ABSTRACT
While previous studies have demonstrated that prenatal exposure to environmental stressors is associated with mitochondrial DNA (mtDNA) methylation, more recent investigations are questioning the accuracy of the methylation assessment and its biological relevance. In this study, we investigated placental mtDNA methylation while accounting for methodological issues such as nuclear contamination, bisulphite conversion, and PCR bias. From the ENVIRONAGE birth cohort, we selected three groups of participants (n = 20/group). One group with mothers who smoked during pregnancy (average 13.2 cig/day), one group with high air pollutant exposure (PM2.5: 16.0 ± 1.4 µg/m3, black carbon: 1.8 ± 0.3 µg/m3) and one control group (non-smokers, PM2.5: 10.6 ± 1.7 µg/m3, black carbon: 0.9 ± 0.1 µg/m3) with low air pollutant exposure. DNA methylation levels were quantified in two regions of the displacement loop control region (D-loop and LDLR2) by bisulphite pyrosequencing. Additionally, we measured DNA methylation on nuclear genes involved in mitochondrial maintenance (PINK1, DNA2, and POLG1) and assessed mtDNA content using qPCR. Absolute D-loop methylation levels were higher for mothers that smoked extensively (+0.36%, 95% CI: 0.06% to 0.66%), and for mothers that were highly exposed to air pollutants (+0.47%, 95% CI: 0.20% to 0.73%). The relevance of our findings is further supported, as D-loop methylation levels were correlated with placental mtDNA content (r = −0.40, p = 0.002) and associated with birth weight (−106.98 g, 95% CI: −209.60 g to −4.36 g for an IQR increase in D-loop methylation). Most notably, our data demonstrates relevant levels of mtDNA methylation in placenta tissue, with significant associations between prenatal exposure to environmental stressors and D-loop methylation.
Background
Mitochondria perform multiple functions in the maintenance of cellular energy homeostatis and have crucial roles in intracellular signalling pathways. Therefore, malfunction of these organelles can be harmful to proper physiological function at multiple biological levels. To characterize mitochondrial function in observational research, many studies have focused on measuring mitochondrial DNA (mtDNA) content [Citation1], mitochondrial reactive oxygen species [Citation2], ATP generation [Citation3,Citation4], and epigenetic marks such as mtDNA methylation [Citation5]. Changes in mtDNA methylation have been associated with pathologies such as cancer [Citation6,Citation7], brain ageing [Citation8] and non-alcoholic fatty liver disease [Citation9]. Yet, despite the existence of a mitochondrial isoform of nuclear-encoded DNA methyltransferase enzyme 1 (mtDNMT1) [Citation10], methylation on the mtDNA is a highly debated matter. Recent findings indicate that mtDNA methylation values derived by bisulphite pyrosequencing have been prone to overestimation, either due to incomplete bisulphite conversion of circular supercoiled mtDNA, or due to the presence of nuclear mitochondrial sequences (NUMTS, copies of specific regions of mtDNA found in the nuclear genome) [Citation11–14]. Several authors even suggest the absence of methylation on the mtDNA, as the absolute methylation levels appear close to the limit of detection [Citation11–13]. As a result, the existence and biological relevance of mtDNA methylation is a controversial topic.
