. Indeed, the importance of miRNA in growth of Nile tilapia has already been shown [Citation68] and some miRNA features (e.g., arm-switching) can be sex-dependent in this species [Citation69].
Several genes related to epigenetic mechanisms of gene expression regulation were also associated with DM CpGs found in males. For example, hdac9 was linked with one intronic CpG hypomethylated in large males, xpo5 related to exonic hypermethylated CpG, and PHD finger protein 20 (phf20 gene) associated with two hypomethylated exonic CpGs. PHF20 is an epigenetic reader involved in histone acetylation [Citation41].
Atg14, a gene found previously related to four CpGs hypomethylated in males when compared to females independently of weight, is also DM between large and small males. Indeed, 20 out of 23 CpGs available in the RRBS dataset (87%) were hypomethylated in large males and situated in the tbp2 (TATA-box binding protein 2) gene body [Citation42] (). Either tbp2 or atg14 or both genes could be affected by methylation. Tbp2 is required for transcription initiation of promoters that contain a TATA-box [Citation42]. Thus, if the C-terminal region of Tbp2 is affected by tbp2 methylation, it could have drastic consequences on a large set of TATA-box-containing genes. Although autophagy-related genes have never been studied in Nile tilapia, atg14 should receive particular attention in the future studies related to male-specific growth rate. It suggests an attractive pathway related to self-renewal of cells that can be epigenetically regulated in males but not in females, since the atg14 is highly methylated in the latter regardless of their growth rate.
Uni-parental mouth brooding in cichlids has many physiological and anatomical consequences. Mouth-brooding females have different immune defence and buccal microbiota [Citation70], larger mouth [Citation71], smaller gill size, higher metabolic rate, higher aquatic surface respiration threshold than non-brooding females or males [Citation72]. Indeed, brood protection has an additional energy cost, especially under hypoxia conditions [Citation73]. Mouth brooding time in Nile tilapia is approximately 1–2 weeks, and generally speaking, such a short period of food deprivation should not necessarily induce a strong physiological response in many fish species. However, in the case of Nile tilapia, a longer food restriction occurs exactly at the moment of increased energy demand in females (for example, due to churning behaviour) and has to be considered in addition to the fasting period due to reproduction itself. It is therefore plausible that female Nile tilapia rely on starvation-related autophagy mechanisms during longer periods of fasting, which could be related to the differential CpG methylation of atg14 that we observed between males and females.
Five strongly hypomethylated CpG sites in large males (42% of all available gene-specific CpG sites) were found in the promoter region of the gene encoding for HADH, a marker of fatty acid beta oxidation (catabolic process) occurring in muscle after exercise [Citation74]. Finally, a group of three related genes, gadd45g, gdf3, and ppargc1a, was associated with hypomethylated CpG sites in large males. GADD45G is encoded by a member of a family of genes affected by agents damaging DNA, involved in DNA demethylation [Citation75] or causing growth arrest and muscle atrophy due to stress [Citation43]. Indeed, gadd45 gamma isoform induces the transcription of p21 through the activation of the p38 MAPK (mitogen-activated protein kinase) pathway [Citation76]. An alpha isoform of gadd45 gene has been studied in hybrid tilapia species, although not in muscle tissue and only in the context of bacterial infection [Citation77]. Environmentally induced methylation patterns of gadd45g have been studied in human cells [Citation78] and found to be associated with different carcinomas [Citation79]. Interestingly, the gadd45a gene promoter is transcribed by pparγ gene (peroxisome proliferator-activated receptor gamma) [Citation80,Citation81], regulating gdf3 [Citation82] and closely related to ppargc1a (PGC-1alpha), both hypomethylated in large males. Ppargc1a is associated with diabetes [Citation83], and is considered as a key regulator of energy metabolism [Citation45] and mitochondrial biogenesis [Citation84]. Its decreased expression in muscle is related to abnormal insulin function [Citation85] and hypermethylation of its non-CpG sites is associated with silenced DNA methyltransferase 3B (dnmt3b) and with reduced mitochondrial density [Citation86]. Gdf3 is involved in muscle metabolism [Citation44]. Taken together, these data suggest that epigenetic modulation associated with growth is sex-specific in Nile tilapia. The analysis of the common locations of weight-related DM CpG sites found separately in males and females shows that the total number of sites is not only very low (57), but also that half of them display opposite patterns of growth-specific methylation in males compared in females.
