ABSTRACT
The biological mechanisms through which adherence to Mediterranean Diet (MD) protects against colon cancer (CC) are poorly understood. Evidence suggests that chronic inflammation may be implicated in the pathway. Both diet and CC are related to epigenetic regulation.
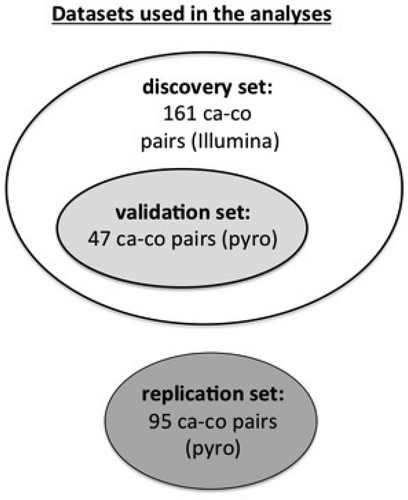
We performed a nested case-control study on 161 pairs from the Italian component of the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort, in which we looked for the methylation signals in DNA extracted from leucocytes associated with both CC and MD in 995 CpGs located in 48 inflammation genes. The DNA methylation signals detected in this analysis were validated in a subgroup of 47 case-control pairs and further replicated (where validated) in 95 new pairs by means of pyrosequencing.
Among the CpG sites selected a-priori in inflammation-related genes, seven CpG sites were found to be associated with CC status and with MD, in line with its protective effect. Only two CpG sites (cg17968347-SERPINE1 and cg20674490-RUNX3) were validated using bisulphite pyrosequencing and, after replication, we found that DNA methylation of cg20674490-RUNX3 may be a potential molecular mediator explaining the protective effect of MD on CC onset.
The use of a ‘meet-in-the-middle’ approach to identify the overlap between exposure and predictive markers of disease is innovative in studies on the relationship between diet and cancer, in which exposure assessment is difficult and the mechanisms through which the nutrients exert their protective effect is largely unknown.
Introduction
Epidemiological studies have unveiled a protective association between Mediterranean diet (MD) and incidence of cancer and of other major chronic diseases [Citation1,Citation2]. In the Italian branch of the European Prospective Investigation into Cancer and Nutrition (EPIC) study, increasing values of the Italian Mediterranean Index (IMI) were associated with a decreased risk of colon cancer (CC) as well as rectal cancer in both men and women [Citation3]. A pooled analysis of three Italian case-control studies [Citation4] and two cohort studies [Citation5,Citation6] confirmed the favourable role of MD in reducing CC incidence. The protective mechanisms of MD have been attributed to a high content of antioxidants, olive oil, fibres, and a moderate alcohol intake [Citation7]. The biological mechanisms through which MD protects against CC remain nonetheless poorly understood.
Adherence to MD has been previously hypothesized to protect against abdominal adiposity [Citation8], and obesity is, in turn, a well-known risk factor for chronic diseases, specifically CC [Citation9]. In a previous study [Citation10] we analysed adiposity as a potential mediator of the association between adherence to MD and CC, but we concluded that its reduction, due to MD, does not explain the protective effect.
An alternative hypothesis suggests that the causal pattern of the association between MD and CC could pass through chronic inflammation. In fact, randomized control trials [Citation11] and observational studies [Citation12] have shown that MD can attenuate the level of systemic inflammation while chronic inflammation has been shown to be itself a possible causative factor for a variety of cancer types, including CC. People with chronic inflammatory bowel diseases show indeed an increased risk of CC [Citation13] and chronic aspirin use seems to reduce the risk of CC [Citation14].
Both diet and CC are related to epigenetic regulation. Diet can perturb the way genes are controlled by epigenetic signals, particularly DNA methylation [Citation15,Citation16], which is, in turn, one of the regulatory mechanisms of systemic and local inflammation [Citation17].
The aim of this investigation was to explore methylation changes of inflammation genes associated with both MD and CC status in peripheral blood cells of members of an Italian epidemiological cohort. Our purpose was to test the hypothesis that DNA methylation could be a mediator of the CC triggered by non-adherence to MD.