Previously, we investigated how placental mtDNA methylation is influenced by exposure to environmental pollutants during pregnancy [Citation15,Citation16]. The placenta was chosen as the tissue of interest, because of its critical importance during foetal development. Cellular energy in the form of ATP is required to fuel developmental processes in this metabolically active organ [Citation17–19], stressing the key role of placental mitochondria during the prenatal period. Prenatal exposure to environmental stressors has been associated with decreased telomere length [Citation20] and lower mtDNA content in the placenta [Citation21,Citation22], which may result in foetal adaptations that form the origins of diseases later in life (as proposed with the concept of the Developmental Origins of Health and Diseases (DOHaD) [Citation23]. However, recent methodological literature on mtDNA methylation has raised concerns about the validity of our previous findings on the impact of prenatal exposure to tobacco smoke and high levels of ambient air pollution on mtDNA methylation in placental tissue due to incomplete bisulphite conversion and the existence of NUMTs. Therefore, in this study, we revised our previous findings and set up a different research strategy using a stratified sampling approach and by using linear, completely converted bisulphite mtDNA and assays specifically designed to avoid NUMT contamination. We targeted both the heavy (D-loop) and light strand (LDLR2) of the displacement loop region to retrieve as much information as possible, since this region is highly relevant for mitochondrial gene transcription. We selected 60 placenta samples from the ENVIRONAGE birth cohort (ENVIRonmental influence ON early AGEing), based on maternal levels of exposure to tobacco smoke and air pollution during pregnancy. We measured mtDNA methylation in placental tissue, as well as other molecular signatures (nuclear gene methylation, mtDNA content) and newborn birth weight to examine the existence and potential biological relevance of this epigenetic marker. By performing this validity study, we show that conclusions of previously published findings based on bisulphite pyrosequencing are legitimate and remain robust, but effect estimates may be inaccurate.
Methods
An in-depth description of the methods can be found in the supplement material (section Supplemental Information Methods).
Study population
Participants were selected from the ENVIRONAGE birth cohort located in Belgium as previously described [Citation24]. A selection of 60 mothers-newborn pairs was made based on their level of exposure to tobacco smoke and air pollution during the pregnancy. Twenty mothers with the highest absolute number of reported pack-years (packs per day multiplied by the number of years they reported smoking, 15.1 ± 3.5 on average) and with an average number of cigarettes smoked per day during the pregnancy above the third quartile (10 cigarettes per day) were selected (hereafter referred to as ‘Smokers’ group). The ‘Smokers’ group was matched on age and newborn sex with a group of 20 non-smoking mothers with the absolute lowest exposure to PM2.5 and black carbon (BC) during the third trimester (BC: 0.7 ± 0.2 µg/m3, PM2.5: 7.3 ± 1.0 µg/m3) as well as during the entire pregnancy (BC: 0.9 ± 0.1 µg/m3, PM2.5: 10.6 ± 1.7 µg/m3) (hereafter referred to as ‘Control’ group). Finally, the ‘Control’ group was matched on age and newborn sex with a group of 20 non-smoking mothers that were exposed to the absolute highest PM2.5 and BC levels during the third trimester (BC: 1.9 ± 0.3 µg/m3, PM2.5: 22.5 ± 3.0 µg/m3) as well as during the entire pregnancy (BC: 1.8 ± 0.3 µg/m3, PM2.5: 16.0 ± 1.4 µg/m3) (hereafter referred to as ‘Air Pollution’ group).
Ambient air pollution assessment
Residential addresses were geocoded with ArcGIS10 software. Daily prenatal exposure levels of PM2.5 and BC (μg/m3) were modelled for each mother’s address using a high-resolution spatiotemporal model that takes into account land cover data obtained from satellite images (CORINE land cover data set) [Citation24] and pollution data from fixed monitoring stations in combination with a dispersion model [Citation25]. Exposures were estimated for the entire pregnancy as well as for the third trimester (27 weeks after conception until delivery).
Sample collection and processing
Within 10 minutes of delivery, a placental biopsy (approximately 1–2 cm3 in size) was taken from the foetal side of the placenta, deep-frozen and stored at −80°C until DNA extraction [Citation21]. DNA was extracted from homogenized tissue using the QIAamp DNA Mini Kit (Qiagen Inc., Venlo, the Netherlands), according to the manufacturer’s instructions. The concentration and purity of the DNA extracts was assessed using Nanodrop and stored at −80°C until further use.
DNA methylation assessment
Using 600 ng of extracted total genomic DNA, mtDNA was linearized with BamHI enzyme (Thermo Fisher Scientific, Geel, Belgium) and bisulphite-treated using the EZ-96 DNA methylation Gold kit (Zymo Research, Orange, USA) according to the manufacturer’s instructions.