The transcription factor gata6 gene, involved in regulation of smooth muscle growth [Citation47], was associated with exonic DM CpG sites that consistently showed the same patterns of methylation in large fish but was also detected in both growth- and sex-specific comparisons (), indicating that gata6 may be involved in transcriptional regulation related to both growth and sex in Nile tilapia.
The importance of sex in growth-related methylation patterns in Nile tilapia is highlighted again by a very low number of DM CpG identified in the sex-independent comparison between large and small fish. Indeed, when large females and large males are grouped together and compared to small fish, only 117 DM CpG sites are found, 10 times less than in the weight-specific comparison between large and small males. Only a few muscle growth-related genes associated with DM CpG sites were unique to the comparison between large versus small fish regardless of sex, such as cdh2 (cadherin 2) [Citation46] ().
Conclusions
This study provides a genome-wide reduced representation profiling of cytosine methylation at a single nucleotide level in Nile tilapia muscle. The growth-related methylome in fast muscle differed significantly between males and females, thus confirming our initial hypothesis of sex-related differences in methylation patterns between large and small fish. For example, in males, genes related to stress-induced inhibition of growth rate, fatty acid metabolism, and autophagy were associated with many cytosines showing differential methylation between large and small individuals. However, none of these genes was DM between large and small females, suggesting a sex-specific epigenetic regulation of different gene networks. The autophagy-related gene, atg14, is particularly interesting, since its epigenetic regulation or alternative splicing may affect growth in a sex-dependent manner and whose methylation could be affected during domestication via selective breeding. Several growth-related genes involved in epigenetic regulation of gene expression such as DNA methylation, miRNA, or histone modifications were DM, which indicates that the growth phenotype in Nile tilapia may depend on the interplay between epigenetic mechanisms at different levels involving a large array of biological processes that could be affected during the process of domestication-related growth improvement. In addition to providing novel insight into the epigenetic regulation of muscle growth in fish, the potential epigenetic marks associated with growth rate may be used in marker-assisted selection, thus improving selective breeding and increasing sustainability of fish farming in the future.
Material and methods
Sampling
Nile tilapia eggs were collected from the mouth of wild females caught in the Nile river, Egypt (location GPS: 25°39ʹ56″ N, 32°37ʹ07″ E) and transported to our research station at Nord University (Norway) where they hatched and were reared in a recirculating aquaculture system (pH = 7.6, temperature = 28°C, 11 h dark/13 h light cycle) during 5 months at a density of 27 fish/m3, which corresponded to a maximum density of 6 kg/m3 at the end of the experiment. The fish were fed ad libitum with 0.15–0.8 mm Amber Neptun pellets (Skretting, Sjøhagen 3, 4016 Stavanger, Norway) and kept in the same tank, so as to avoid any potential bias in methylation patterns due to environmental differences (tank effect). They were euthanatized with clove oil (Sigma Aldrich, 3050 Spruce St, St. Louis Missouri, 63103, USA) using a 1:10 mix of 15 mL clove oil with 95% ethanol diluted in 10 L of water and then fast muscle was carefully excised from 24 fish (12 biggest and smallest individuals from each sex, ) just prior to snap-freezing and then stored at −80°C until DNA extraction. DNA was extracted with the DNEasy Blood & Tissue kit (Qiagen GmbH, Qiagen Strasse 1, 40724 Hilden, Germany) according to the manufacturer’s recommendations. DNA quality check was performed by NanoDrop and DNA quantification was performed with Qubit (Thermofisher Scientific, 168 Third Avenue, Waltham, MA USA 02451) and Tape Station (AgilentTechnologies, 5301 Stevens Creek Blvd, Santa Clara, CA 95051, USA) Assays.
Library preparation
Library preparation for RRBS analysis was performed with the Premium Reduced Representation Bisulfite Sequencing (RRBS) Kit (Diagenode, Rue Bois Saint-Jean, 34102 Seraing (Ougrée), Belgium), essentially following the manufacturer’s instructions. The protocol was optimized by including a double digestion (Taqα1 + MspI) instead of single MspI digestion, which produced more fragments and of more appropriate length (Supplementary Figure 1). In short, normalized DNA samples (100 ng, 26 μL) were sequentially digested with MspI (16 h at 37°C) in CutSmart® Buffer and Taqα1 (8 h at 65°C, followed by deactivation 20 min at 80°C). The digested fragments were then end-repaired, followed by addition of dA. Next, 5ʹ and 3ʹ adapters were ligated, and each sample was size-selected (discarding all reads below 140 bp) with AMPure XP beads (Beckman Coulter, Miami Campus 11800, S.W. 147th Avenue, Miami, Florida 33196, USA), PCR-amplified (13 cycles) and purified again (AMPure XP beads). Finally, the libraries were pooled at 1.6 nM, diluted and single-end sequenced with the NextSeq 500/550 High Output v2 Kit, 75 cycles (Illumina, 5200 Illumina Way, San Diego, CA 92122, USA), according to the manufacturer’s recommendations. Raw reads were converted to FASTQ format, demultiplexed using bcl2fastq V 2.16 (Illumina) and their quality was examined with the FastQC tool (Babraham Bioinformatics). All reads are available at the SRA database under the reference PRJNA506143.