Methods
Study sample and discovery set
Data from the Italian component of the EPIC study (EPIC-Italy) [Citation18], a total of 42,894 volunteers recruited from four centres in Italy (Varese, Turin, Naples, and Ragusa) in 1993–1998 were considered. Over a mean follow-up of 11.0 years (range 1.0–14.8), 313 cases of CC were diagnosed. Colon cancers were primary incident cases, identified as proximal (International Classification of Diseases for Oncology, third Edition [ICD-O-3] codes C18.0-C18.5; N = 121), distal (ICD-O-3 codes C18.6-C18.7; N = 154), and over-lapping or unspecified (ICD-O-3 codes C18.8-C18.9; N = 36) sites.
All participants in the study sample completed a validated semiquantitative food-frequency questionnaire at enrolment. Blood samples were collected at recruitment and sent to local laboratories for processing and aliquot preparation.
A nested case-control study was conducted within EPIC-Italy cohort employing 169 incident CC cases diagnosed within follow-up and 169 matched controls selected at random from the participants at risk of CC at the time of the case diagnoses.
Controls were matched to cases by sex, date of birth (within 5 years), seasonality of blood sampling (autumn-winter/spring-summer) and study centre. Members of the cohort treated for diabetes or with a diagnosis of intestinal polyps at baseline were excluded from the case-control study, as well as those with a prevalent diagnosis of cancer (except non-melanoma skin cancer).
Epigenome-wide DNA methylation analysis
Genomic DNA was extracted from buffy coats using the QIAsymphony DNA Midi Kit (Qiagen, Hilden, Germany). Five hundred nanogram of DNA were bisulphite-converted using the EZ-96 DNA Methylation-Gold™ Kit (Zymo, California, USA) and hybridised to Infinium HumanMethylation450 BeadChips (Illumina, California, USA). Each chip was subsequently scanned using the Illumina HiScanSQ system, and sample quality was assessed using control probes on the microarrays. Raw intensity data were finally exported from Illumina GenomeStudio (version 2011.1). Each case-control pair was arranged randomly on the same chip.
Data pre-processing was carried out using an in-house software written for the R statistical computing environment (see [Citation19] for a short description of the procedure). One hundred and sixty-one case-control pairs passed the pre-processing step for the successive analysis. DNA methylation was expressed as the ratio between the intensities of methylated cytosines and the total intensities (ß-values).
Main exposure and other variables
Italian Mediterranean Index (IMI), a summary measure of adherence to the MD, was used as a measure of the exposure [Citation20]. Briefly, the computation of this index is based on the intake of 11 food items: intakes of the Mediterranean foods pasta, Mediterranean vegetables (raw tomatoes, cooked leafy vegetables, raw leafy vegetables, onion or garlic, mixed salad, or mixed vegetables), fruits, legumes, olive oil, and fish of four ‘non Mediterranean’ foods (soft drinks, butter, red meat and potatoes); and of alcohol. If consumption of typical Mediterranean foods is in the third tertile of the distribution, the person receives 1 point; all other intakes receive 0 points. If consumption of non-Mediterranean foods is in the first tertile of the distribution the person receives 1 point. Alcohol receives 1 point for intake up to 12 g/day; abstainers and persons who consume >12 g/day receive 0 points. For the analysis, IMI was categorized in the following three classes: 0–2 (low adherence to MD), 3–4 (middle adherence to MD), and 5–11 (high adherence to MD).
We considered as further covariates: age, sex, centre, smoking status (never, former and current), total physical activity (inactive, moderately inactive, moderately active and active [Citation21]), level of education (tertiles of the relative index of inequality RII [Citation22]), body mass index (BMI) and cell types. The latter were estimated according to Houseman method [Citation23].
Selection of CpG sites
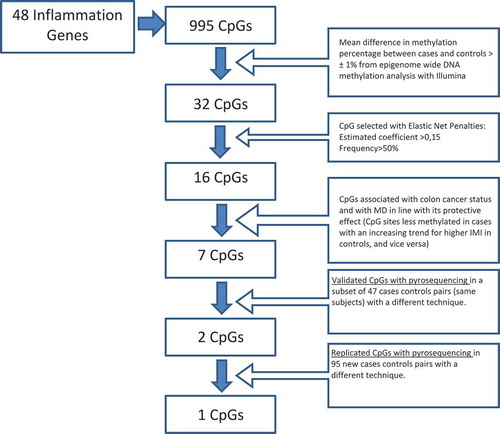
To study the methylation status of inflammation genes, data obtained from the epigenome-wide study were pruned selecting the CpGs located in a set of 48 inflammation genes (list in Supplementary Table 1). Such genes were selected through an extensive literature review to identify those related to inflammatory mechanisms involved in colon carcinogenesis.