Methylation on multiple CpG sites in both nuclear (POLG1, PINK1, DNA2) and two mitochondrial (D-loop, LDLR2) DNA regions was determined. Both mitochondrial regions were located on the displacement loop, one on the heavy (D-loop) and one on the light (LDLR2) chain. The displacement loop region is highly relevant to mitochondrial gene transcription, and is one of the few regions on mtDNA that does not overlap with nuclear DNA segments. Therefore, to retrieve as much information as possible on the D-loop region, we chose to target both the heavy and light strands. Detailed information regarding primer sequences can be found in Supplemental Table 1. Prior to pyrosequencing, the regions of interest were amplified by PCR using PyroMark PCR kits (Qiagen Inc., Venlo, The Netherlands) in a thermal cycling device (C1000 Thermal Cycler, Bio-Rad Laboratories, Temse, Belgium). Amplified PCR products were sequenced by pyrosequencing using the PyroMark Q48 Autoprep system (Qiagen, Inc., Venlo, The Netherlands). The degree of methylation was defined as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines. The samples were measured in duplicate (correlation of average CpG methylation between technical replicates: r = 0.82 for D-loop and r = 0.99 for LDLR2). Efficiency of the bisulphite conversion process was assessed using non-CpG cytosine residues within the sequence to analyse. Samples that did not pass pyrosequencing quality control were excluded from the analyses (D-loop n = 1, LDLR2 n = 3).
mtDNA content
Placental mtDNA content was defined as the ratio between a mitochondrial gene (MT-ND1) to one single-copy nuclear control gene (RPLP0) (primers sequences in Supplemental Table 2) and measured using a real-time quantitative polymerase chain reaction assay as previously published [Citation21].
Statistical analysis
All statistical analyses were performed using R version 3.5.3 (R Core Team, Vienna, Austria). Using linear mixed effect model analysis, we took into account each CpG position of one gene and tested the associations between gene-specific DNA methylation and exposure group. All linear mixed models were constructed using the lme4 and lmerTest packages [Citation26,Citation27], with two main models constructed for every target region: one for the comparison between the ‘Control’ and ‘Smokers’ group and one for the comparison between the ‘Control’ and ‘Air Pollution’ group. Main models included exposure group, CpG site, maternal education status, maternal parity status and newborn birth weight as fixed effects. All models for the ‘Smokers’ group additionally included exposure to PM2.5 during the third trimester as a fixed effect. Random intercepts were included for individuals. Interactions between exposure group and CpG site were tested, but only included in the main models when the interaction term was significant (p < 0.05). Pearson correlation coefficient was used to assess the correlation between average mtDNA methylation and mtDNA content. To determine significant differences in mtDNA content between the exposure groups, linear regression models were used, with adjusted models including exposure group, maternal education status, maternal parity status and newborn birth weight as covariates. The relationship between newborn birthweight and mtDNA methylation was assessed using linear models, with adjustments for maternal age, gestational age, and parity.
Results
Study population characteristics
The characteristics of the study population by exposure group are summarized in . Population characteristics of the groups did not differ from the characteristics of the general ENVIRONAGE birth cohort [Citation24]. On average, mothers from all groups were 31 years old (ranging from 23 to 40 years old). Each group included 13 male and 7 female newborns. Average maternal pre-gestational BMI was comparable between the ‘Control’ and ‘Smoker’ groups (23.9 kg/m2 and 24.0 kg/m2 respectively), while being one unit lower in the air pollution group (22.9 kg/m2). Mothers from the ‘Smokers’ group had a lower educational status compared with mothers from the ‘Control’ group. Average birth weight was lower in the ‘Smokers’ group (3149 g, p = 0.08) compared with the “Control’ group (3394 g) while no difference was observed for the ‘Air Pollution’ group (3390 g, p = 0.98). All groups were highly comparable regarding the average newborn’s gestational age (ranging between 35 and 41 weeks). For ethnicity, there were slight differences between the groups, with both the ‘Control’ (25%) and ‘Air Pollution’ (15%) groups including more non-European newborns than the ‘Smokers’ group (5%), although this difference was not significant (p = 0.18).
