 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Although underlying mechanisms of long-term exposure to air pollution and cardiovascular disease remain obscure, effects might partially act through changes in DNA methylation. We examined the associations between long-term ambient fine particulate matter (PM2.5) and methylation, considering both a global measure and methylation at several specific inflammation-related loci, in two random sub-cohorts selected from a nationwide prospective study of US women. In one sub-cohort we measured long interspersed nucleotide element (LINE-1); in the other, we measured methylation at three candidates CpG loci related to inflammatory pathways [tumour necrosis factor-alpha (TNF-α) and toll-like receptor-2 (TLR-2)]. Annual average contemporaneous ambient PM2.5 concentrations were estimated for the current residence. We used both classical least-squares and quantile regression models to estimate the long-term effects. The women in sub-cohorts 1 (n = 491) and 2 (n = 882) had mean ages of 55.8 and 56.7, respectively. Neither modelling approach showed an association between long-term PM2.5 and LINE-1 methylation or between PM2.5 and either of the two CpG sites in TLR-2. Using linear regression, there was an estimated change of −6.5% (95% confidence interval CI: −13.34%, 0.35%) in mean methylation of TNF-α per 5 µg/m3 increase in PM2.5. Quantile regression showed that the downward shift was mainly in the lower half of the distribution of DNA methylation. Long-term residence in regions with higher ambient PM2.5 may be associated with increased TNF-α through a reduction in methylation, particularly in the lower tail. Epigenetic markers and quantile regression might provide insight into mechanisms underlying the relationship between air pollution and cardiovascular disease.
Introduction
The Global Burden of Disease study has estimated that long-term exposure to ambient fine particulate air pollution (particulate matter with aerodynamic diameter less than or equal to 2.5 µm, PM2.5) caused 4.2 million deaths and 103.1 million lost years of healthy life in 2015, representing 7.6% of total global mortality, and making it the fifth-ranked global risk factor in 2015 [Citation1]. Cardiovascular disease accounted for most excess deaths [Citation1]. Previous studies provided compelling evidence that long-term exposure to particulate matter air pollution is associated with cardiovascular morbidity and mortality [Citation2–Citation5].
Although the precise mechanisms underlying this association remain unclear, the induction of systemic inflammation following particle inhalation is regarded as a plausible pathway [Citation6–Citation9]. Increases in inflammatory biomarkers [e.g., tumour necrosis factor-alpha (TNF-α), C-reactive protein] have been linked to cardiovascular disease [Citation10,Citation11]. In considering biological pathways by which air pollution could induce inflammation, changes in DNA methylation in the inflammatory system could play an important role [Citation12–Citation18], as such changes are also associated with cardiovascular diseases [Citation19–Citation22].
DNA methylation involves the addition of a methyl group to cytosine and predominantly occurs at cytosine-guanine dinucleotide (CpG) sites [Citation23]. The degree of methylation is quantified as methylated cytosines over the sum of methylated and unmethylated cytosines at the position of 5-cytosine, ranging from 0 to 1. Reduced methylation in inflammatory genes is often associated with increased expression. Evidence directly linking air pollution exposure with DNA methylation is limited, and primarily focused on short-term effects [Citation12–Citation15,Citation18,Citation24]. Previously, we carried out a panel study [Citation18,Citation24] to evaluate short-term associations between PM2.5 and LINE-1 methylation and DNA methylation of several inflammation genes, whose regulatory proteins were targets in other environmental health studies [Citation14,Citation25–Citation29]. The panel study examined biomarkers including TNF-α (two loci). intercellular adhesion molecule-1 (ICAM-1, three loci), cluster of differentiation 40 ligand (CD40l, two loci), interleukin-6 (two loci), and Toll-like receptor-2 (TLR-2, five loci) (Details provided in supplementary material, Table S1). We found that acute increases in PM2.5 corresponded to significant decreases in LINE-1 methylation and DNA methylation for TNF-α, ICAM-1 and TLR-2 at specific loci. Few have investigated health effects on DNA methylation of medium- or long-term time intervals of exposure [Citation16,Citation17].
Hence, we were interested in looking at the long-term associations between PM2.5 and LINE-1 methylation and DNA methylation of the same genes at the same loci. Our panel study used personal monitors to measure PM2.5 concentration, but it is not feasible to ask participants to wear the monitors for a long time. Therefore, we conducted the long-term association analyses in the Sister Study, which already had available annual PM2.5 estimates and DNA methylation levels (Illumina’s Infinium HumanMethylation450 BeadChip) as well. After comparison, we only found three common CpG sites (TNF-α at cg21370522; TLR-2 at cg16547110 and cg06405222). Moreover, methylation level of LINE-1 was also available in the Sister Study [Citation30]. We hypothesized that long-term exposure to air pollution is associated with changes in methylation globally, and at cg21370522 of TNF-α, at cg16547110, cg06405222 of TLR-2. Such effects could cause an overall shift in the distribution or could vary by quantiles, reflecting heterogeneity in susceptibility. Thus, we carried out both classical regression to assess the association between mean methylation and the annual average ambient PM2.5 exposure, and quantile regression [Citation31].