Among the resulting 995 inflammation-related CpGs, only those with a mean difference (both positive and negative) in methylation percentage between cases and controls higher than 1% were considered in the association analysis (32 CpGs). The cutoff of 1% was chosen to increase the probability that the difference in methylation reflected an effective biological change.
A conditional logistic regression model with elastic net penalties [Citation24] was employed to select the most important CpG sites among the 32 CpGs related to CC. DNA methylation levels of the CpGs were standardized to 1 standard deviation computed on the control group. Elastic net (EN) [Citation25] is a regularization and variable selection method, which retains the parsimony property of Lasso regression method [Citation26], but encourages at the same time the grouping effect like Ridge regression [Citation27]. We applied the cyclic coordinate descent algorithm [Citation28], and we set the parameter controlling the trade-off between Lasso and Ridge penalties to 0.5. We used 10-fold cross-validation (CV) for the choice of the regularization parameter that characterizes the best model. To assess whether the associations found were stable in random subsets of the sample, 1000 EN models were fitted using each time 63.4% of the initial data. At the end, we obtained a ranked list of probes based on how many times they were included in models based on data subsets.
The CpG sites considered for further detailed analyses had to satisfy both the following criteria: i) CpGs selected by EN, with an estimated coefficient of the association with case-control status higher than 0.15 in absolute value (median of the distribution of the estimated coefficients) and ii) CpGs in the list of the most associated sites with a frequency higher than 50%. A diagram illustrating the selection of CpG for the analysis is shown in .
Detailed analysis of the selected signals
For each selected CpG, a conditional logistic regression model was fitted to estimate odds ratio of CC with DNA methylation levels included as independent variable (model A). A second model (model B) was fitted including additional adjustment for BMI, smoking status, total physical activity, level of education and IMI. The possible effect of cell composition on the results was assessed by further adding to model B the proportions of cell counts (model C). Sensitivity analyses were performed excluding cases where time elapsed between blood collection and diagnosis of CC was lower than 2 years or higher than 10 years.
The association between M-values of methylation (the logarithmic transform of ß-values, M = log2(ß/(1- ß)) [Citation29]) and IMI was evaluated in the control group by fitting a linear mixed effect model with chip as random effect and IMI, sex, age, centre, BMI, smoking status, total physical activity, level of education and differential cell types as fixed effects. Linearity of trends across categories of IMI was tested by treating the categorical variable as continuous in the linear mixed effect model.
Validation, replication and meta-analysis
A random selection of 47 case-control pairs from EPIC-Turin (validation set) was performed for validation with the pyrosequencing methodology. The CpGs for which adherence to MD conferred methylation levels that were protective on CC were selected for validation. This means that among the CpGs whose hypermethylation was protective on CC (odds ratio<1 for 1 standard deviation increase in methylation percentage) only those for which higher methylation levels corresponded to higher adherence to MD were validated. Similarly, CpGs whose hypomethylation was associated with decreased CC risk and for which lower methylation levels were associated with higher adherence to MD were validated.
Another nested case-control study (replication set) was finally conducted employing 95 independent case-control pairs from EPIC-Italy. The validated signals were replicated in this sample using pyrosequencing.
Both in the validation and in the replication, the associations with DNA methylation levels obtained by pyrosequencing were analysed using models similar to those employed in the discovery phase (see details in the Tables). Additionally, a random effects meta-analysis of the discovery and replication studies was performed on the validated signals.
