ABSTRACT
The metabolism of DNA methylation is reported to be sensitive to oxidant molecules or oxidative stress. Hypothesis: early-life oxidative stress characterized by the redox potential of glutathione influences the DNA methylation level. The in vivo study aimed at the impact of modulating redox potential of glutathione on DNA methylation. Newborn guinea pigs received different nutritive modalities for 4 days: oral nutrition, parenteral nutrition including lipid emulsion Intralipid (PN-IL) or SMOFLipid (PN-SF), protected or not from ambient light. Livers were collected for biochemical determinations. Redox potential (p < 0.001) and DNA methylation (p < 0.01) were higher in PN-infused animals and even higher in PN-SF. Their positive correlation was significant (r2 = 0.51; p < 0.001). Methylation activity was higher in PN groups (p < 0.01). Protein levels of DNA methyltransferase (DNMT)-1 were lower in PN groups (p < 0.01) while those of both DNMT3a isoforms were increased (p < 0.01) and significantly correlated with redox potential (r2 > 0.42; p < 0.001). The ratio of SAM (substrate) to SAH (inhibitor) was positively correlated with the redox potential (r2 = 0.36; p < 0.001). In conclusion, early in life, the redox potential value strongly influences the DNA methylation metabolism, resulting in an increase of DNA methylation as a function of increased oxidative stress. These results support the notion that early-life oxidative stress can reprogram the metabolism epigenetically. This study emphasizes once again the importance of improving the quality of parenteral nutrition solutions administered early in life, especially to newborn infants.
Abbreviation of Title: Parenteral nutrition and DNA methylation
Introduction
Extensive experimental and epidemiological data suggest that early postnatal nutrition plays a critical role in health status throughout life as a result of a process called ‘developmental programing.’ This concept stems from the hypothesis that environmental stresses experienced in utero or early in life can lead to the later-life development of chronic non-communicable diseases [Citation1,Citation2]. Epigenetic modulation of gene expression, particularly DNA methylation, is one of the privileged mechanisms to explain the reprogramming means of metabolisms associated with diseases. The persistence of this modulation occurring in the perinatal (pre and early postnatal) environment may evolve into chronic diseases later in life [Citation3,Citation4]
The methylation profile of DNA results from the balance between methylation by DNA methyltransferases (DNMTs) and demethylation by the Ten Eleven Translocation enzymes (TETs). In mammals, at least three isoforms of DNMTs transfer the methyl group of S-adenosylmethionine (SAM) preferentially to a cytosine of a CpG island [Citation5]. DNMT1, which has a preference towards hemimethylated DNA, is known to perpetuate the methylation pattern during cell replication [Citation5], while DNMT3a and DNMT3b are required for de novo methylation [Citation6]. In vitro and in vivo evidence has demonstrated that oxidative stress down-regulate the activity and expression of DNMT1 [Citation7–9], whereas oxidant molecules promote the expression of DNMT3a [Citation7]. On the other hand, it is suspected that the antioxidant ascorbate level may modulate the activity of TETs [Citation10,Citation11]. These enzymes responsible for the demethylation of DNA have a dioxygenase domain [Citation12]. Such a domain is dependent on the presence of ascorbate. Thus, oxidative stress could alter the level of DNA methylation, and if this stress is induced early in life, it may lead to health issues later in life. Therefore, our hypothesis was that an early-life oxidative stress characterized by the redox potential of glutathione influences the DNA methylation level. The objective of the study was to determine the impact of in vivo modulation of the redox potential of glutathione on DNA methylation. The animal model was the newborn guinea pig exposed or not to parenteral nutrition (PN). This mode of nutrition induces an oxidative stress caused by the contamination of PN with oxidant molecules such as H2O2 [Citation13], lipid peroxides [Citation13,Citation14] and aldehydes [Citation14]. PN induces an oxidative shift of the glutathione redox potential in the lungs [Citation15] and livers of animals [Citation16], and in the blood of premature infants [Citation17,Citation18]. Hydrogen peroxide is the result of the reduction of dissolved oxygen by vitamin C present in parenteral solution, i.e., a reaction by which vitamin C is oxidized into dehydroascorbate that is rapidly hydrolysed into diketogulonate [Citation19]. This reaction is halved by an adequate photoprotection limiting the catalytic action of photo-excited riboflavin [Citation20]. Photoprotection also prevents the breakdown of vitamin C in parenteral solutions [Citation19] and its decreased concentrations in the livers and lungs of animals [Citation21,Citation22]. Oxidation of the glutathione redox potential may also result from the presence of aldehydes. The level and type of aldehydes vary according to the richness of the parenteral lipid emulsion in omega-3 or omega-6 fatty acids. Omega-3 fatty acids are more prone to oxidization [Citation14,Citation23]. Therefore, the animals received regular oral nutrition or parenteral nutrition including Intralipid (low ratio of omega-3/omega-6 fatty acids) or SMOFLipid (high ratio of omega-3/omega-6 fatty acids), protected or not from ambient light. Glutathione levels, glutathione redox potential status, global DNA methylation levels, methylation activity, and protein levels of DNMT1, 3a, and 3b were determined in the liver.
Results
Glutathione levels and redox potential of glutathione
Our hypothesis relied on the fact that PN is contaminated with peroxides and aldehydes. Therefore, the non-radical oxidative stress state was assessed by the measurement of the redox potential (), which was higher (oxidized) in the PN groups compared to the ON group (p < 0.001). The redox potential was higher in animals receiving PN, which included SMOFLipid (PN-SF), than in animals infused with PN containing Intralipid (PN-IL) (p < 0.05). The impact of photoprotection was not significant.