Materials and methods
Study population
The Sister Study is a nationwide prospective cohort of 50,884 women, aged 35–74 years, who each had a sister with breast cancer but were free of breast cancer themselves (https://sisterstudy.niehs.nih.gov/English/index1.htm). Participants enrolled in this study between 2003 and 2009, when they completed computer-assisted telephone interviews and were visited at home by a trained examiner. The visit included blood collection and measurements of height and weight. The Sister Study was approved by the Institutional Review Boards of the National Institute of Environmental Health Sciences and the Copernicus Group. All women provided written informed consent. Because effects and methylation patterns could differ by race/ethnicity [Citation32], we restricted our analysis to the major subpopulation, i.e., non-Hispanic white women.
Study sample
The study samples were from two random sub-cohorts, which were originally designed to study the association between DNA methylation and breast cancer. The first sub-cohort [Citation30] was identified earlier, and included smaller number of participants. It was used in study of the association between global methylation and breast cancer. The second sub-cohort [Citation33] included more participants and was used to study the association between breast cancer and individual CpGs. Here we have utilized the methylation data from these two studies to investigate the effect of air pollution on DNA methylation changes.
LINE-1 analysis
The original random sub-cohort selected for LINE-1 analyses included 721 participants. We then excluded women if they had lived in their current residence less than 5 years, or if they were current smokers or past smokers who had quit smoking less than 5 years prior to baseline. After those exclusions, 491 participants remained.
Locus-specific analyses
The original random sub-cohort selected for gene-specific methylation analyses included 1,295 women. After applying the same exclusion criteria as above for duration of residence and smoking status, 882 participants remained. There were 151 women selected in both sub-cohorts. The flowchart in describes the selection of participants for the current analyses.
DNA methylation measures
LINE-1 methylation
Global DNA methylation was assessed by LINE-1 methylation pyrosequencing. Briefly, genomic DNA was extracted from frozen whole blood samples and bisulphite converted. Then the bisulphite-converted DNA was amplified and bound to a single-strand template. The pyrosequencing run was carried out using the PSQ HS 96 System (Biotage, Charlotte, North Carolina, USA). Further details are provided elsewhere [Citation30]. The methylation level was expressed as the proportion of methylated cytosines over the sum of methylated and unmethylated cytosines at position 5-cytosine, as described above.
Locus-specific methylation
We used Illumina’s Infinium HumanMethylation450 BeadChip, which measures methylation levels at 485,577 CpG sites, with an average coverage of 17 CpG sites per gene. For this analysis, we targeted three specific CpG sites that had been measured in both the HumanMethylation450 BeadChip and our previous panel study [Citation18]. We selected the candidate loci based on published literature [Citation34–Citation36]. They included one locus in TNF-α (cg21370522) and two loci (cg16547110 and cg06405222) in TLR-2. The details of methods for methylation measurements have been previously described [Citation37]. We list the genes and each CpG position in our previous panel study in the supplementary material, Table S1, where we marked the three specific CpG sites in the present study (see supplementary material, Table S1).
Exposure assessment
We applied a regionalized national universal kriging model using partial least squares to estimate annual PM2.5 concentrations [Citation38] for residences. We assigned the concentration of PM2.5 in the year preceding blood draw to approximate long-term residential exposure to PM2.5. More specifically, for women who had their blood draw in the first half of the year, the PM2.5 level was taken to be the annual average of the preceding year; otherwise, the PM2.5 level was taken as the annual average in the enrolment year.
Statistical analyses
DNA methylation levels were log-transformed to better approximate normal distributions. For the DNA methylation distribution, see supplementary material, Figure S1. First, we used least-squares regression to examine the associations of long-term ambient PM2.5 exposure and LINE-1, and methylation at our selected inflammation-related loci. Second, we applied quantile regression to estimate the associations between PM2.5 and quantiles of LINE-1, and methylation at gene-specific loci, applying the model:
where is the τ th quantile of the ln(DNA methylation) distribution;
is between 0 and 100. Also, PM2.5 is the one-year average concentration, as specified above, X2, X3, … are covariates, and
is an error term. We specified nine quantiles, which were 10th, 20th, 30th, 40th, 50th, 60th, 70th, 80th, and 90th.