Details on the pyrosequencing methodology
Pyrosequencing assay was performed on a PyroMark Q24 MDx system using PyroMark Gold Q24 Advanced reagents (Qiagen, Hilden Germany). Primers were designed according to PyroMark Assay Design software version 2.0 (Qiagen). PCR reaction was performed in a total volume of 35 μl using the PyroMark PCR kit (Qiagen) containing 1X PCR Master Mix, 1X CoralLoad Concentrate, 0.2 μM of each primer, and 1 μl of bisulphite-converted DNA with the following cycling profile: 95°C for 10 min followed by 45 cycles of denaturation at 95°C for 30 sec, annealing at specific temperature for each gene (55°C for RUNX3; 50°C for SERPINE1) for 30 sec, extension at 72°C for 1 min. Extension at 72°C for 10 min was finally performed. The PCR product (15 μl) was added to 19 μl of distilled water and incubated under shaking with 40 μl of binding buffer pH 7.6, containing 10mM Tris-HCl, 2 M NaCl, 1mM EDTA, and 1 μl of sepharose beads covered by streptavidin. The PCR product was washed with ethanol 70%, denatured with NaOH 0.2 M and re-washed with Tris-Acetate 10 mM pH 7.6. Pyrosequencing reaction was performed in a total volume of 20 μl, including 19.85 μl of 20 mM Tris-Acetate, 5 mM MgAc2 and 0.15 μl of 50 μM sequencing primer. Assays were created according to manufacturer’s instruction. The nucleotide dispensation order was suggested by the software PyroMark Q24 Advanced version 3.0.0.
Methylation quantification was achieved using the provided software, and expressed for each DNA locus as percentage ratio of methylated cytosines of the sum of methylated and unmethylated cytosines. Positive controls for methylated [EpiTect Control DNA (human), methylated (Qiagen)] and unmethylated status [EpiTect Control DNA (human), unmethylated (Qiagen)] were included in each pyrosequencing run. Each sample was analysed twice in different runs and the average of the two results was computed. Adequacy of the results for each sample was achieved when difference in methylation percentage between runs was ≤2% and pyrograms resulted as ‘passed’.
Software for analysis
All statistical analyses were performed using R Statistical Software (The R Foundation for Statistical Computing, Vienna, Austria) version 3.2.3 (2015–12-10) and Stata version 13 (StataCorp, College Station, TX, USA). A diagram illustrating the data sets used in the different stages of the analysis is shown in .
Results
shows the baseline characteristics of the samples. Cases and controls showed differences only with respect to BMI. For completeness, the features of the validation and replication samples are reported in the Supplementary materials. Cases and controls showed differences in BMI and educational level in the validation set, while no differences were detected in the replication set.
Table 1. Descriptive statistics of the case-control study nested in the EPIC-Italy cohort.
The profile of parameter estimates plotted against the value of the regularization parameter and the CV curve of the EN conditional logistic model in the discovery analysis are reported in Supplementary Figures 1 and 2. In particular, the CV error was minimized for a model with 26 predictors. shows the 16 predictors with an absolute value of the estimated coefficient higher than 0.15. The mean differences in methylation percentages were small for all these CpG sites (<3%), except for cg12195446-IRS2 (7.6%) and cg12252547-MAL2 (5.1%). Ten CpGs were hypomethylated in cases. At least two probes were detected for each gene except for RUNX1, STAT3, RUNX3 and PTX3 for which only one probe was found.
Table 2. CpG sites selected by EN with 10-fold CV with an estimated coefficient higher than 0.15 in absolute value in the case-control study nested in the EPIC-Italy cohort.
Table 3. Detailed analysis of the CpG sites Selected for Validation (those reported in with a frequency higher than 50% and an association with MD in the control group in line with its protective effect).
A detailed analysis of the seven CpG sites selected for validation is reported in (results for the remaining CpG sites are reported in Supplementary Table 4). Evidence emerged of an association between all the CpGs and the case-control status also after adjustment for cell types and other confounding variables. In particular, for all the CpGs except cg08053846-SERPINE1, hypomethylation was associated with CC and methylation levels were increased for higher IMI categories.
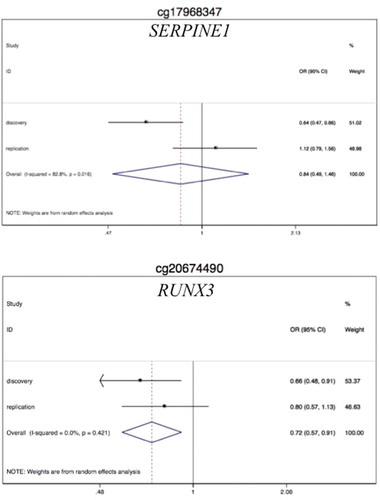
Considering the validation dataset, only cg17968347-SERPINE1 and cg20674490-RUNX3 showed coherent associations with both CC and IMI ( and ). Although these associations were at the limit of significance, their direction and magnitude were essentially the same. Of the two validated CpG sites, only for cg20674490-RUNX3 the association with CC showed coherent direction and similar magnitude in the replication sample (OR = 0.80, 95% CI 0.57, 1.13 in the replication set versus OR = 0.59, 95% CI 0.29,1.19 in the validation set; and , model B). For this CpG the association with MD showed an increasing trend in the replication sample not found in the discovery and validation phases (–). The meta-analysis strengthened the association of CC with RUNX3 higher methylation levels (ORMETA = 0.72, 95% CI 0.57, 0.91, P-valueMETA = 0.006; see ).