Figure 1. Potential redox of glutathione and vitamin C concentrations depending of nutritive modalities
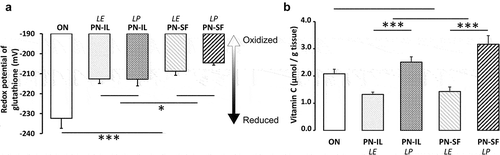
Oxidized glutathione (GSSG) levels () were increased (p < 0.001) in the PN groups compared to the ON group. There was no significant difference between PN groups. In contrast, the GSH levels () of all PN groups did not differ from the ON group. However, GSH was lower (p < 0.01) in the PN-SF groups than in the PN-IL groups. Photoprotection had no impact.
Table 1. Hepatic GSH and GSSG levels depending on nutritive modalities
Table 2. Characteristics of lipid emulsions
Vitamin C
Vitamin C is the main source of peroxides in PN [Citation24] by giving its electrons to dissolved oxygen [Citation25,Citation26]. During the process, ascorbate is oxidized into dehydroascorbate, followed by rapid hydrolysis of its lactam ring, forming the diketogulonic acid. This is the first step in the breakdown of vitamin C. This generation of peroxides is catalysed by photo-excited riboflavin [Citation25,Citation27]. Therefore, the lack of adequate photoprotection of PN induces a loss of vitamin C (ascorbate + dehydroascorbate). shows that hepatic vitamin C levels were 50% lower in animals infused with PN without photoprotection (p < 0.001). There was no significant difference between PN-IL and PN-SF groups. Because the vitamin C levels measured in the ON group were similar to the mean levels between the PN groups exposed (LE) or not (LP) to ambient light, the difference between the ON group and the PN groups did not reach the statistical difference.
Methylation of DNA
DNA methylation profile
Compared to the ON group, the levels of methylated cytosine (overall methylation of DNA, ) were higher (p < 0.01) in the PN groups. The difference between PN-IL and PN-SF groups did not reach statistical significance (p = 0.06). There was no impact of light protection. The overall DNA methylation levels were positively correlated to the redox potentials of glutathione (y = 0.16 pmol •μg DNA−Citation1•mV−Citation1 •x + 41 pmol •μg DNA−Citation1; r2 = 0.51, p < 0.0001, ) regardless of the type of nutrition.
Figure 2. DNA methylation depending of nutritive modalities
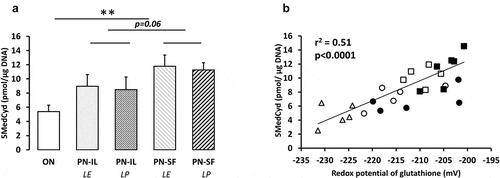
DNA methylation activity
DNA methylation activity is under the control of DNMTs for methylation and TETs for demethylation. DNA methylation activities () were higher (p < 0.01) in the PN groups compared to the ON group, without a significant difference between PN groups. The correlation of DNA methylation activities with the redox potential was not statistically significant (rCitation2 = 0.10). TET activities (1.6 ± 0.3 pmol/min/mg protein) did not differ between all groups, including the ON group (not shown).
Figure 3. DNA methylation activity, levels of SAM and SAH depending of nutritive modalities
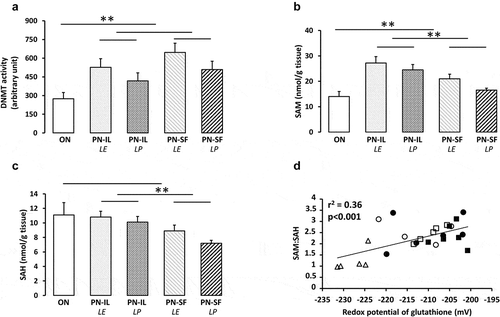
SAM (substrate) and SAH (inhibitor)
Levels of SAM, the DNMT substrate (), were higher (p < 0.01) in the PN groups compared to the ON group, and higher in the PN-IL groups (p < 0.01) relative to the PN-SF groups. The influence of light exposure or protection was not statistically significant. There was no relationship between SAM and the redox potential (rCitation2 = 0.07). The enzymatic pathways transferring the methyl moiety of SAM generate the S-adenosyl homocysteine (SAH). SAH is a competitive inhibitor of DNMTs [Citation28]. The SAH level () was lower (p < 0.05) in the PN-SF groups compared to the PN-IL groups. Comparison between PN groups and ON group did not reach statistical significance. The relationship between SAH levels and redox potential did not reach significance (y = −0.11 nmol g tissue−Citation1 mV−Citation1 x − 14.1 nmol g tissueCitation1; r2 = 0.14, p < 0.06). The impact of the SAM and SAH interaction can be illustrated by their ratio (SAM/SAH), for which the correlation with redox potential () is strong (y = 0.045 mV−Citation1 • x – 11.8; r2 = 0.36, p < 0.001).