Table 1. Characteristics of the selected mother-newborn pairs, categorized by exposure group
Average exposure to ambient air pollution during the whole pregnancy was significantly lower for the ‘Control’ group (PM2.5: 10.6 µg/m3; BC: 0.9 µg/m3) than for the ‘Air Pollution’ group (PM2.5: 16.0 µg/m3; BC: 1.8 µg/m3). Notably, air pollution exposure in the ‘Control’ group was also lower as compared with the ‘Smokers’ group (PM2.5: 13.3 µg/m3; BC: 1.3 µg/m3). Similar trends regarding exposure levels were observed during the third trimester (). In the ‘Smokers’ group, mothers had an average of 15 pack-years and smoked 13 cigarettes per day on average.
Environmental exposure and mtDNA methylation
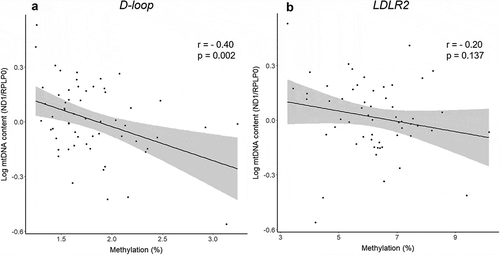
In the total sample population of 60 individuals, the absolute mtDNA methylation levels were close to the limit of detection in the D-loop region (ranging between 1% and 4%), whereas average methylation levels in the LDLR2 where slightly higher (ranging between 3% and 10%) ().
Figure 1. Mean mitochondrial DNA methylation levels as determined by bisulphite pyrosequencing (n = 20/group). The horizontal line is equivalent to the median. The lower and upper hinges represent the first and third quartiles. The upper whisker extends from the hinge to the largest value no further than 1.5 * IQR (= interquartile range) from the hinge, whereas the lower whisker extends from the hinge to the smallest value no further than 1.5 * IQR of the hinge. Data outside this range is represented as dots
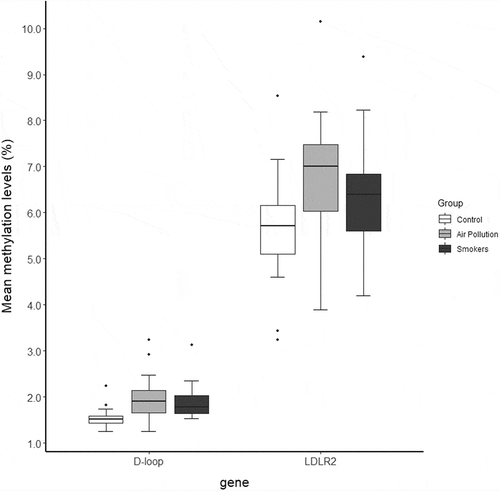
Using adjusted linear mixed effect models, a significant association was found between in utero environmental exposures and methylation levels on the D-loop region of the mitochondrial DNA (). D-loop methylation levels were higher in both exposure groups as compared with the control group (β = +0.47%, 95% CI: 0.20% to 0.73% for the ‘Air Pollution’ group, β = +0.36%, 95% CI: 0.06% to 0.66% for the ‘Smokers’ group). For LDLR2, no significant difference was observed in the ‘Smoker’ (p = 0.45) or ‘Air pollution’ (p = 0.11) group compared with the control group. We observed no significant interaction effects between CpG site and exposure group for both mtDNA target regions (p ≥ 0.09).