Table 4. Validation analysis: associations between colon cancer/MD and DNA methylation levels at SERPINE1 and RUNX3 CpG sites obtained by pyrosequencing in the validation set.
Table 5. Replication analysis: Associations between colon cancer/MD and DNA methylation levels at SERPINE1 and RUNX3 CpG sites obtained by pyrosequencing in the replication set.
Discussion
The adherence to MD is known to have beneficial effects on human health, and in particular, a preventive effect on cancer. Several biological mechanisms have been hypothesized to explain such protective effect, but the specific cellular pattern involved has not been experimentally found yet.
DNA methylation is likely to play a major role in carcinogenesis and it is probably influenced by diet [Citation15,Citation16]. Since it has been previously shown that MD may attenuate the level of chronic inflammation, which in turn has been related to an increased risk of CC, we hypothesized that the protective effect of adherence to the MD in CC is mediated by DNA methylation in genes that are involved in inflammation.
To test this hypothesis, we used the ‘meet-in-the-middle’ approach [Citation30,Citation31]. This approach is based on the assumption that identifying biomarkers associated with both a particular exposure and a certain disease strengthens the causal links between such exposure and the disease. The approach involves to combine, within a prospective study, a search for biomarkers modified in subjects who develop the disease and a search for links of such biomarkers to past environmental exposures. In our study, we considered MD as the exposure, CC as the disease and DNA methylation levels of inflammation genes as the candidate molecular biomarkers. Among the 995 inflammation CpGs studied, we identified probes in the IL1B, SERPINE1, RUNX1, STAT3, NFATC1 and RUNX3 genes as good candidate molecular biomarkers. We implemented the discovery phase with an epigenome-wide assay that is a powerful tool to cover an increasing number of CpG sites but is inherently imprecise and noisy [Citation32]. To overcome this problem we performed validation and replication using a locus-specific methylation technique. Among the seven probes selected for validation, only two were confirmed and only one of the two showed associations in line with our hypothesis in an independent sample (RUNX3).
The human runt-related transcription factor 3 (RUNX3) maps in chromosomal locus 1p36 in a region that is frequently deleted in many types of cancers. RUNX3 has important functions in innate and adaptive immune cell types, in particular in inactivating IL-23A transcription, and has been associated with several immune-related diseases, in particular with an increased risk of ulcerative colitis [Citation33] and Chron’s disease [Citation34]. Recent studies demonstrated that the control of immunity and inflammation exerted by RUNX3 influences epithelial tumour development [Citation35]. Furthermore, RUNX3 is a tumour suppressor gene whose promoter hypermethylation was shown to be a key mechanism of its inactivation [Citation36]. In a study performed on 184 South Korean patients affected by gastric cancer, Zhang at al. found an association between the intake of different foods, in particular, fruit and egg, and RUNX3 methylation [Citation37]. At the opposite, no association among dietary items and RUNX3 gene methylation has been found in a sample of 276 USA healthy women enrolled in a randomized controlled trial on breast cancer prevention [Citation38]. For this reason, we think that RUNX3 is a good candidate mediator between MD and CC, but further functional studies are needed to confirm this promising observation.
Few studies can be found in literature comparing different DNA methylation assays for biomarker development [Citation39,Citation40]. These studies show a good concordance between measurements of the two arrays, but they consider DNA methylation assessed in solid tissues or specific cell lines. To our knowledge, in only one study [Citation41] based on DNA methylation of blood leukocyte, a validation of Illumina 450K array results with pyrosequencing technique is performed, but only five samples were analysed. In our study, we considered 94 samples for validation and 190 samples for replication. The fact that only one signal was confirmed emphasizes the importance of the validation and replication phases employing an alternative methodology. These phases are essential in order to exclude technical errors and false-positive findings especially when the differences in methylation percentages are small most probably due to background noise.