In both regression models, we adjusted for several covariates: age at baseline, body mass index [BMI, computed as weight (in kilograms) divided by height (in metres squared)], alcohol consumption (never/former drinker, current drinker <1 drink/day, and current drinker ≥1 drink/day), education (high school or less, some college, bachelor’s degree, graduate degree), mother’s smoking status when participant was in utero (definitely or probably, definitely not or probably not), and cell-type proportions (CD8 + T-cells, CD4 + T-cells, B-cells, granulocytes, monocytes, and natural killer cells), which were estimated using a method described by Houseman et al. [Citation39].
We report the linear and quantile regression coefficient as the percentage change associated with a 5 µg/m3 difference in PM2.5. We provide the corresponding 95% confidence intervals, without correction for multiple testing. All analyses were conducted with SAS (version 9.4), using the ‘glm’ (for linear regression), and ‘quantreg’ (for quantile regression) packages.
Sensitivity analyses were performed to investigate whether the associations between PM2.5 exposure and DNA methylation were sensitive to time spent outdoors and age. We stratified by age based on the median in each sub-cohort.
Results
Main results
summarizes characteristics of the sub-cohorts. At baseline, more than two-thirds were current drinkers who drank less than 1 drink per day, and more than half had a bachelor’s degree or higher.
Table 1. Demographic characteristics of the two random sub-cohorts in the Sister Study.
Characteristics of the methylation levels and one-year mean PM2.5 are presented in . LINE-1 mean methylation level was 0.763. The methylation levels of TNF-α and TLR-2 had means of 0.082, 0.020, and 0.042 for cg21370522, cg16547110, and cg06405222, respectively. The variation in TNF-α methylation at cg21370522 was higher than that for other loci and LINE-1. The mean annual PM2.5 level was 9.3 µg/m3 in both sub-cohorts, with low coefficients of variation across participants (see supplementary material, Figure S2).
Table 2. Distribution of DNA methylation and prior year mean PM2.5 concentration.
Least-squares regression showed that the long-term ambient PM2.5 level was associated with a small reduction in TNF-α methylation at cg21370522 (−6.50%, 95% CI: −13.34%, 0.35%, per 5 μg/m3 increase in ambient PM2.5). The estimated associations between PM2.5 and LINE-1, cg16547110 (TLR-2), and cg06405222 (TLR-2) were weaker ()), with estimated changes of −0.13% (95% CI: −0.51%, 0.25%), −2.62% (95% CI: −7.25%, 2.00%), and 0.91% (95% CI: −3.46%, 5.28%), respectively.
Figure 2. (a) Percentage change in DNA methylation associated with a 5 µg/m3 increase in ambient PM2.5, results from linear regression. The X-axis refers to different genes; the Y-axis refers to the corresponding changes (mean and 95% confidence interval). (b) Percentage change in DNA methylation associated with a 5 µg/m3 increase in ambient PM2.5 at each decile of the methylation. The X-axis refers to different quantiles; the Y-axis refers to the corresponding changes (mean and 95% confidence interval). (Number of subjects in LINE-1 analyses is 491; number of subjects in locus-specific analysis is 882).
LINE-1: long interspersed nucleotide element; TNF-α: tumour necrosis factor-alpha; TLR-2: toll-like receptor-2; PM2.5: particulate matter with aerodynamic diameter less than or equal to 2.5 µm.
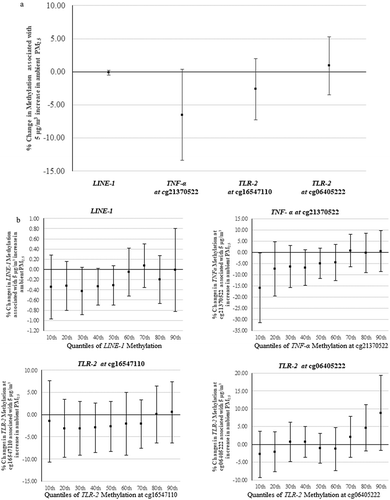
The quantile analysis yielded a range of estimates: the associations between PM2.5 and LINE-1 methylation and gene-specific methylation varied some across quantiles ()). The association of PM2.5 with quantiles of TNF-α methylation at cg21370522 showed a trending pattern of that was consistent with the results of linear regression. The associations of PM2.5 with LINE-1 and TLR-2 showed more irregular fluctuation across deciles of the corresponding outcomes. All estimated effects from linear and quantile regressions are shown in supplementary material, Table S1.