Table 2. Differences in mtDNA methylation levels between exposure groups as determined by linear mixed models
Associations between mtDNA methylation and biologically relevant markers
mtDNA methylation should be interpreted within the context of its biological relevance. In this study, we demonstrate subtle changes in mtDNA methylation levels that may have a functional meaning in the placenta. Therefore, we explored how other molecular markers behave in these exposure groups. First, mtDNA content was negatively correlated with D-loop methylation levels (r = −0.40, p = 0.002), whereas no correlation was observed for LDLR2 (p = 0.14) (). Significantly lower placental mtDNA content was observed in the ”Smokers” group compared with the ‘Control’ group (Supplemental Table 3), while no significant difference was observed in the ‘Air Pollution’ group. In addition to methylation levels on mtDNA, we also determined methylation levels on several nuclear DNA regions relevant to mitochondrial maintenance (POLG1 promotor, POLG1 exon 2, PINK1 and DNA2). We observed a moderate negative correlation between mtDNA methylation and PINK1 promotor methylation (r = −0.33, p = 0.01). Methylation levels on PINK1 were also significantly lower for mothers in the ‘Air Pollution’ group as compared with the ‘Control’ group (β = −0.42%, 95% CI: −0.60% to −0.24%). No significant correlations were observed for LDLR2 methylation and nuclear gene methylation. For relevant clinical outcomes, we investigated the association between mtDNA methylation and newborn birth weight and observed that for an IQR increase in D-loop methylation levels, birth weight decreased by 106.98 g (95% CI: −209.60 g to −4.36 g) (). No association with newborn birth weight was observed for LDLR2.
Table 3. Association between placental mtDNA methylation levels and newborn birth weight
Discussion
Our investigation has highlighted that conclusions of previously published findings based on bisulphite pyrosequencing are reliable but effect estimates might be overestimated when not accounting for key methodological strategies. We observed significant associations between placental mtDNA methylation with prenatal exposure to ambient air pollution and tobacco smoke, as well as with other molecular signatures of mitochondrial health. We also demonstrate a significant association between mtDNA methylation and newborn birth weight, a highly relevant clinical outcome.
In recent years, the existence and biological relevance of mtDNA methylation has been an important topic of debate, as investigations into the epigenetic marker have resulted in inconsistent findings. The earliest studies made use of methyl-sensitive restriction-enzyme based assays to quantify methylation levels, and reported low levels of mtDNA methylation (2–5% on CCGG sites) [Citation28,Citation29]. By contrast, studies using methylated DNA immunoprecipitation followed by PCR, mass spectrometry and ELISA report higher values, with some reporting methylation levels as high as 13–25% [Citation30]. The variation in methylation levels between these studies is not surprising, given the inherent differences in sensitivity of the used techniques. Bisulphite sequencing is considered the gold standard method to quantify DNA methylation because of its high reliability and sensitivity. Using this method, some studies report relatively high levels [Citation15,Citation31], whereas others present evidence that suggests the absence of biologically relevant levels of mtDNA methylation [Citation12,Citation13]. Methodological issues such as insufficient bisulphite conversion and NUMT contamination likely explain these inconsistencies.
NUMTS are large sections of the mitochondrial genome that occur as transcriptionally-inactive pseudogenes on the nuclear DNA. These sequences cover a large portion of the mitochondrial genome. Given that methylation levels on nuclear genes tends to be higher, it is necessary to avoid NUMT amplification when designing pyrosequencing assays. In our study, we developed primers to specifically amplify mtDNA with the intent to avoid NUMT contamination (hence why only a small number of mitochondrial targets were selected for this study). Detailed information on primer design and specificity is described in the Supplemental Information Methods and Supplemental Figure 1. Secondly, it has been demonstrated that the circular structure of mtDNA can lead to incomplete bisulphite conversion, resulting in overestimations of mtDNA methylation. Furthermore, research indicates that the extent to which bisulphite conversion efficiency is affected by mtDNA structure is both region and tissue dependent [Citation11,Citation14]. Our findings align with those results, as we observed that methylation levels in the LDLR2 and D-loop regions were significantly affected by poor bisulphite conversion efficiency on non-linearized mtDNA in the placenta (Supplemental Figure 2). However, the effect was less pronounced for the D-loop region, implying that the extent to which the mtDNA is affected by poor bisulphite conversion is also region dependent. Therefore, region-specific variances in secondary mtDNA structure across the mitochondrial