Despite some limitations, this study has several strengths. First of all, we mention the use of the ‘meet-in-the-middle’ approach that we already applied in a similar study suggesting oxidative stress as the mediator in the association of air pollution and cardiovascular diseases [Citation42]. In a study investigating the relation between diet and cancer, exposure assessment is always difficult and the specific mechanisms through which the nutrients exert their protective effect is largely unknown. By assessing the mediation role of intermediate biomarkers we elucidated the biological mechanisms explaining this association.
Secondly, DNA methylation levels were measured in peripheral blood, providing a valuable source of information for low-grade inflammation. However, we were aware that heterogeneity in white blood cells could potentially confound DNA methylation measurements [Citation43]. To address this problem we applied Houseman correction for cell composition in the association analyses [Citation23].
Finally, we performed a case-control study nested in a prospective cohort, in which DNA methylation was assessed in peripheral blood collected at recruitment before the onset of cancer. The analysis of blood sample drawn years before the onset of disease prevents from reverse causality biases.
Conclusions
In conclusion, our study is a first attempt to identify the biological mechanism behind the protection of MD on CC investigating the methylation levels of genes in circulating lymphocytes years before the onset of the disease. The results of the study indicate that DNA methylation of RUNX3 gene may be a potential molecular mediator explaining the protective effect of MD on CC.
Supplemental Material
Download MS Word (55.2 KB)Supplemental Material
Download MS Word (56.8 KB)Acknowledgments
The authors thank all participants in the Italian section of the EPIC study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Trichopoulou A, Costacou T, Bamia C, et al. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med. 2003;348:2599–2608.
- Sofi F, Cesari F, Abbate R, et al. Adherence to Mediterranean diet and health status: meta-analysis. BMJ. 2008;337:a1344.
- Agnoli C, Grioni S, Sieri S, et al. Italian Mediterranean Index and risk of colorectal cancer in the Italian section of the EPIC cohort. Int J Cancer. 2013;132:1404–1411.
- Rosato V, Guercio V, Bosetti C, et al. Mediterranean diet and colorectal cancer risk: a pooled analysis of three Italian case-control studies. Br J Cancer. 2016;115:862–865.
- Torres Stone RA, Waring ME, Cutrona SL, et al. The association of dietary quality with colorectal cancer among normal weight, overweight and obese men and women: a prospective longitudinal study in the USA. BMJ Open. 2017;7:e015619.
- Park SY, Boushey CJ, Wilkens LR, et al. High-quality diets associate with reduced risk of colorectal cancer: analyses of diet quality indexes in the multiethnic cohort. Gastroenterology. 2017;153:386–394.
- Kontou N, Psaltopoulou T, Panagiotakos DB, et al. The Mediterranean diet in cancer prevention: a review. J Med Food. 2011;14:1065–1078.
- Buckland G, Bach A, Serra-Majem L. Obesity and the Mediterranean diet: a systematic review of observational and intervention studies. Obes Rev. 2008;9(6):582–593.
- Arnold M, Pandeya N, Byrnes G, et al. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. Lancet Oncol. 2015;16:36–46.
- Fasanelli F, Zugna D, Giraudo MT, et al. Abdominal adiposity is not a mediator of the protective effect of Mediterranean diet on colorectal cancer. Int J Cancer. 2017;140:2265–2271.
- Casas R, Sacanella E, Urpí-Sardà M, et al. Long-term immunomodulatory effects of a Mediterranean diet in adults at high risk of cardiovascular disease in the PREvención con DIeta MEDiterránea (PREDIMED) randomized controlled trial. J Nutr. 2016;146:1684–1689.
- Koloverou E, Panagiotakos DB, Pitsavos C, et al.; ATTICA Study Group. Adherence to Mediterranean diet and 10-year incidence (2002–2012) of diabetes: correlations with inflammatory and oxidative stress biomarkers in the ATTICA cohort study. Diabetes Metab Res Rev. 2016;32:73–81.
- Cannon J. Colorectal neoplasia and inflammatory bowel disease. Surg Clin North Am. 2015;95:1261–1269.