Protein levels of DNMT1, 3a1, 3a2 and 3b
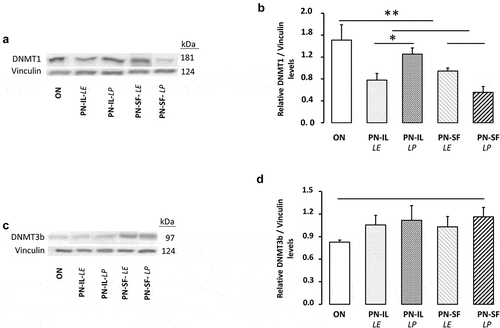
shows the protein determinations of DNMT1 and 3b. The western blot revealed a molecular weight of 181 kDa for DNMT1 (,b) and 97 kDa for DNMT3b (,d), in concordance with predicted weights [Citation29]. DNMT1 level was lower in the PN groups (p < 0.01) compared to the ON group. Exposure to light was significant (p < 0.05) only in the PN-IL groups. Differences between groups for DNMT3b did not reach the statistical significance.
Figure 4. DNMT1 and DNMT3b protein levels depending of nutritive modalities
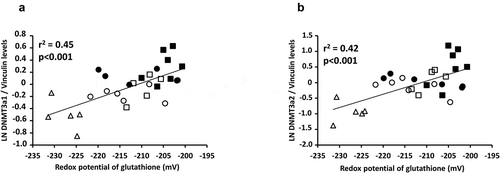
The method used made possible to separate the two isoforms of DNMT3a () with a molecular weight of 117 and 135 kDa, as expected [Citation30]. The levels of the heaviest (135 kDa) isoform () were higher in the PN groups (p < 0.01) compared to the ON group, higher in the PN-SF groups (p < 0.05) compared to the PN-IL groups, and higher in the photoprotected PN groups (p < 0.05). The relationship between the natural logarithm of the protein expression of the heavier isoform of DNMT3a and redox potential was significant (: y = 0.025 pixel mV−Citation1 x + 5.19 pixel; r2 = 0.45, p < 0.001). For the lightest isoform (117 kDa), protein levels () were higher in the PN groups (p < 0.001) compared to the ON group, higher in the PN-SF groups (p < 0.05) compared to the PN-IL groups, and higher in the photoprotected PN-SF group (p < 0.01). The relationship between the natural logarithm of the protein expression of the light isoform of DNMT3a and the redox potential was significant (: y = 0.043 pixel mV−Citation1 + 8.98 pixel; r2 = 0.42, p < 0.001).
Figure 5. DNMT3a protein levels depending of nutritive modalities
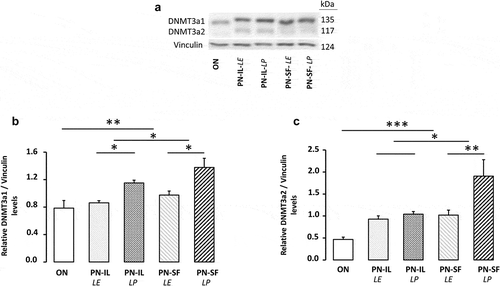
Figure 6. Relationships between DNMT3a and redox potential of glutathione
Discussion
The main contribution of the study is its demonstration of a strong in vivo relationship between the redox potential of glutathione and the DNA methylation level, strengthening the hypothesis that early-life oxidative stress influences the DNA methylation level. The main metabolic effects of the glutathione redox potential were the protein expression of both isoforms of DNMT3a and the equilibrium between the availability of both substrate (SAM) and competitive inhibitor (SAH) of DNMTs.
The methylation level of DNA results from the balance between methylation by DNMTs and demethylation by TETs. The lack of difference between groups for TET activity, even with a significant difference in ascorbate levels between groups, excludes the impact of demethylation by these enzymes. The mode of nutrition used affected the DNA methylation activity, but the method used did not allow the impact of each methyltransferase (DNMT1, 3a, and 3b) to be identified separately. The in vitro enzymatic activity is determined at its Vmax, which is obtained in saturation condition by its substrate. Under this condition, the activity depends on the enzyme level. The mode of nutrition used affected the protein expression of these enzymes differently. We found a decrease of DNMT1 protein in the PN groups compared to the ON group. DNMT3b protein levels were not different between groups. By contrast, we showed that the two isoforms of DNMT3a were higher in the PN groups and PN compounded with SF than in IL, and even higher in animals receiving the photoprotected PN. The fact that the relationship with the redox potential was significant only with the two isoforms of DNMT3a suggests that they played a predominant role in the methylation of DNA. The opposite effect of PN on DNMT1 and DNMT3a, without change for DNMT3b, has also been reported by Armstrong et al. [Citation31] in the liver of growth hormone-deficient mice; this observation was reversed by the administration of growth hormone.
Under in vivo conditions, the substrate concentration of DNMTs catalysed reactions is generally close to Km [Citation32]. Thus, a small change in its concentration can have a significant impact on the enzymatic activity. Here, among the in vivo factors affecting DNA methylation activity are the availability of the substrate SAM as well as of the competitive inhibitor SAH [Citation28]. A kinetic characteristic of a competitive inhibition is a higher Km value. Thus, in an environment where the concentration of SAH is lower, as here in animals receiving the PN compounded with SF, the DNMT affinity for SAM is increased (lower Km), resulting in a greater impact of increasing levels of SAM, as observed here, on DNA methylation activity. The enzymatic kinetics are described by the following equation: vi = Vmax • [S]/([S] + Km • {1+ [I]/ki}); where vi is the initial rate, Vmax is the maximum rate at substrate saturation, [S] is the concentration of substrate (SAM), [I] is the concentration of the inhibitor (SAH), Km is the Michaelis-Menten constant between SAM and DNMT, ki is the inhibition constant. Thus, the SAM ratio on SAH can illustrate the complex interaction between the substrate and the inhibitor on methylation activity. The strong and positive influence of the redox potential on the SAM ratio on SAH (r2 = 0.36) suggests that the DNA methylation activity was also affected in our model by the availability of SAM and SAH. This relationship could be explained, at least in part, by the oxidative environment that promotes GSH synthesis [Citation33]. Indeed, SAH is transformed into cysteine whose availability is the main limiting factor of GSH synthesis [Citation34].