genome may result in differences in bisulphite conversion efficiency. While methylation levels were relatively low on the D-loop (ranging between 1% and 4%), average LDLR2 methylation levels were higher. The CpG site of LDLR2 (bp 545, UCSC, Gr18/hg38 genome assembly) averages 10%, which is consistent with other studies observing similarly high methylation levels on the same CpG site [Citation14]. Despite the fact that previous studies may have overestimated mtDNA methylation, our current findings suggest that significant levels of methylation can occur on specific sites within the mtDNA. This conclusion is further supported by research regarding the activity of mitochondria-localized DNA methyltransferase 1 (mtDNMT1) enzymes, which have been shown to bind mtDNA and may provide the mechanism by which mtDNA is methylated [Citation10]. Recent research has shown that mtDNA is extensively methylated at non-CpG sites [Citation32,Citation33] which could lead to pyrosequencing related PCR biases [Citation34]. In short, primers are generated in the assumption that all non-CpG cytosine residues are converted to thymine residues by bisulphite conversion. If non-CpG methylation is present, these primers are less likely to bind highly methylated mtDNA sequences. As a result, DNA fragments with low methylation levels are selectively amplified, likely leading to an underestimation of the actual methylation levels on CpG sites. This might be a reason why we – and others – observe low methylation levels on mtDNA when using PCR-based pyrosequencing methods.
With regard to the influence of environmental exposures, we observed increased absolute D-loop methylation levels in both the ‘Smokers’ (+0.36%) and ‘Air Pollution’ (+0.47%) group as compared with the ‘Control’ group. This observation is consistent with the findings from Janssen et al. [Citation15], who found that for each increment of 3 µg/m3 in PM2.5 exposure during the entire pregnancy the absolute methylation in D-loop methylation increased by 0.21%. If we extrapolate these findings to the 6 µg/m3 difference in entire pregnancy PM2.5 between the ‘Control’ and ‘Air pollution’ group, we observe an absolute increase of 0.42%, which is reasonably comparable with the effect estimate we observed in our study (+0.47%), considering the overestimation of methylation levels in that study. Similarly, while previously no association was observed between smoking during pregnancy and placental D-loop methylation [Citation16], we observed significant differences. Thus, overestimation of mtDNA methylation levels may not necessarily disprove significant associations between environmental exposures and DNA methylation levels. Nonetheless, results from studies that do not consider bisulphite conversion efficiency should be interpreted with caution. We did not observe significant effects for the LDLR2 region. This suggests that the effects of environmental exposure may affect the strands in the displacement loop differently, as LDLR2 is located on the light strand (as opposed to the D-loop region, which is located on the heavy strand).
When interpreting DNA methylation data, it is also crucial to consider the biological relevance. The overall methylation levels of the mitochondrial genome that we observed were relatively low (ranging between 1% and 10%), thus leading to seemingly small effect estimates. However, it should be noted that small increases in methylation at the lower end of the scale (e.g. from 2% to 2.5%) may have a proportionally bigger impact compared with small increases in methylation in the higher end of the scale (e.g. from 70% to 71%). It should be noted that similar small effect sizes are commonly accepted as relevant nuclear methylation changes that exert biological effects [Citation35]. As mentioned before, other studies have questioned the biological relevance of low mtDNA methylation levels for the regulation of mitochondrial gene expression and mtDNA replication [Citation13]. Nonetheless, while no associations were found between the induction of high levels of CpG methylation and mitochondrial gene expression, metabolic activity or superoxide production, significant effects on mtDNA content have been demonstrated previously in HCT116 cells [Citation14]. Mitochondrial DNA content is an established marker of mitochondrial damage and function [Citation1] and has been identified as an aetiological determinant in a variety of human diseases [Citation36]. In our study, we demonstrated a strong correlation between D-loop methylation and mtDNA content but not for LDLR2, suggesting that methylation at the heavy strand of the mitochondrial genome is linked with the amount of mtDNA copy numbers. Furthermore, our results indicate that mtDNA methylation may be related to mitophagy, as D-loop methylation levels were correlated with methylation on the PINK1 promotor. PINK1 encodes for a protein kinase that localizes to mitochondria and is involved in mitochondrial quality control and degradation of damaged mitochondria [Citation37]. Microarray data of placental tissue is available in the ENVIRONAGE birth cohort database [Citation24], but unfortunately did not overlap with our sample subset.