Sensitivity analyses
As secondary analysis, we examined the associations after restricting to women who reported spending more than 4 h per week outdoors. As shown in ), PM2.5 exposure was associated with a 7.33% decrease in methylation for TNF-α at cg21370522 (95% CI: −14.63%, −0.03%) per 5 μg/m3 increase in ambient PM2.5. Figure S3A shows the corresponding results from quantile regression (see supplementary material, Figure S3A).
Figure 3. (a) Percentage change in DNA methylation associated with a 5 µg/m3 increase in ambient PM2.5 in women who spent more than 4 h per week outside, results from linear regression. The X-axis refers to different genes; the Y-axis refers to the corresponding changes (mean and 95% confidence interval). (Number of women in LINE-1 analysis is 429; number of women in locus-specific analyses is 786.). (b) The percentage changes in DNA methylation associated with a 5 µg/m3 increase in ambient PM2.5 in younger (age ≤ median) and older women (age > median), using linear regression. The X-axis refers to different genes; the Y-axis refers to the corresponding changes (mean and 95% confidence interval). (The median age in LINE-1, and locus-specific analysis is 56.2, and 57.0, respectively,).
LINE-1: long interspersed nucleotide element; TNF-α: tumour necrosis factor-alpha; TLR-2: toll-like receptor-2; PM2.5: particulate matter with aerodynamic diameter less than or equal to 2.5 µm.
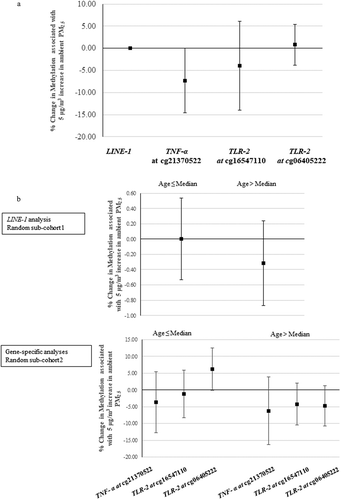
We also assessed the associations between one-year average PM2.5 and DNA methylation for women older and younger than the median age, in the two random sub-cohorts, separately, shown in ). PM2.5 was not strongly associated with our measures of DNA methylation either in younger or older women. Considering quantiles, the relationship between PM2.5 and DNA methylation was inverse in older women across almost all percentiles (see supplementary material, Figure S3B).
Discussion
We examined the associations between long-term PM2.5 exposure and LINE-1 methylation and DNA methylation at specific loci associated with inflammation, using both linear and quantile regression. Our findings suggest that the prior year’s exposure to ambient PM2.5 may be negatively associated with TNF-α methylation at cg21370522, with displacement showing predominantly in the lower tail. We did not find compelling evidence of associations between long-term PM2.5 and methylation of LINE-1 or CpGs in TLR-2, though older women were estimated to be more susceptible than younger women to reduction in methylation associated with increased PM2.5 at cg06405222 (TLR-2).
Acute exposure to air pollution and DNA methylation been reported in both the environmental [Citation12], and the occupational literature [Citation40,Citation41]. For instance, in the Normative Ageing Study, which included a large sample of elderly men, LINE-1 methylation was inversely associated with ambient levels of black carbon and PM2.5 measured across the 7 days before the examination [Citation12]. However, few studies have examined whether medium- or long-term exposure to air pollution influences LINE-1 methylation, and the results have been somewhat inconsistent [Citation16,Citation17]. For example, Madrigano et al., found that an interquartile range (0.25 μg/m3) increase in black carbon over a 60-day period was associated with a decrease of 0.0029 (95% confidence interval (CI): −0.0056, −0.0002) in long interspersed nucleotide element (LINE-1) [Citation17], whereas Gloria et al., did not find substantial changes in LINE-1 associated with the previous 1 year of air pollution exposure [Citation16].
To date, no studies have estimated chronic effects of air pollution on DNA methylation of inflammation-specific genes such as TNF-α and TLR-2. Our findings only showed associations between long-term PM2.5 and TNF-α at cg21370522, which is located upstream of the TNF-α promoter, and has been targeted in several studies [Citation42,Citation43]. For example, Shinozake et al. found that the DNA methylation level at cg21370522 had a negative correlation with chronological age in the Grady Trauma Project cohort, which suggested that it is involved in gene expression regulation in ageing [Citation42]. Moreover, lower TNF-α methylation at cg21370522 was shown to be significantly correlated with its mRNA expression increment in several diseases, such as pheochromocytoma and paraganglioma, based on Broad Institute TCGA Genome Data Analysis Centre [Citation44–Citation47].