The approach used to obtain a gradation of the redox potential in vivo was to feed the animal with a different mode of nutrition, oral (ON) versus parenteral (PN), to vary the quality of lipid emulsion of these PNs (higher level of omega-6 fatty acids in Intralipid than in SMOFLipid), and protect or not these PNs from ambient light (which promotes the generation of peroxides). The oxidative nature of the PN [Citation19], particularly if the PN contained SMOFLipid instead of Intralipid has already been reported in the lungs of guinea pigs [Citation15]. Although exposure to light affected hepatic vitamin C levels, it had no influence on the redox potential. PN exposure to light is known to affect the redox potential of glutathione in the lungs but not in the liver [Citation22]. However, both the redox potential and the exposure to light influenced the level of DNMT3a. This dichotomy of the effect of light suggests that in addition to the change in the redox potential of glutathione other components of PN, nutrients or by-products generated during exposure to light, could also influence methylation of DNA. In our model, parenteral nutrition induces a great oxidative stress. This stress has proven to be powerful enough to induce pulmonary remodelling [Citation10]. It is this magnitude of oxidative stress in vivo that has been used to establish the correlation between DNA methylation levels and the severity of oxidative stress (glutathione redox potential) and that explains the extent of methylation levels obtained between the groups. However, the individual variation within each group is similar to what has been reported by other teams using different modes of analysis and different animal models [Citation35-41].
In summary, the oxidized state of the redox potential of glutathione helped to reduce the hepatic concentration of SAH, an inhibitor of DNMTs, and to increase the protein expression of DNMT3a, two factors that explain the close relationship between the redox potential and DNA methylation level. The influence of oxidative stress, or oxidant molecules, on the increase in the protein content of DNMT3a, has already been reported [Citation8,Citation42]. But, to our knowledge, this is the first time that a linear relationship between the redox potential of glutathione and the DNA methylation is established in vivo, in addition to the association of the redox potential with the SAM:SAH ratio and the protein expression of the two isoforms of DNMT3a. However, these associations with the redox potential do not exclude a possible impact of PN or specific parenteral nutriments on DNA methylation. The study focused on guinea pig liver during its first week of life. Further studies must be undertaken to investigate the existence of such a relationship in other tissues and other ages, as well as the stability of DNA methylation over time.
The increased levels of DNA methylation and redox potential (oxidized) measured in PN groups confirm that this mode of nutrition is likely to induce epigenetic modifications, at least at DNA methylation level. This corroborates the concern over the long-term impact of PN-induced oxidative stress received early in life [Citation43]. The results highlight the importance of protecting the newborn or young child from an oxidative environment, specifically by improving the quality of PN solutions or/and improving the antioxidant defence of the newborn.
Materials and methods
Experimental design
Thirty-six male Hartley guinea pigs were purchased from Charles River Laboratories (St-Constant, Québec, Canada) on their third day of life and housed in the institutional animal facility at constant temperature and humidity as well as 12 h light–12 h dark cycle. The study was approved by the Institutional Committee for Good Practice with Animals in Research of the CHU Sainte-Justine in accordance with the Canadian Council on Animal Care guidelines. Upon arrival, the animals were separated into five groups, an oral nutrition group (ON) and four parenteral nutrition groups (PN).
Animals on PN were nourished through a catheter inserted in the jugular vein under anaesthesia with a ketamine/xylazine cocktail on the third day of life, as previously described [Citation9,Citation15,Citation16]. PN solutions, freshly prepared daily, consisted of two separate bags. Bag 1 contained dextrose, amino acids, and multivitamins and bag 2, dextrose, amino acids, and lipid emulsion. Both solutions met at the last Y near the infusion site. The final solutions contained 8.2% (w/v) dextrose, 2.0% (w/v) amino acids (Primene, Baxter, Mississauga, ON, Canada), 1% (v/v) multivitamin preparation (Multi-12 paediatrics, Sandoz, Boucherville, QC, Canada), electrolytes, 1 U/mL heparin, and 1.6% (w/v) lipid emulsion. The continuous infusion rate was 200 mL/kg/day.
The studied groups were:
ON: animals fed orally ad libitum with regular chow for guinea pigs.
PN-IL-LE: Animals fed exclusively with PN compounded with 1.6% (w/v) IntraLipid-20%® (Pharmacia Upjohn, Baie D’Urfé, QC, Canada). The solution was without light protection.
PN-IL-LP: Animals received fully photoprotected PN.
PN-SF-LE: Animals fed exclusively with PN compounded with 1.6% (w/v) SMOFLipid-20%® (Fresenius Kabi Canada Ltd., Richmond Hill, ON, Canada). The solution was without light protection.
PN-SF-LP: Animals received fully photoprotected PN compounded with SmofLipid.
The composition of the lipid emulsions is presented in .
There was no difference in body weight (103 ± 2 g) between groups. After four days, animals were anaesthetized and sacrificed for collection, preparation, and preservation (−80°C) of the liver until biochemical determinations. There was no difference in liver weight (3.9 ± 0.1 g) between groups.