Bioinformatics and statistical analyses
The significance of weight differences between small and large males and females was determined by Student’s t-test, after verifying normality and equal variance assumptions.
Raw sequencing reads were adapter-trimmed with Cutadapt (Babraham Bioinformatics) [Citation87] and aligned to the reference genome ASM185804v2 (orenil3, NCBI, https://www.ncbi.nlm.nih.gov/) using the Bismark v0.19.1 pipeline (Babraham Bioinformatics) [Citation88]. The two strands of oreNil3 were modified in silico (conversion of all C’s to T’s) and indexed according to Bowtie2 [Citation89] requirements. Reads were mapped to the original and in silico-modified genome using Bismark (with parameters: -q – p – N1 – bowtie2). The resulting CpG coverage files were used as input in MethylKit package [Citation90] in R to calculate the coverage, methylation levels of each sample, and differences in methylation among samples. The statistical method used to detect DM cytosines between groups was logistic regression, and the significance thresholds were FDR <0.01 and a minimum difference in methylation level of 25%. Small fish and females were used as reference in size and sex comparisons, respectively. For each CpG site, if the group compared to the reference has a methylation level at least 25% higher, this CpG site was considered as hypermethylated. Conversely, if the methylation level is at least 25% lower in the target than in the reference group, the CpG site was considered as hypomethylated. Integrated Genome Viewer [Citation91] was used to visualize the methylation profile of all CpG from each sample and Circos [Citation92] was used to represent and locate all DM CpG sites. The O. niloticus annotation release 103 was used as input in Homer [Citation93] to locate CpG sites within genomic context (introns, exons, promoters, intergenic regions) and to assign the gene reference in RefSeq format with the closest transcription start site to each DM cytosine. Gene references were then converted to gene symbols and full gene names using the NCBI browser and manually associated with function using the GeneCards browser [Citation94] and the literature. In addition, Functional Enrichment Analysis was performed with DAVID [Citation95] using a custom background gene list corresponding to all the CpG sites analysed in RRBS dataset, and specific to each comparison.
Authors’ contributions
TP carried out the wet lab and bioinformatics analysis, interpreted the results, and wrote the article. SB contributed significantly to wet lab analysis (DNA extraction and RRBS protocol) and revised the article. IK contributed significantly to sampling, to bioinformatics analysis and revised the article. JMOF conceived the study, designed the experiment, provided reagents, and contributed significantly to wet lab analysis, bioinformatics analysis, interpretation of results, and article revision. All authors read and approved the manuscript.
Ethics statement
All procedures involving animals were performed according to the guidelines of the Norwegian Animal Research Authority (FOTS ID 1042) and approved by the Nord University (Norway) ethics committee.
Supplemental Material
Download MS Excel (420.8 KB)Supplemental Material
Download MS Word (3.9 MB)Acknowledgments
We are thankful to Hilde Ribe, Steinar Johnsen, Øivind Torslett and particularly to Kaspar Klaudiussen (Nord University, Norway) for their assistance with fish husbandry.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Johnston IA. Environment and plasticity of myogenesis in teleost fish. J Exp Biol. 2006;209(12):2249–2264.
- van der Most JP, de Jong B, Parmentier KH, et al. Trade-off between growth and immune function: a meta-analysis of selection experiments. Funct Ecol. 2010;25(1):74–80.
- Hagen Ø, Fernandes JMO, Solberg C, et al. Expression of growth-related genes in muscle during fasting and refeeding of juvenile Atlantic halibut, Hippoglossus hippoglossus L. Comp Biochem Physiol Part B Biochem Mol Biol. 2009;152(1):47–53.
- Valente LMP, Moutou KA, Lec C, et al. What determines growth potential and juvenile quality of farmed fish species? Rev Aquac. 2013;5:S168–S193.
