ABSTRACT
Merkel cell carcinoma (MCC) is a very rare, but highly aggressive skin cancer which occurs mainly in elderly patients. MCC cells show an expression pattern of three cell lineages: epithelial, neuroendocrine, and B-cell progenitor. This trilinear expression pattern suggests stemness activity in MCC. The etiopathogenesis of MCC is either linked to the Merkel cell polyomavirus (MCPyV) or in a smaller proportion (20%) to high levels of UV-induced somatic mutations. Both viral presence and accumulation of mutations have been shown to be associated with accelerated DNA methylation Age (DNAmAge) compared to chronological age. The MCC DNAmAge was significantly lower compared to the chronological age, which was irrespective of the viral presence or mutational burden. Although these features indicate some aspects of stemness in MCC cells, gene-expression-based pluripotency testing did not provide evidence for pluripotency of MCC cells.
To the editor
Merkel cell carcinoma (MCC) is a highly aggressive non-melanoma skin cancer of mainly elderly and immunosuppressed patients. The survival of patients with MCC in advanced clinical stages is only 50% after 9 months [Citation1]. The etiopathogenesis of the majority of MCC is closely linked to the recently discovered Merkel cell polyomavirus (MCPyV) [Citation1]. The remaining approximately 20% of MCPyV-negative MCCs are associated with a very high burden of UV-induced somatic mutations [Citation2]. Interestingly, infection of blood cells with Epstein-Barr virus (EBV), cytomegalovirus (CMV) as well as human immunodeficiency virus (HIV) and expression of the E6/E7 human papillomavirus (HPV) 16/18 proteins in cervical squamous cell carcinoma have been associated with accelerated DNA methylation Age (DNAmAge) as compared to the patients' chronological age [Citation3–5]. In addition, Horvath described an inverse relationship between somatic mutations and DNAmAge acceleration [Citation3]. In contrast, a low DNAmAge is found in stem cells and induction of pluripotency is associated with ‘juvenescence’ of cells.
Remarkably, MCC reveals a trilinear differentiation characterized by concurrent neuroendocrine, epithelial, and pre/pro B-cell lymphocytic gene expression obscuring its – currently unknown and controversially – debated cellular origin [Citation6,Citation7]. The trilinear differentiation of MCC is suggestive for a certain ‘stemness’ of the cell of origin. Considering these characteristics of MCC and in order to shed further light into their pathogenesis, we here investigated both the DNAmAge and the pluripotency gene-expression profile of MCPyV-positive and negative MCCs.
We isolated DNA and RNA of 14 fresh-frozen MCC tissues (originating from 12 patients) and four MCC cell lines (MKL-1, MKL-2, WaGa, and MCC13) using the AllPrep DNA/RNA Mini Kit (Qiagen). DNA and RNA were further processed by the custom service provided GenomeScan BV, Leiden, the Netherlands. First, the DNA concentration was assessed using the Qubit assay from Invitrogen, Carlsbad, California, USA. The quality was assessed by gel electrophoresis. All samples passed the quality control from GenomeScan BV. Subsequently, 500 ng of each DNA sample was bisulphite converted using the EZ DNA Methylation Gold Kit (Zymo Research, Irvine, California, USA). A bisulphite quality control on the samples consisting of a qPCR reaction and melting curve analysis was performed by GenomeScan BV. Again, all samples passed the GenomeScan BV quality control. Afterwards, each sample was directly utilized for the methylation analysis. For the microarray-based DNA methylation analyses, the bisulphite-converted DNA was applied to the Infinium MethylationEPIC BeadChip targeting over 850,000 methylation sites per sample (850 K) according to the manufacturer’s instructions (Illumina, Inc., San Diego, California, USA) performed by GenomeScan BV. The GenomeScan protocol for sample preparation, hybridization, and washing of the 850 K array was performed without deviations of the Illumina protocol (‘Infinium II Methylation Assay Manual Protocol’). Subsequently, the 850 K arrays were scanned using the Illumina iScan and all samples passed the MethylAid data quality assessment by using the corresponding R script (version R-3.6.2). Raw hybridization signals were processed using the GenomeStudio software (v2011.1; methylation module 1.9.0; Illumina Inc., USA) applying the default settings and internal controls for normalization. Loci with a detection p-value >0.01 in all hybridizations (863,057/865,918) and hybridizations showing a loci call rate above 98% were (18/18) used for further analysis. All hybridizations met the defined quality criteria and entered the next step in the analysis. MCCs were subjected for the DNAmAge calculation as described by Horvath [Citation3]. We extracted from the 863,057 loci passing the quality criteria those 30,084 loci necessary for the DNAmAge calculation given by Horwath in the datMiniAnnotation file. Next, we uploaded the DNA methylation values of these loci in https://dnamage.genetics.ucla.edu/home (February 2020) and used the recommended normalization option. Of note, Horvath used Infinium Methylation450 BeadChip targeting over 450,000 methylation sites per sample (450 K) for the DNAmAge calculation [Citation3]. We are aware that by using 850 K generated data an underestimation of −3.96 DNAmAge years compared to 450 K data and a correlation to the chronological age was observed [Citation4]. For the assessment of the MCC DNAmAge correlation with the chronological age, the following statistical analyses were performed: the Pearson’s correlation coefficient and the two-tailed p-value for the assessment of the scatter plot () and the paired t-test for the assessment of the boxplots () were calculated using Graphpad Prism 8.3.1.