Redox potential of glutathione
At sacrifice, 500 mg of liver was homogenized ex temporani in 5 volumes of metaphosphoric acid (MPA) 5% (w/v) with Polytron tissue homogenizer (BioSpec Products Inc, Bartlesville, OK, USA) and centrifuged at 7 200 g for 4 min. Both supernatants and pellets were stored at −80°C until analysis. Reduced (GSH) and oxidized (GSSG) glutathione contained in supernatants was separated by capillary electrophoresis, into a fused-silica capillary (75 μm internal diameter × 50 cm of effective length) in a P/ACE MDQ system (Beckman Coulter, Mississauga, ON, Canada) [Citation16,Citation44]. Before each run, the capillary was conditioned by sequentially washing with methanol (10 min, 20 psi), 0.1 N HCl (10 min, 20 psi), HPLC water (2 min, 20 psi), 1 N NaOH (20 min, 20 psi), HPLC water (4 min, 20 psi), and finally with running buffer (10 min, 20 psi). We achieved the separation of both glutathione forms using 75 mM boric acid + 25 mM Bis–Tris, pH 8.4 as running buffer. Samples were injected under 0.5 psi pressure for 10 sec, followed by a water plug at 0.2 psi for 10 seconds. The migration time was 10 minutes at 28°C and 18 kV. Absorbance was monitored at 200 nm. The concentrations of GSH and GSSG are extrapolated from standard curves of GSH (0 to 100 μM) and GSSG (0 to 10 μM) and were used to calculate redox potential by using the Nernst equation (25°C, pH 7.0) [Citation45].
Global DNA methylation levels
The global DNA methylation levels were determined with a competitive enzyme-linked immunosorbent assay kit by measuring 5ʹ-methyl-2ʹ-deoxycytidine (5-MedCyd). First, genomic DNA was isolated from 200 mg liver using the E.Z.N.A.®HP Tissue DNA Kit (OMEGA Bio-tek, Norcross, GA, USA), following the manufacturer’s instructions. Then, DNA samples (0.4 μg/μL) were denatured into single-strand DNA by heating samples at 95°C for 5 min, followed by a quick cooling on ice. To digest DNA into nucleosides, 5 μL of 200 mM sodium acetate, pH 5.2, and 5 units (5 μL) of nuclease P1 (N8630, Sigma–Aldrich, St. Louis, Missouri, USA) were added and samples were incubated 2 h at 37° C. Thereafter, 5 μL of 1 M Tris, pH 7.5, and 5 units (5 μL) of alkaline phosphatase (P5931, Sigma–Aldrich, St. Louis, Missouri, USA) were added and the mixture incubated for an additional 1 h at 37° C. The level of 5-MedCyd was quantified using the Global DNA methylation enzyme-linked immunosorbent assay kit (Cell Biolabs, San Diego, CA, USA) following the protocol assay of the manufacturer. Briefly, standards and samples were added in duplicate to a plate coated with 5-MedCyd DNA conjugate. After 10 min incubation on orbital shaker, an anti-5-MedCyd monoclonal antibody was added, followed by a horseradish peroxidase-conjugated secondary antibody. The concentration of 5-MedCyd in samples was determined by comparing their absorbance at 450 nm to the absorbance of a predetermined 5-MedCyd standard curve ranging from 0 to 80 μM.
S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) levels
Two hundred milligram of liver was homogenized in four volumes perchloric acid by using Polytron tissue homogenizer (BioSpec Products Inc, Bartlesville, OK, USA). Samples were centrifuged at 10 000 g for 10 min and the supernatant was collected after filtration through 0.45 μm syringe filters and stored at −80°C until use. SAM and SAH separations were carried out by capillary electrophoresis with P/ACE MDQ system (Beckman Coulter, Mississauga, ON, Canada), into a fused-silica capillary (75 μm internal diameter × 50 cm of effective length) [Citation46]. The capillary was equilibrated before each run by washing sequentially with methanol (10 min, 20 psi), 0.1 N HCl (10 min, 20 psi), HPLC water (2 min, 20 psi), 1 N NaOH (20 min, 20 psi), HPLC water (4 min, 20 psi) and finally with 200 mM glycine, pH 1.8, as running buffer (10 min, 20 psi). Samples were diluted 1:1 (v/v) with HPLC water before the analysis. SAM and SAH standards were prepared freshly in 0.167 N HClO4 to equal the perchloric acid concentration in the 1:1 dilution of samples. Samples and standards were injected at 0.5 psi for 30 sec at 25° C and migrated for 15 min at 22.5 kV. SAM and SAH peaks were identified at 254 nm and peak area were used to measure their concentrations in samples by comparing to standards.
Vitamin C determination
Animals from ON group received approximately 10 mg vitamin C per day while those from PN groups received 3.2 mg vitamin C per day.
Liver samples were taken out from −80° C and 100 mg were homogenized in 5 volumes of metaphosphoric acid 5% (w/v) with Polytron tissue homogenizer (BioSpec Products Inc, Bartlesville, OK, USA). Previous studies have found metaphosphoric acid adequate to prevent the conversion of ascorbate to L-dehydroascorbic acid [Citation47,Citation48]. All steps were carried out at 4°C. The homogenate was immediately centrifuged at 7 200 g for 4 min and both supernatants and pellets were stored at −80°C until analysis. Total vitamin C (ascorbate + dehydroascorbate) was obtained following reduction with 50 mM dithiothreitol for 40 min in the dark. Vitamin C was isolated using an Agilent 7100 capillary electrophoresis system (Santa Clara, CA, USA) into a bare fused-silica capillary (50 μm internal diameter x 40 cm of effective length). Before sample injection, the system was conditioned as follows: methanol (Flush, 600 sec), 0.1 N HCl (Flush, 600 sec), HPLC water (Flush, 120 sec), 1 N NaOH (Flush, 1200 sec), HPLC water (Flush, 240 sec), running buffer (200 mM borate, 20% acetonitrile, pH 9.6; Flush, 600 sec). Samples and standards were appropriately diluted, injected at 100 mBar for 8 sec, and migrated for 15 min at 30 kV, 28° C. Ascorbate was detected at 268 nm and its concentration was calculated using an ascorbate standard curve (5–130 μM).