- Nagasawa K, Giannetto A, Fernandes JMO. Photoperiod influences growth and MLL (mixed-lineage leukaemia) expression in Atlantic cod. PLoS One. 2012;7:5.
- Biro PA, Abrahams MV, Post JR, et al. Behavioural trade-offs between growth and mortality explain evolution of submaximal growth rates. J Anim Ecol. 2006;75(5):1165–1171.
- Runemark A, Brydegaard M, Svensson EI. Does relaxed predation drive phenotypic divergence among insular populations? J Evol Biol. 2014;27(8):1676–1690.
- Huntingford FA. Implications of domestication and rearing conditions for the behaviour of cultivated fishes. J Fish Biol. 2004;65(s1):122–142.
- Elsbeth McPhee M. Generations in captivity increases behavioral variance: considerations for captive breeding and reintroduction programs. Biol Conserv. 2004;115(1):71–77.
- Teletchea F, Fontaine P. Levels of domestication in fish: implications for the sustainable future of aquaculture. Fish Fisheries. 2014;15(2):181–195.
- Solberg MF, Ø S, Nilsen F, et al. Does domestication cause changes in growth reaction norms? A study of farmed, wild and hybrid Atlantic salmon families exposed to environmental stress. PLoS One. 2013;8(1):e54469.
- Christie MR, Marine ML, Fox SE, et al. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun. 2016;7. DOI:10.1038/ncomms10676.
- Swain DP, Riddell BE, Murray CB. Morphological differences between hatchery and wild populations of coho salmon (Oncorhynchus kisutch): environmental versus genetic origin. Can J Fisheries Aquat Sci. 1991;48(9):1783–1791.
- Barros SP, Offenbacher S. Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res. 2009;88(5):400–408.
- Le Luyer J, Laporte M, Beacham TD, et al. Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc Natl Acad Sci USA. 2017;114(49):12964–12969.
- Sun LX, Wang YY, Zhao Y, et al. Global DNA methylation changes in Nile Tilapia gonads during high temperature-induced masculinization. PLoS One. 2016;11(8):e0158483.
- Wan ZY, Xia JH, Lin G, et al. Genome-wide methylation analysis identified sexually dimorphic methylated regions in hybrid tilapia. Sci Rep. 2016;6:35903.
- Burgerhout E, Mommens M, Johnsen H, et al. Genetic background and embryonic temperature affect DNA methylation and expression of myogenin and muscle development in Atlantic salmon (Salmo salar). PLoS One. 2017;12(6):e0179918.
- Campos C, Valente LM, Conceicao LE, et al. Temperature affects methylation of the myogenin putative promoter, its expression and muscle cellularity in Senegalese sole larvae. Epigenetics. 2013 Apr;8(4):389–397. PubMed PMID: 23538611; PubMed Central PMCID: PMCPMC3674048. eng.
- Moghadam H, Mørkøre T, Robinson N. Epigenetics-potential for programming fish for aquaculture? J Mar Sci Eng. 2015;3(2):175–192.
- Soltan MA, Hassaan MS, Meshrf RN. Response of Nile tilapia (Oreochromis niloticus) to diet acidification: effect on growth performance and feed utilization. J Appl Aquacult. 2017;29(3–4):207–219.
- Charo-Karisa H, Komen H, Reynolds S, et al. Genetic and environmental factors affecting growth of Nile tilapia (Oreochromis niloticus) juveniles: modelling spatial correlations between hapas. Aquaculture. 2006;255(1–4):586–596.
- Lind CE, Safari A, Agyakwah SK, et al. Differences in sexual size dimorphism among farmed tilapia species and strains undergoing genetic improvement for body weight. Aquacult Rep. 2015;1:20–27.
- Byun SK, An TH, Son MJ, et al. HDAC11 inhibits myoblast differentiation through repression of MyoD-dependent transcription. Mol Cells. 2017;40(9):667–676.
- Yu T, Luo G, Zhang L, et al. Leptin promotes proliferation and inhibits differentiation in porcine skeletal myoblasts. Biosci Biotechnol Biochem. 2008;72(1):13–21.
- Yang JT, Rando TA, Mohler WA, et al. Genetic analysis of α4 integrin functions in the development of mouse skeletal muscle. J Cell Biol. 1996;135(3):829–835.
- Mendias CL, Gumucio JP, Davis ME, et al. Transforming growth factor-beta induces skeletal muscle atrophy and fibrosis through the induction of atrogin-1 and scleraxis. Muscle Nerve. 2012;45(1):55–59.
- Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435.
- Seiliez I, Gutierrez J, Salmerón C, et al. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part B Biochem Mol Biol. 2010;157(3):258–266.
- Giorgetti E, Yu Z, Chua Jason P, et al. Rescue of metabolic alterations in AR113Q skeletal muscle by peripheral androgen receptor gene silencing. Cell Rep. 2016;17(1):125–136.
- Marino JS, Tausch BJ, Dearth CL, et al. β2-Integrins contribute to skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol. 2008;295(4):C1026–C1036.
- Mourikis P, Tajbakhsh S. Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev Biol. 2014;14(1):2.
- Bae G-U, Yang Y-J, Jiang G, et al. Neogenin regulates skeletal myofiber size and focal adhesion kinase and extracellular signal-regulated kinase activities in vivo and in vitro. Mol Biol Cell. 2009;20(23):4920–4931.
- Tran P, Ho SM, Kim BG, et al. TGF-β-activated kinase 1 (TAK1) and apoptosis signal-regulating kinase 1 (ASK1) interact with the promyogenic receptor Cdo to promote myogenic differentiation via activation of p38MAPK pathway. J Biol Chem. 2012;287(15):11602–11615.
- Rangwala SM, Wang X, Calvo JA, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem. 2010;285(29):22619–226129.
- Li Z, Gilbert JA, Zhang Y, et al. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23(6):1176–1188.
- Huang H, Weng H, Sun W, et al. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295.
- Okada C, Yamashita E, Lee SJ, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326(5957):1275–1279.
- Lee GE, Kim JH, Taylor M, et al. DNA methyltransferase 1-associated protein (DMAP1) is a co-repressor that stimulates DNA methylation globally and locally at sites of double strand break repair. J Biol Chem. 2010;285(48):37630–37640.
- Kirsh O, Seeler J-S, Pichler A, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. Embo J. 2002;21(11):2682–2691.
- Sanchez R, Zhou -M-M. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36(7):364–372.
- Jallow Z, Jacobi UG, Weeks DL, et al. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA. 2004;101(37):13525–13530.
- Liebermann DA, Tront JS, Sha X, et al. Gadd45 stress sensors in malignancy and leukemia. Crit Rev Oncog. 2011;16(1–2):129–140.
- Hays TT, Patsalos A, Hammers D, et al. A novel role of growth differentiation factor 3 for the regulation of muscle metabolism. Diabetes. 2018;67:1.
- Ahmetov II, Rogozkin VA. The role of PGC-1α in the regulation of skeletal muscle metabolism. Human Physiol. 2013;39(4):441–449.
- Redfield A, Nieman MT, Knudsen KA. Cadherins Promote Skeletal Muscle Differentiation in Three-dimensional Cultures. J Cell Biol. 1997;138(6):1323.
- Yin F, Herring BP. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J Biol Chem. 2005;280(6):4745–4752.
- Chen X, Wang Z, Tang S, et al. Genome-wide mapping of DNA methylation in Nile tilapia. Hydrobiologia. 2016;791(1):1–11.
- Chatterjee A, Stockwell PA, Horsfield JA, et al. Base-resolution DNA methylation landscape of zebrafish brain and liver. Genom Data. 2014;2:342–344.
- Martinez-Arguelles DB, Lee S, Papadopoulos V. In silico analysis identifies novel restriction enzyme combinations that expand reduced representation bisulfite sequencing CpG coverage. BMC Res Notes. 2014;7(1):534.
- Jones PA. The DNA methylation paradox. Trends Genet. 1999;15(1):34–37.
- Brenet F, Moh M, Funk P, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6(1):e14524.
- Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79.
- Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257.
- Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455.
- Chen J, Fan Z, Tan D, et al. A review of genetic advances related to sex control and manipulation in tilapia. J World Aquacult Soc. 2018;49(2):277–291.
- Schreiber S, Focken U, Becker K. Individually reared female Nile tilapia (Oreochrornis niloticus) can grow faster than males. J Appl Ichthyol. 2007;14(1–2):43–47.
- Yanginlar C, Logie C. HDAC11 is a regulator of diverse immune functions. BBA-Gene Regul Mech. 2018;1861(1):54–59.
- Hirose K, Ito T, Fukawa K, et al. Evaluation of effects of multiple candidate genes (lep, lepr, mc4r, pik3c3, and vrtn) on production traits in Duroc pigs. Anim Sci J. 2014;85(3):198–206.