Our null findings for LINE-1 are in line with some scientific reports [Citation16,Citation48,Citation49]. For instance, Chi et al. found no association between one-year average exposure to ambient air pollution and LINE-1 methylation in 1,207 participants in the Multi-Ethnic Study of Atherosclerosis [Citation16]. Using data from the Italian and Dutch components of the European Prospective Investigation into Cancer and Nutrition Cohort Study, Plusquin et al., reported that long-term exposure to PM2.5 did not result in statistically significant global changes in methylation (as measured by the arithmetic mean of beta-values across all somatic probes) [Citation49]. However, some studies did find an association between medium- or long-term air pollution and lower LINE-1 methylation [Citation17,Citation41,Citation48]. For instance, based on 706 elderly males in the Normative Ageing Study, Madrigano et al., found that an interquartile range increase (0.83 μg/m3) in SO4 over a 90-day period was associated with a small decrease of 0.0027 (95%CI: −0.0052, −0.0002) in LINE-1 [Citation17] (less than 1% of the mean). In an electric furnace steel plant, Tarantini et al., measured LINE-1 methylation in 63 workers, and found that PM10 exposure levels were negatively associated with methylation in LINE-1 (PM10 Coefficient −0.0034; p = 0.04), presumably reflecting long-term PM10 effects [Citation41]. Barchitta et al. enrolled 299 healthy women in a cross-sectional study, and found that the monthly mean PM10 level was statistically significantly and negatively associated with LINE-1 methylation (PM10 Coefficient −0.0012; p = 0.037) [Citation48].
Because of its high representation throughout the genome, LINE-1 methylation has been shown to correlate with global genomic DNA methylation content [Citation50], and is regarded as a measure of global DNA methylation [Citation51]. Some investigations have linked changes in LINE-1 methylation to inflammation [Citation52,Citation53]. Lower LINE-1 methylation was associated with higher serum vascular Cell Adhesion Molecule-1 [Citation54], which has been identified as an independent marker of cardiovascular disease [Citation55]. Moreover, lower levels of LINE-1 methylation were associated with an adverse lipid profile – higher levels of fasting low-density lipoprotein (and lower levels of fasting high-density lipoprotein), which is a risk factor for cardiovascular disease [Citation20]. Also, lower LINE-1 methylation has recently been associated with incident ischaemic heart disease and stroke, and total mortality [Citation19].
There is increasing evidence for a role of inflammation in cardiovascular disease. For example, higher levels of TNF-α protein have been shown to correlate with severity of peripheral arterial disease [Citation56], and are also linked with the degree of early atherosclerosis in healthy middle-aged men [Citation11]. Although we did not measure the TNF-α protein, hypo-methylation in TNF-α might lead to an incremental increase in the levels of TNF-α protein, as we saw in our previous panel study [Citation18]. Our sensitivity analysis showed that methylation at a locus related to TNF-α (cg21370522) was particularly reduced in women who spent more than 4 h outdoors per week (see )).
We did not find any overall associations between long-term PM2.5 and TLR-2 at two specific loci (cg16547110 and cg06405222) in our main analyses. In our previous panel study, we also did not find associations between short-term PM2.5 and TLR-2 at those loci. But in that panel study, we did find a statistically significant decrease of DNA methylation in two other CpG sites (not available for the current analysis) near TLR-2 [Citation18]. No previous study has examined the long-term influence of PM2.5 on TLR-2 DNA methylation. But several studies investigated short-term effects. For example, Bind et al. found that the previous 4-week exposure to black carbon was associated with a statistically significant mean decrease in TLR-2 methylation, which would lead to higher TLR-2 protein levels [Citation14]. TLR-2 is a member of the Toll-like receptor family, which serves as first-line defence in innate immunity. They interpreted these findings as evidence that TLR-2 might play an important role in the development and/or progression of atherosclerosis [Citation57].
In contrast to linear regression, quantile regression can be used to estimate the associations between air pollution and specific percentiles of the outcome distribution. It allows one to identify whether the associations suggest that responses are heterogeneous, increasing the spread in the methylation distribution. Another advantage of quantile regression is that it does not require one to assume normality for the residuals and can therefore be used to estimate associations between air pollution and biomarkers that are not normally distributed. For consistency with the linear regression, we also log-transformed methylation in quantile regression in the present study. The larger reductions in methylation at cg21370522 that we saw in the lower quantiles of TNF-α methylation distribution, are consistent with the possibility that a subpopulation responds to chronic exposure to PM2.5 (and perhaps to other noxious exposures) by reducing methylation at this site.