DNA methyltransferases (DNMTs) activity
The analysis of DNMTs activity was performed in the nuclear extract with a modified method adapted from previous studies [Citation9,Citation49,Citation50]. Briefly, approximately 200 mg of liver was homogenized in a high concentration sucrose (1.8 M) – glycerol (9%) buffer, pH 7.6 (containing 10 mM Hepes, 25 mM KCl, 0.15 mM spermine and 0.5 mM spermidine, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol) by using a dounce homogenizer, then centrifuged at 79 000 g for 30 min. The nuclei (pellet) were washed in a 25% glycerol-based buffer pH 7.9 (25 mM Na-Hepes, 100 mM KCl, 1.5 mM MgCl2 and 1 mM dithiothreitol) and recovered after centrifugation at 74 700 g for 45 min. (NH4)2SO4 (0.3 g/ml) was added to the supernatant, mixed for 30–45 min and centrifuged at 111 000 g for 45 min. The pellet was resuspended in 250 μL of dialysis buffer consisting of 25 mM Na-Hepes, 40 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol then filtered through a 100 K Amicon Ultra filter. All steps were carried out at 4° C. The activity of DNA methyltransferases was measured in nuclear extract with the EpiQuik DNMT activity assay kit (Epigentek®, Farmingdale, NY, USA) following the manufacture’s instructions. The nuclear extracts (1.5 μg) were added into microplate wells coated with a cytosine-rich DNA subtract. Samples were added in duplicate with 0.16 mM of S-adenosinemethionine (SAM). After 60 min of incubation, samples were mixed and incubated with first, a capture antibody, followed by a detection antibody, then an enhancer solution and finally a developer solution. The reaction was stopped with the provided stop solution. The methylated DNA level that is proportional to the DNA methyltransferase (DNMT) activity was measured at 450 nm and the results of samples were compared to a standard curve and expressed in arbitrary unit.
Ten eleven translocation protein (TET) activity
The nuclear extraction of TET enzymes was executed with an adapted method used for DNMTs nuclear extraction as described in the above section ‘DNA methyltransferases activity.’ The modified medium was EDTA, phenylmethylsulfonyl fluoride, spermine, and spermidine free, but contained a cocktail of protease inhibitors (cOmplete mini EDTA free® Cat # 11836170001, Roche Diagnostics Deutschland, Mannheim, Germany). TETs activity was measured with the Epigenase™ 5mC Hydroxylase TET Activity/Inhibition Colorimetric Assay Kit (Epigentek®, Farmingdale, NY, USA). Protocol from the manufacturer was adapted to our samples. Briefly, 6 μl of the sample (containing 6.5 μg of 100KDa-filtered nuclear extract) was added in a microplate coated with methylated cytosine which served as subtract and incubated 90 min with TET assay buffer mixed with cofactors: ascorbate (1.7 mM), 2-ketoglutarate (0.85 mM) and Fe++ (85 μM). A capture antibody was added, followed by a detection antibody, an enhancer solution, and finally a developer solution. The reaction was stopped with the provided stop solution. The amount of hydroxymethylated cytosine, which is proportional to TET enzyme activity was measured by reading the absorbance at 450 nm. The activity was quantified using a standard hydroxymethylcytosine curve.
Protein levels of DNMT1, 3a and 3b
Briefly, 50 mg of liver was homogenized in 250 μL of RIPA buffer, incubated at 4°C and centrifuged at 5,000 rpm/30 min/4°C. The supernatants were collected then centrifuged at 60,000 rpm/45 min/4°C. Fifty micrograms of protein from the supernatant were boiled in Laemmli buffer 2X for 5 min then loaded onto a 5% and 6% SDS polyacrylamide gel, run for 1 h/150 V and transferred onto a polyvinylidene difluoride membrane (PDVF, Bio-Rad Laboratories, CA, USA) in Tris-glycine buffer for 80 min/100 V. Membranes were blocked for 1 h at room temperature (R.T.) with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) then incubated with primary antibodies for DNMT1 (1:1000, ab188453, Abcam, Cambridge, MA, USA), DNMT3a (1:1500, ab2850, Abcam), DNMT3b (1:1000, ab227942, Abcam). Antibodies were diluted in 5% milk-TBST (DNMT1 and DNMT3a) or 2.5% milk-TBST (DNMT3b) at 4°C overnight. Membranes were washed and incubated with anti-rabbit IgG-HRP (1:2000, W401B, Promega, Madison, WI, USA) for 1 h at R.T. Proteins were detected using SuperSignalTM West Pico Plus (Thermo Scientific, Rockford, IL, USA) with an imaging system (G: BOX, Synoptics, Cambridge, UK). Densitometry was analysed using the ImageJ program. To detect vinculin (loading control), membranes were stripped with the RestoreTM Western Blot Stripping Buffer (Thermo Scientific, Rockford, IL, USA) and incubated with primary antibody for vinculin (1:1000, ab188453, Abcam), diluted in 5% milk-TBST. The membranes were washed then incubated with anti-mouse IgG-HRP secondary antibody (1:2000, HAF007, R&D Research, Minneapolis, MN, USA) for 1 h at R.T.