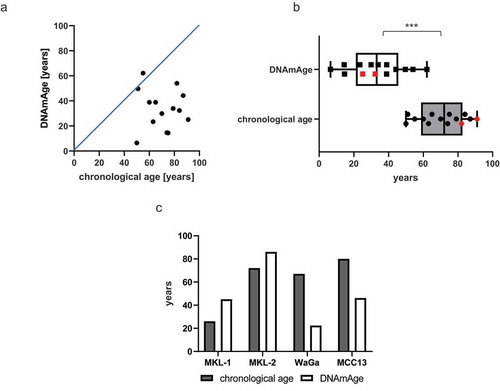
Figure 1. Epigenetic age of MCC cells is significantly younger compared to the chronological age of MCC patients
For the PluriTest approach [Citation8], the RNA of all samples was sequenced by GenomeScan BV. Briefly, the RNA quality of all samples was assessed using the Fragment Analyzer (Agilent, Santa Clara, California, USA). All samples passed the quality control. The tissue sample preparation was performed according to the protocol ‘NEBNext Ultra Directional RNA Library Prep Kit for Illumina’ (NEB, Hitchin, Great Britain). The NEBNext Ultra II Directional RNA Library Prep Kit for Illumina was used to process the cell line samples. From the total tissue RNA, rRNA was depleted using the rRNA depletion kit (NEB, Hitchin, Great Britain). The cell line mRNA was isolated from total RNA using oligo-dT magnetic beads. Afterwards from all RNAs, cDNA synthesis was performed, which was used for ligation with the sequencing adapters and PCR amplification of the resulting product. The quality and yield of all samples matched the company’s expectation. The tissue cDNAs were sequenced using the next generation sequencing (NGS) platform Illumina NextSeq 500 (read length of 1 * 75 bp gained in an average output of 1.2 giga base pairs reads per sample) according to the manufacturer’s protocols (Illumina Inc., San Diego, California, USA). The cell line cDNAs were sequenced by using the NGS platform Novaseq 6000 (read length of 2 * 150 bp gained in an average output of 17 giga base pairs reads per sample) (Illumina Inc., San Diego, California, USA). After alignment to GRCh37.87 (hg19) using the STAR aligner (v. 2.4.5b), the bam-files were subjected to the program featureCounts (v 1.6.3) to assign the reads to the necessary regions for the PluriTest approach [Citation8]. These regions were chosen to match the positions of probes on expression arrays which were defined in the original PluriTest publication [Citation8]. The raw read counts were then used to address pluripotency by adopting the R script published by Müller et al. 2011 [Citation8] (https://github.com/jhsiao999/pluritest). This script was run using the outdated R version 2.15.1 because it was not runnable with newer R versions. For the PluriTest approach, five randomly chosen publicly available RNAseq-datasets from induced pluripotent stem cells (iPSC) (GSE107654) were used as positive controls. The raw data fastq-files were downloaded and processed using the same protocol.
The mean DNAmAge for the tested 14 MCC tissues was 33.0 ± 15.0 y with a Pearson’s correlation coefficient of 0.05. Thus, the DNAmAge was significantly younger and did not match with the patients' chronological age (mean 70.0 ± 13.0 y, p < 0.0001) (, b, ). This significant lower DNAmAge was observed irrespective of the presence of MCPyV. MCPyV at least in the context of MCC is, thus, not associated with DNAmAge acceleration as it has been described for other viral infections including HPV-associated cancers [Citation5]. This negative DNAmAge is in the range of what has been reported in uterine endometroid cancer with −30 y, colorectal cancer tissue with −20 y and head and neck cancers with approximal −17 y [Citation9]. Comparing two different MCC metastases (ID L-MCC8 and L-MCC16, ) which were resected within 1 y from patient number 5 revealed no substantial difference of the DNAmAge. Of interest, we observed a 43-y DNAmAge increase between the primary tumour (ID L-MCC7) and its metastasis (ID L-MCC10) of one patient (number 4) which was both resected within 1 y (). Only one of the analysed MCCs (ID L-MCC5) and two of the analysed MCC cell lines (MKL-1 and MKL-2) revealed an accelerated DNAmAge compared to the chronological age ( and ).