- Santilli F, Boccatonda A, Davì G. Aspirin, platelets, and cancer: the point of view of the internist. Eur J Intern Med. 2016;34:11–20.
- Scoccianti C, Ricceri F, Ferrari P, et al. Methylation patterns in sentinel genes in peripheral blood cells of heavy smokers: influence of cruciferous vegetables in an intervention study. Epigenetics. 2011;6:1114–1119.
- Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013;34:753–764.
- Wang J, Hodes GE, Zhang H, et al. Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice. Nat Commun. 2018;9:477–491.
- Palli D, Berrino F, Vineis P, et al. EPIC-Italy. A molecular epidemiology project on diet and cancer: the EPIC-Italy prospective study. Design and baseline characteristics of participants. Tumori. 2003;89:586–593.
- Fasanelli F, Baglietto L, Ponzi E, et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun. 2015 Dec;15(6):10192.
- Agnoli C, Krogh V, Grioni S, et al. A priori-defined dietary patterns are associated with reduced risk of stroke in a large Italian cohort. J Nutr. 2011;141:1552–1558.
- Wareham NJ, Jakes RW, Rennie KL, et al. Validity and repeatability of a simple index derived from the short physical activity questionnaires used in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Public Health Nutr. 2003;6:407–413.
- Ricceri F, Sacerdote C, Giraudo MT, et al. The association between educational level and cardiovascular and cerebrovascular diseases within the EPICOR study: new evidence for an old inequality problem. PLoS One. 2016;11:e0164130.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86–102.
- Reid S, Tibshirani R. Regularization paths for conditional logistic regression: the clogitL1 package. J Stat Softw. 2014;58:1–23.
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005;67:301–320.
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol. 1996;58:267–288.
- Hoerl A, Kennard R. Ridge regression. Encyclopedia of Statistical Sciences. Vol. 8. New York: Wiley; 1998. p. 129–136.
- Freidman HJ, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22.
- Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587–596.
- Vineis P, Perera F. Molecular epidemiology and biomarkers in etiologic cancer research: the new in light of the old. Cancer Epidemiol Biomarkers Prev. 2007;16:1954–1965.
- Chadeau-Hyam M, Athersuch TJ, Keun HC, et al. Meeting-in-the-middle using metabolic profiling - a strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers. 2011;16:83–88.
- Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–955.
- Guo C, Yao F, Wu K, et al. Chromatin immunoprecipitation and association study revealed a possible role of Runt-related transcription factor 3 in the ulcerative colitis of Chinese population. Clin Immunol. 2010;135:483–489.
- Yamazaki K, Umeno J, Takahashi A, et al. A genome-wide association study identifies 2 susceptibility Loci for Crohn’s disease in a Japanese population. Gastroenterology. 2013;144:781–788.
- Lotem J, Levanon D, Negreanu V, et al. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta. 2015;1855:131–143.
- Goel A, Arnold CN, Tassone P, et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer. 2004;112:754–759.
- Zhang YW, Eom SY, Yim DH, et al. Evaluation of the relationship between dietary factors, CagA-positive Helicobacter pylori infection, and RUNX3 promoter hypermethylation in gastric cancer tissue. World J Gastroenterol. 2013;19:1778–1787.
- Gillman AS, Gardiner CK, Koljack CE, et al. Body mass index, diet, and exercise: testing possible linkages to breast cancer risk via DNA methylation. Breast Cancer Res Treat. 2018;168:241–248.
- Roessler J, Ammerpohl O, Gutwein J, et al. Quantitative cross-validation and content analysis of the 450k DNA methylation array from Illumina, Inc. BMC Res Notes. 2012;5:210–217.
- BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–737.
- Milenkovic D, Vanden Berghe W, Boby C, et al. Dietary flavanols modulate the transcription of genes associated with cardiovascular pathology without changes in their DNA methylation state. PLoS One. 2014;9:e95527.
- Fiorito G, Vlaanderen J, Polidoro S, et al. EXPOsOMICS consortium‡. Oxidative stress and inflammation mediate the effect of air pollution on cardio- and cerebrovascular disease: A prospective study in nonsmokers. Environ Mol Mutagen. 2018;59:234–246.
- Adalsteinsson BT, Gudnason H, Aspelund T, et al. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS One. 2012;7:e46705.