Statistical analysis
The significant effect of PN, lipid emulsion, and light exposure was searched by running the following orthogonal comparisons by ANOVA after verification of homoscedasticity by Bartlett’s χ2. 1) PN effect: ON group vs. all PN groups; 2) lipid emulsion: all PN-IL vs. all PN-SF; 3) Light exposure: PN-IL-LE vs. PN-IL-LP and PN-SF-LE vs. PN-SF-LP. Pearson linear regression (r2) was used for correlation analysis. Results are expressed as means ± s.e.m. The threshold of significance was set at p < 0.05.
List of abbreviations
| 5-mC: 5-methylcytosine | = | |
| 5-MedCyd: 5’-methyl-2’ -deoxycytidine | = | |
| CpG dinucleotide: Cytosine-phosphate-Guanine dinucleotide | = | |
| DNA: Deoxyribonucleic acid | = | |
| DNMT: DNA methyltransferase | = | |
| GSH: Reduced glutathione | = | |
| GSSG: Oxidized glutathione | = | |
| IL: Intralipid emulsion | = | |
| LE: Light-exposed parenteral nutrition | = | |
| LP: Light-protected parenteral nutrition | = | |
| MAT: Methionine adenosyltransferase | = | |
| SAH: S-adenosyl homocysteine | = | |
| SAM: S-adenosyl methionine | = | |
| SF: SMOFlipid emulsion | = | |
| PN: Parenteral nutrition | = | |
| TET: Ten-eleven translocation protein | = |
Acknowledgments
We recognize the important participation of Mrs. Thérèse Rouleau who realized, among others, the catheterization of animals.
Disclosure statement
The authors declare no conflict of interest.
This work was supported by a grant from the Canadian Institutes of Health Research (PJT-148522).
Additional information
Funding
- Barker DJ. The fetal and infant origins of adult disease. BMJ. 1990;301:1111.
- Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311:171-174.
- Godfrey KM, Sheppard A, Gluckman PD, et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes. 2011;60:1528–1534.
- Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049.
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222.
- Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257.
- Miozzo F, Arnould H, de Thonel A, et al. Alcohol exposure promotes DNA methyltransferase DNMT3A upregulation through reactive oxygen species-dependent mechanisms. Cell Stress Chaperones. 2018;23:115–126.
- Remely M, Ferk F, Sterneder S, et al. Vitamin E Modifies High-Fat Diet-Induced Increase of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Nutrients. 2017;9:607.
- Yara S, Levy E, Elremaly W, et al. Total parenteral nutrition induces sustained hypomethylation of DNA in newborn guinea pigs. Pediatr Res. 2013;73:592–595.
- Elremaly W, Mohamed I, Rouleau T, et al. Adding glutathione to parenteral nutrition prevents alveolar loss in newborn Guinea pig. Free Radic Biol Med. 2015;87:274–281.
- Yin R, Mao SQ, Zhao B, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–10403.
- Koivunen P, Laukka T. The TET enzymes. Cell Mol Life Sci. 2018;75:1339–1348.
- Lavoie JC, Belanger S, Spalinger M, et al. Admixture of a multivitamin preparation to parenteral nutrition: the major contributor to in vitro generation of peroxides. Pediatrics. 1997;99:E6.
- Miloudi K, Comte B, Rouleau T, et al. The mode of administration of total parenteral nutrition and nature of lipid content influence the generation of peroxides and aldehydes. Clin Nutr. 2012;31:526–534.
- Lavoie JC, Mohamed I, Nuyt AM, et al. Impact of SMOFLipid on pulmonary alveolar development in newborn guinea pigs. JPEN J Parenter Enteral Nutr. 2018;42:1314–1321.
- Elremaly W, Rouleau T, Lavoie JC. Inhibition of hepatic methionine adenosyltransferase by peroxides contaminating parenteral nutrition leads to a lower level of glutathione in newborn Guinea pigs. Free Radic Biol Med. 2012;53:2250–2255.
- Mohamed I, Elremaly W, Rouleau T, et al. Oxygen and parenteral nutrition two main oxidants for extremely preterm infants: ‘It all adds up’. J Neonatal Perinatal Med. 2015;8:189–197.
- Mohamed I, Elremaly W, Rouleau T, et al. Ascorbylperoxide contaminating parenteral nutrition is associated with bronchopulmonary dysplasia or death in extremely preterm infants. JPEN J Parenter Enteral Nutr. 2017;41:1023–1029.
- Knafo L, Chessex P, Rouleau T, et al. Association between hydrogen peroxide-dependent byproducts of ascorbic acid and increased hepatic acetyl-CoA carboxylase activity. Clin Chem. 2005;51:1462–1471.
- Khashu M, Harrison A, Lalari V, et al. Impact of shielding parenteral nutrition from light on routine monitoring of blood glucose and triglyceride levels in preterm neonates. Arch Dis Child Fetal Neonatal Ed. 2009;94:F111–5.
- Lavoie JC, Chessex P, Rouleau T, et al. Shielding parenteral multivitamins from light increases vitamin A and E concentration in lung of newborn guinea pigs. Clin Nutr. 2007;26:341–347.
- Lavoie JC, Rouleau T, Tsopmo A, et al. Influence of lung oxidant and antioxidant status on alveolarization: role of light-exposed total parenteral nutrition. Free Radic Biol Med. 2008;45:572–577.