- Hao Y, Cui Y, Gu X. Genome-wide DNA methylation profiles changes associated with constant heat stress in pigs as measured by bisulfite sequencing. Sci Rep. 2016;6:27507.
- Guerra B, Fuentes T, Delgado-Guerra S, et al. Gender dimorphism in skeletal muscle leptin receptors, serum leptin and insulin sensitivity. PLoS One. 2008;3(10):e3466.
- Meadows E, Cho J-H, Flynn JM, et al. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev Biol. 2008;322(2):406–414.
- Xiong X, Tao R, DePinho RA, et al. The autophagy-related gene 14 (atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287(46):39107–39114.
- Ruszkowska A, Ruszkowski M, Dauter Z, et al. Structural insights into the RNA methyltransferase domain of METTL16. Sci Rep. 2018;8(1):5311.
- Kudou K, Komatsu T, Nogami J, et al. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 2017;7(9):170119.
- Printen JA, Brady MJ, Saltiel AR. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275(5305):1475–1478.
- Rönn T, Volkov P, Davegårdh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6):e1003572.
- Huang CW, Li YH, Hu SY, et al. Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus). J Anim Sci. 2012;90(12):4266–4279.
- Pinhal D, Bovolenta LA, Moxon S, et al. Genome-wide microRNA screening in Nile tilapia reveals pervasive isomiRs’ transcription, sex-biased arm switching and increasing complexity of expression throughout development. Sci Rep. 2018;8(1):8248.
- Keller IS, Bayer T, Salzburger W, et al. Effects of parental care on resource allocation into immune defense and buccal microbiota in mouthbrooding cichlid fishes. Evolution. 2018;72(5):1109–1123.
- Van Wassenbergh S, Potes NZ, Adriaens D. Hydrodynamic drag constrains head enlargement for mouthbrooding in cichlids. J Royal Soc Interface. 2015;12:109.
- O’Connor CM, Reardon EE, Chapman LJ. Shorter gills in mouth-brooding females of the cichlid Pseudocrenilabrus multicolor. Copeia. 2012;2012(3):382–388.
- Corrie LW-C, Chapman LJ, Reardon EE. Brood protection at a cost: mouth brooding under hypoxia in an African cichlid. Environ Biol Fishes. 2008;82(1):41–49.
- Tremblay A, Simoneau JA, Bouchard C. Impact of exercise intensity on body fatness and skeletal muscle metabolism. Metabolism. 1994;43(7):814–818.
- Niehrs C, Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22(4):220–227.
- Ishida K, Yuge Y, Hanaoka M, et al. Gadd45g regulates dental epithelial cell proliferation through p38 MAPK-mediated p21 expression. Genes Cells. 2013;18(8):660–671.
- Shen Y, Ma K, Liu F, et al. Characterization of two novel gadd45a genes in hybrid tilapia and their responses to the infection of Streptococcus agalactiae. Fish Shellfish Immunol. 2016;54:276–281.
- Ying J, Srivastava G, Hsieh WS, et al. The stress-responsive gene gadd45g is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res. 2005;11(18):6442–6449.
- Tamura RE, de Vasconcellos JF, Sarkar D, et al. GADD45 proteins: central players in tumorigenesis. Curr Mol Med. 2012;12(5):634–651.
- Ebert SM, Dyle MC, Kunkel SD, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287(33):27290–27301.
- Bruemmer D, Yin F, Liu J, et al. Regulation of the growth arrest and DNA damage-inducible gene 45 (gadd45) by peroxisome proliferator-activated receptor gamma in vascular smooth muscle cells. Circ Res. 2003;93(4):e38–47.
- Varga T, Mounier R, Patsalos A, et al. Macrophage PPARγ, a lipid activated transcription factor controls the growth factor GDF3 and skeletal muscle regeneration. Immunity. 2016;45(5):1038–1051.
- Kelstrup L, Hjort L, Houshmand-Oeregaard A, et al. Gene expression and DNA methylation of PPARGC1A in muscle and adipose tissue from adult offspring of women with diabetes in pregnancy. Diabetes. 2016;65(10):2900–2910.
- Benton CR, Wright DC, Bonen A. PGC-1alpha-mediated regulation of gene expression and metabolism: implications for nutrition and exercise prescriptions. Appl Physiol Nutr Metab. 2008;33(5):682–843.
- Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–735.
- Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–198.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–12.
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9. DOI:10.1038/nmeth.1923
- Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87–R87.
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192.
- Krzywinski M, Schein J, Birol İ, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645.
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589.
- Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13(4):163.
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.