One major limitation in our study is that we used the individual’s home address as basis for ambient PM2.5 exposure estimates, which inevitably causes some degree of exposure misclassification at the individual level. Although most people spend 85–90% of their time indoors [Citation48], our sensitivity analyses excluding individuals with very limited outdoor time did not show pronounced changes. Another serious limitation is that our study population comprised individuals living in the United States, which has relatively low air pollution concentrations compared to many parts of the world. The variation of PM2.5 across the women in our sub-cohorts was also very low in our study period, which makes it difficult to detect any association. Finally, this was a cross-sectional study, and we were unable to infer causality between PM2.5 and DNA methylation. Studies of migrants would be particularly useful for confirming the temporal effects that would be needed to infer causality.
This study was, to our knowledge, the first to examine the associations between long-term PM2.5 and DNA methylation using quantile regression. Unlike linear regression analysis, quantile regression allows one to explore effects on the overall shape of the outcome distribution, rather than shifts in the mean only. Another strength of our study was that we had a complete residential history for all participants, allowing us to exclude woman who had recently changed their address.
Conclusions
Though we did not observe clear associations between estimated PM2.5 exposure and methylation levels of LINE-1 or specific inflammation-related genes, our results suggest that long-term ambient PM2.5 exposure may influence TNF-α through reduced methylation at cg21370522, especially in the lower quantiles of that distribution. It is possible that detrimental effects of PM2.5 are more pronounced for women who already have reduced methylation at that locus, perhaps due to other exposures, health conditions or inherent susceptibility. These results require replication, ideally in a population with a greater range of chronic exposure to PM2.5.
Declarations of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download MS Word (669 KB)Acknowledgments
We thank Dr Kaitlyn Gam and Dr Jacob Kresovich for their critical review of this manuscript. We also thank all women who volunteered to participate in this study. We would also like to thank the National Institute of Environmental Health Sciences (NIEHS) Microarray Core for their technical support. This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, which provided funding for design and conduct of the study; collection, management, analysis and interpretation of the data, and preparation, review and approval of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Cohen AJ, Brauer M, Burnett R, et al. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the global burden of diseases study 2015. Lancet. 2017;389(10082):1907–1918. PubMed PMID: 28408086; PubMed Central PMCID: 5439030.
- Brook RD, Rajagopalan S, Pope CA, et al. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American heart association. Circulation. 2010;121(21):2331–2378. PubMed PMID: 20458016.
- Hoek G, Krishnan RM, Beelen R, et al. Long-term air pollution exposure and cardio-respiratory mortality: a review. Environ Health. 2013;12(1):43. PubMed PMID: 23714370; PubMed Central PMCID: 3679821.
- Newby DE, Mannucci PM, Tell GS, et al. Expert position paper on air pollution and cardiovascular disease. Eur Heart J. 2015;36(2):83–93b. PubMed PMID: 25492627; PubMed Central PMCID: 6279152.
- Pun VC, Kazemiparkouhi F, Manjourides J, et al. Long-term PM2.5 exposure and respiratory, cancer, and cardiovascular mortality in older US adults. Am J Epidemiol. 2017;186(8):961–969. PubMed PMID: 28541385.
- Green R, Broadwin R, Malig B, et al. Long- and short-term exposure to air pollution and inflammatory/hemostatic markers in midlife women. Epidemiology. 2016;27(2):211–220. PubMed PMID: 26600256; PubMed Central PMCID: 4841679.
- Mostafavi N, Vlaanderen J, Chadeau-Hyam M, et al. Inflammatory markers in relation to long-term air pollution. Environ Int. 2015;81:1–7. PubMed PMID: 25898227.
- Ostro B, Malig B, Broadwin R, et al. Chronic PM2.5 exposure and inflammation: determining sensitive subgroups in mid-life women. Environ Res. 2014;132:168–175. PubMed PMID: 24792413; PubMed Central PMCID: 4314307.
- Viehmann A, Hertel S, Fuks K, et al. Long-term residential exposure to urban air pollution, and repeated measures of systemic blood markers of inflammation and coagulation. Occup Environ Med. 2015;72(9):656–663. PubMed PMID: 26163546.
- Li Y, Zhong X, Cheng G, et al. Hs-CRP and all-cause, cardiovascular, and cancer mortality risk: a meta-analysis. Atherosclerosis. 2017;259:75–82. PubMed PMID: 28327451.
- Skoog T, Dichtl W, Boquist S, et al. Plasma tumour necrosis factor-alpha and early carotid atherosclerosis in healthy middle-aged men. Eur Heart J. 2002;23(5):376–383. PubMed PMID: 11846495.
- Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. PubMed PMID: 19136372; PubMed Central PMCID: 2720123.
- Bellavia A, Urch B, Speck M, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. 2013;2(3):e000212. PubMed PMID: 23782920; PubMed Central PMCID: 3698788.
- Bind MA, Lepeule J, Zanobetti A, et al. Air pollution and gene-specific methylation in the normative aging study: association, effect modification, and mediation analysis. Epigenetics. 2014;9(3):448–458. PubMed PMID: 24385016; PubMed Central PMCID: 4053463.
- Chen R, Qiao L, Li H, et al. Fine particulate matter constituents, nitric oxide synthase DNA methylation and exhaled nitric oxide. Environ Sci Technol. 2015;49(19):11859–11865. PubMed PMID: 26372312.
- Chi GC, Liu Y, MacDonald JW, et al. Long-term outdoor air pollution and DNA methylation in circulating monocytes: results from the multi-ethnic study of atherosclerosis (MESA). Environ Health. 2016;15(1):119. PubMed PMID: 27903268; PubMed Central PMCID: 5131503.
- Madrigano J, Baccarelli A, Mittleman MA, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119(7):977–982. PubMed PMID: 21385671; PubMed Central PMCID: 3222977.
- Wang C, Chen R, Shi M, et al. Possible mediation by methylation in acute inflammation following personal exposure to fine particulate air pollution. Am J Epidemiol. 2017. PubMed PMID: 29020142; PubMed Central PMCID: 5860518. DOI:10.1093/aje/kwx277
- Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21(6):819–828. PubMed PMID: 20805753; PubMed Central PMCID:3690659.
- Cash HL, McGarvey ST, Houseman EA, et al. Cardiovascular disease risk factors and DNA methylation at the LINE-1 repeat region in peripheral blood from Samoan Islanders. Epigenetics. 2011;6(10):1257–1264. PubMed PMID: 21937883; PubMed Central PMCID: 3225843.
- Kim M, Long TI, Arakawa K, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5(3):e9692. PubMed PMID: 20300621; PubMed Central PMCID: 2837739.
- Turunen MP, Aavik E, Ylä-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790(9):886–891. PubMed PMID: 19233248.
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204. PubMed PMID: 23400093.
- Wang C, Chen R, Cai J, et al. Personal exposure to fine particulate matter and blood pressure: a role of angiotensin converting enzyme and its DNA methylation. Environ Int. 2016;94:661–666. PubMed PMID: 27397929.
- Schneider A, Neas LM, Graff DW, et al. Association of cardiac and vascular changes with ambient PM2.5 in diabetic individuals. Part Fibre Toxicol. 2010 June 2;7:14. 20525188.Pmc2896918.
- Chen R, Zhao Z, Sun Q, et al. Size-fractionated particulate air pollution and circulating biomarkers of inflammation, coagulation, and vasoconstriction in a panel of young adults. Epidemiology. 2015;26(3):328–336.
- Donaldson GC, Seemungal TA, Patel IS, et al. Airway and systemic inflammation and decline in lung function in patients with COPD. Chest. 2005;128(4):1995–2004.
- Willerson JT, Ridker PM. Inflammation as a cardiovascular risk factor. Circulation. 2004;109(21_suppl_1):II-2-II-10.
- Peng C, Bind MC, Colicino E, et al. Particulate air pollution and fasting blood glucose in nondiabetic individuals: associations and epigenetic mediation in the normative aging study, 2000–2011. Environ Health Perspect. 2016 Nov;124(11):1715–1721. 27219535.PMC5089881 represent the views of the U.S. Department of Veterans Affairs. The authors declare they have no actual or potential competing financial interests. DOI:10.1289/ehp183
- Deroo LA, Bolick SC, Xu Z, et al. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the Sister Study. Carcinogenesis. 2014;35(2):333–338. PubMed PMID: 24130171; PubMed Central PMCID: 3908748.
- Bind MA, Peters A, Koutrakis P, et al. Quantile regression analysis of the distributional effects of air pollution on blood pressure, heart rate variability, blood lipids, and biomarkers of inflammation in elderly American men: the normative aging study. Environ Health Perspect. 2016;124(8):1189–1198. PubMed PMID: 26967543; PubMed Central PMCID: 4977045.
- Zhang FF, Cardarelli R, Carroll J, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6(5):623–629. PubMed PMID: 21739720; PubMed Central PMCID: 3230547.
- Xu Z, Sandler DP, Taylor JA. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the Sister Study. J Natl Cancer Inst. 2019. PubMed PMID: 30989176. DOI:10.1093/jnci/djz065
- Zhang S, Barros SP, Moretti AJ, et al. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013;84(11):1606–1616. PubMed PMID: 23368949; PubMed Central PMCID: PMC3986590.