- Michalski MC, Calzada C, Makino A, et al. Oxidation products of polyunsaturated fatty acids in infant formulas compared to human milk–a preliminary study. Mol Nutr Food Res. 2008;52:1478–1485.
- Lavoie JC, Chessex P, Rouleau T, et al. Light-induced byproducts of vitamin C in multivitamin solutions. Clin Chem. 2004;50:135–140.
- Laborie S, Lavoie JC, Chessex P. Paradoxical role of ascorbic acid and riboflavin in solutions of total parenteral nutrition: implication in photoinduced peroxide generation. Pediatr Res. 1998;43:601–606.
- Laborie S, Lavoie JC, Pineault M, et al. Contribution of multivitamins, air, and light in the generation of peroxides in adult and neonatal parenteral nutrition solutions. Ann Pharmacother. 2000;34:440–445.
- Lavoie JC, Rouleau T, Chessex P. Interaction between ascorbate and light-exposed riboflavin induces lung remodeling. J Pharmacol Exp Ther. 2004;311:634–639.
- Hoffman DR, Marion DW, Cornatzer WE, et al. S-Adenosylmethionine and S-adenosylhomocystein metabolism in isolated rat liver. Effects of L-methionine, L-homocysteine, and adenosine. J Biol Chem. 1980;255:10822–10827.
- Zhou J, Yi Z, Fu Q. Dynamic decreased expression and hypermethylation of secreted frizzled-related protein 1 and 4 over the course of pulmonary fibrosis in mice. Life Sci. 2019;218:241–252.
- Chen T, Ueda Y, Xie S, et al. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754.
- Armstrong VL, Rakoczy S, Rojanathammanee L, et al. Expression of DNA methyltransferases is influenced by growth hormone in the long-living Ames dwarf mouse in vivo and in vitro. J Gerontol A Biol Sci Med Sci. 2014;69:923–933.
- Svedruzic ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44:9472–9485.
- Higgins LG, Kelleher MO, Eggleston IM, et al. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol Appl Pharmacol. 2009;237:267–280.
- Turcot V, Rouleau T, Tsopmo A, et al. Long-term impact of an antioxidant-deficient neonatal diet on lipid and glucose metabolism. Free Radic Biol Med. 2009;47:275–282.
- Awazu M, Hida M, Bader M. Folic acid supplementation alleviates reduced ureteric branching, nephrogenesis, and global DNA methylation induced by maternal nutrient restriction in rat embryonic kidney. PLoS One. 2020;15:e0230289.
- Jarmasz JS, Stirton H, Davie JR, et al. DNA methylation and histone post-translational modification stability in post-mortem brain tissue. Clin Epigenetics. 2019;11(5). DOI:10.1186/s13148-018-0596-7
- Kronfol MM, Jahr FM, Dozmorov MG, et al. DNA methylation and histone acetylation changes to cytochrome P450 2E1 regulation in normal aging and impact on rates of drug metabolism in the liver. Geroscience. 2020;3:819–832. DOI:10.1007/s11357-020-00181-5
- Lim U, Flood A, Choi SW, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55.
- Stach D, Schmitz OJ, Stilgenbauer S, et al. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res. 2003;31:E2.
- Stach D, Schmitz OJ, Stilgenbauer S, Benner A, Dohner H, Wiessler M, Lyko F. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Research. 2003;31:E2. 2 doi:10.1093/nar/gng002
- Teng M, Chen X, Wang C, et al. Life cycle exposure to propiconazole reduces fecundity by disrupting the steroidogenic pathway and altering DNA methylation in zebrafish (Danio rerio). Environ Int. 2020;135:105384.
- Parsanathan R, Jain SK. Glutathione deficiency induces epigenetic alterations of vitamin D metabolism genes in the livers of high-fat diet-fed obese mice. Sci Rep. 2019;9:14784.
- Kleiber N, Chessex P, Rouleau T, et al. Neonatal exposure to oxidants induces later in life a metabolic response associated to a phenotype of energy deficiency in an animal model of total parenteral nutrition. Pediatr Res. 2010;68:188–192.
- Maghdessian R, Cote F, Rouleau T. Ben Djoudi Ouadda A, Levy E, Lavoie JC. Ascorbylperoxide contaminating parenteral nutrition perturbs the lipid metabolism in newborn guinea pig. J Pharmacol Exp Ther. 2010;334:278–284.
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212.
- Uthus EO. Simultaneous detection of S-adenosylmethionine and S-adenosylhomocysteine in mouse and rat tissues by capillary electrophoresis. Electrophoresis. 2003;24(78):1221–1226.
- Lykkesfeldt J. Ascorbate and dehydroascorbic acid as reliable biomarkers of oxidative stress: analytical reproducibility and long-term stability of plasma samples subjected to acidic deproteinization. Cancer Epidemiol Biomarkers Prev. 2007;16:2513–2516.
- Sławomir Dresler WM. Capillary zone electrophoresis for determination of reduced and oxidised ascorbate and glutathione in roots and leaf segments of Zea mays plants exposed to Cd and Cu. Acta Scientiarum Polonorum - Hortorum Cultus. 2013;12:143–145.
- Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47(5):767–776.
- Rose KM, Stetler DA, Jacob ST. Phosphorylation of RNA polymerases: specific association of protein kinase NII with RNA polymerase I. Philos Trans R Soc Lond B Biol Sci. 1983;302:135–142.