Table 1. Summary of the clinicopathological data of the MCC patients and tissues including DNAmAge
The observed approximation of 50-y DNAmAge to 51-y chronological age of patient number 4 within 1 yr might be due to the increase of DNAmAge during disease progression. This has previously been reported for haematopoietic stem cells revealing a DNAmAge deceleration after transplantation and a DNAmAge acceleration compared to the original donor’s chronological age after several months [Citation10]. Interestingly, in contrast to the MCC cell lines, WaGa, as well as MCC13 the DNAmAge of the MCC cell lines, MKL-1 and MKL-2 were accelerated. Horvath described that the DNAmAge correlates with the passage number [Citation3]. Considering this and that the MCPyV-positive cell lines, MKL-1, MKL-2, as well as WaGa, were established earlier than 1987 [Citation11], 2002 [Citation12] as well as 2010 [Citation13], respectively, one might argue that the DNAmAge acceleration of the cell lines MKL-1 and MKL-2 could be explained by cell culture effects. In contrast, MCC13 was established earlier than 1995 [Citation14] and a lower DNAmAge could be calculated. Considering that MKL-1, MKL-2 and the primary MCC tumour L-MCC5 illustrated an acceleration of the DNAmAge it might be that these MCCs possibly belong to a distinct minority subgroup of MCC with accelerated DNAmAge compared to the chronological age. Further studies using larger MCC cohorts are needed to determine in as much MCC consists of different subgroups dividing in accelerated or low DNAmAge.
The highly significant low DNAmAge in combination with the typical trilinear differentiation of MCC gene expression might possibly indicate a pluripotency program to be active in these cells. Thus, we assessed the pluripotency status of the MCC cells and cell lines using RNAseq in combination with PluriTest analyses. Using this approach, the MCC tumour cells and the cell lines did consistently not map at or close to the pluripotency region (). Thus, MCC cells and cell lines are not pluripotent according to PluriTest.
Figure 2. MCCs are not pluripotent
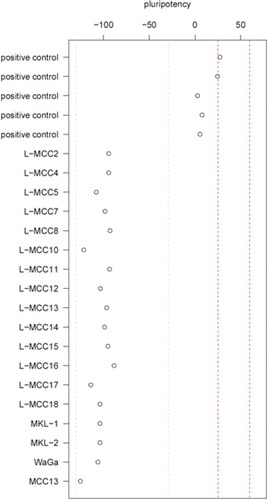
Therefore, we conclude that the majority of MCCs are characterized by epigenetic youth but lack of pluripotency. The understanding of these epigenetic findings in MCC might contribute to the identification of the yet elusive cellular origin of MCC.
List of abbreviations
| Merkel cell carcinoma | = | MCC |
| Merkel cell polyomavirus | = | MCPyV |
| DNA methylation age | = | DNAmAge |
| RNA sequencing | = | RNAseq |
| Pluripotent test | = | PluriTest |
| Induced pluripotent stem cells | = | iPSC |
Acknowledgments
This research was supported by RWTH Aachen University through Graduiertenförderung nach Richtlinien zur Förderung des wissenschaftlichen Nachwuchses (RFwN). We would like to thank Sietse Aukema, MUMC+, for the initiation of the collaboration with Prof. Reiner Siebert, Ulm University Medical Center. The authors thank Prof. Ole Ammerpohl, Ulm University Medical Center, for fruitful discussions.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Schadendorf D, Lebbé C, zur Hausen A, et al. Merkel cell carcinoma: epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer. 2017;71:53–69.
- Harms PW, Harms KL, Moore PS, et al. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat Rev Clin Oncol. 2018;15:763–776.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115–R.
- Dhingra R, Nwanaji-Enwerem JC, Samet M, et al. DNA methylation age-environmental influences, health impacts, and its role in environmental epidemiology. Curr Environ Health Rep. 2018;5:317–327.
- Lu X, Zhou Y, Meng J, et al. Epigenetic age acceleration of cervical squamous cell carcinoma converged to human papillomavirus 16/18 expression, immunoactivation, and favourable prognosis. Clin Epigenetics. 2020;12:23.
- Sauer CM, Haugg AM, Chteinberg E, et al. Reviewing the current evidence supporting early B-cells as the cellular origin of Merkel cell carcinoma. Crit Rev Oncol Hematol. 2017;116:99–105.
- Kervarrec T, Samimi M, Guyétant S, et al. Histogenesis of Merkel cell carcinoma: A comprehensive review. Front Oncol. 2019;9:451.
- Müller F-J, Schuldt BM, Williams R, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317.
- Horvath S. Erratum to: DNA methylation age of human tissues and cell types. Genome Biol. 2015;16:96.
- Stölzel F, Brosch M, Horvath S, et al. Dynamics of epigenetic age following hematopoietic stem cell transplantation. Haematologica. 2017;102:e321–e3.
- Rosen ST, Gould VE, Salwen HR, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab Invest. 1987;56:302–312.
- Van Gele M, Leonard JH, Van Roy N, et al. Combined karyotyping, CGH and M-FISH analysis allows detailed characterization of unidentified chromosomal rearrangements in Merkel cell carcinoma. Int J Cancer. 2002;101:137–145.
- Houben R, Shuda M, Weinkam R, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072.
- Leonard JH, Dash P, Holland P, et al. Characterisation of four Merkel cell carcinoma adherent cell lines. Int J Cancer. 1995;60:100–107.