- Kaut O, Ramirez A, Pieper H, et al. DNA methylation of the TNF-α promoter region in peripheral blood monocytes and the cortex of human Alzheimer’s disease patients. Dement Geriatric Cogn Disord 2014; 38(1–2):10–15. PubMed PMID: 24556805; PubMed Central PMCID: PMC3329504.
- Madrigano J, Baccarelli AA, Mittleman MA, et al. Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics. 2012;7(1):63–70. PubMed PMID: 22207354; PubMed Central PMCID: PMC3329504.
- Kresovich JK, Xu Z, O’Brien KM, et al. Methylation-based biological age and breast cancer risk. J Natl Cancer Inst. 2019;111:1051–1058. PubMed PMID: 30794318.
- Sampson PD, Richards M, Szpiro AA, et al. A regionalized national universal kriging model using partial least squares regression for estimating annual PM2.5 concentrations in epidemiology. Atmos Environ (1994). 2013; 75:383–392. PubMed PMID: 24015108; PubMed Central PMCID: 3763950.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. PubMed PMID: 22568884; PubMed Central PMCID: 3532182.
- Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. PubMed PMID: 17283117.
- Tarantini L, Bonzini M, Apostoli P, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117(2):217–222. PubMed PMID: 19270791; PubMed Central PMCID: 2649223.
- Schneider A, Neas LM, Graff DW, et al. Association of cardiac and vascular changes with ambient PM2.5 in diabetic individuals. Part Fibre Toxicol. 2010;7:14. PubMed PMID: PubMed PMID: 23368949; PubMed Central PMCID: PMC3986590.
- Huang B. Immune dysregulation and pancreatic cancer: overactivity, inflammation and epigenetic modifications of immune function [dissertation]. UCLA; 2019.
- Broad Institute TCGA Genome Data Analysis Center. Correlation between mRNA expression and DNA methylation. Broad Institute of MIT and Harvard. 2016. DOI:10.7908/C1BR8RJ2
- Broad Institute TCGA Genome Data Analysis Center. Correlation between mRNA expression and DNA methylation. Broad Institute of MIT and Harvard. 2016. DOI:10.7908/C1Q23ZM2
- Broad Institute TCGA Genome Data Analysis Center (2016). Correlation between mRNA expression and DNA methylation. Broad Institute of MIT and Harvard. 2016. DOI:10.7908/C1TQ60ZK
- Broad Institute TCGA Genome Data Analysis Center (2016). Correlation between mRNA expression and DNA methylation. Broad Institute of MIT and Harvard. 2016. DOI:10.7908/C1W958NZ
- Barchitta M, Maugeri A, Quattrocchi A, et al. Mediterranean diet and particulate matter exposure are associated with LINE-1 methylation: results from a cross-sectional study in women. Front Genet. 2018;9:514. PubMed PMID: 30425730; PubMed Central PMCID: 6218419.
- Plusquin M, Guida F, Polidoro S, et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int. 2017;108:127–136. PubMed PMID: 28843141; PubMed Central PMCID: 6139298.
- Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by methyLight. Nucleic Acids Res. 2005;33(21):6823–6836. PubMed PMID: 16326863; PubMed Central PMCID: 1301596.
- Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. PubMed PMID: 14973332; PubMed Central PMCID: 373427.
- Carraro JC, Mansego ML, Milagro FI, et al. LINE-1 and inflammatory gene methylation levels are early biomarkers of metabolic changes: association with adiposity. Biomarkers. 2016;21(7):625–632. PubMed PMID: 27098005.
- Gasche JA, Hoffmann J, Boland CR, et al. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer. 2011;129(5):1053–1063. PubMed PMID: 21710491; PubMed Central PMCID: 3110561.
- Baccarelli A, Tarantini L, Wright RO, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5(3):222–228. PubMed PMID: 20305373; PubMed Central PMCID: 3155741.
- Galkina E, Ley K. Vascular adhesion molecules in atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27(11):2292–2301. PubMed PMID: 17673705.
- Signorelli SS, Anzaldi M, Libra M, et al. Plasma levels of inflammatory biomarkers in peripheral arterial disease: results of a cohort study. Angiology. 2016;67(9):870–874. PubMed PMID: 26888895.
- Mullick AE, Tobias PS, Curtiss LK. Modulation of atherosclerosis in mice by Toll-like receptor 2. J Clin Invest. 2005;115(11):3149–3156. PubMed PMID: 16211093; PubMed Central PMCID: 1242192.