In addition to its relevance in biological processes and as an environmental biosensor, mtDNA methylation may also serve as a predictive biomarker for clinical outcomes. Results from Baccarelli et al. indicate that patients with cardiovascular disease show dramatically higher levels of mtDNA methylation in platelets which lack nuclear DNA [Citation38]. In addition, a more recent study showed that platelet methylation of MT-CO1, MT-CO3, and MT-TL1 are, together, strong predictors of future cardiovascular disease incidence in adults with overweight and obesity [Citation39]. The potential clinical relevance of mtDNA methylation that we demonstrate in this study is limited to a significant association with birth weight, which is an important predictor for negative health outcomes such as cardiovascular diseases, hypertension and psychiatric medication use later in life [Citation40–42]. We are currently performing a follow-up study of our participants at the age of 4 to 6 years, which will allow us to study the association between early life environmental factors and mitochondrial effects, and link those relationships with other health outcomes such as cardiovascular function and neurobehavioral performance in childhood. More studies are necessary to conclude on the associations between mtDNA methylation, cardiovascular disease and risk factors such as environmental exposures and obesity, but the potential use of mtDNA methylation as an early predictor and/or diagnostic tool seems promising.
Although our study presents valuable new insights and has key strengths such as the avoidance of NUMT contamination in our assays, it should be noted that it also has several limitations. First, our sample size was relatively small, therefore limiting statistical power and our ability to detect small differences between the groups, which may explain why we were not able to observe significant associations in fully adjusted models between mtDNA content and ambient air pollution, despite the fact that these associations have been demonstrated previously in a larger sample population of the ENVIRONAGE birth cohort [Citation21]. Furthermore, our investigation into mtDNA methylation only represents a small portion of the mitochondrial genome. Future studies may apply whole genome sequencing approaches to cover the entire mtDNA, provided that they use nuclear free DNA by extracting mtDNA from nuclear DNA [Citation43] or by the use platelet-derived mtDNA samples – which are free of nuclear DNA [Citation44].
Another important limitation of our study, is the recent finding that mtDNA is extensively methylated at non-CpG sites [Citation32]. This may have affected our results in two ways. First, non-CpG methylation may have caused a bias in our bisulphite PCR. In standard assays, primers are chosen irrespective of whether the bisulphite sequence contains a thymine corresponding to a converted non-CpG cytosine. Hence, if those cytosine are methylated, these primer pairs will selectively amplify ‘unmethylated’ products which explain the low methylation levels. However, any bias that would result from this assumption would lead to an underestimation (rather than an overestimation) of the actual methylation values, as highly methylated mtDNA segments would be less likely to be amplified. Nonetheless, future studies should also take into account non-CpG methylation when designing bisulphite pyrosequencing mtDNA methylation assays to avoid this potential bias. Second, our bisulphite controls located on non-CpG cytosine residues may have affected our bisulphite efficiency controls. However, we do not believe the use of non-CpG cytosine residues as bisulphite efficiency controls was a problem for our study. Theoretically, the most significant downside would be the occurrence of false negative controls (cases where the bisulphite conversion was adequate, but the control nonetheless indicates failure because the cytosine residue was methylated). In our case, the number of samples where the bisulphite control failed was very low (n = 1 for D-loop). Therefore, we believe that the chance that this introduced bias in our results is negligible.
Finally, the placenta is a naturally heterogeneous tissue featuring many different cell types, the composition of which may be influenced by environmental exposures. This means that cell type composition potentially has a confounding effect on our results. Because we are not using a genome-wide DNA methylation technique and a reference panel for placental cell types does not yet exist, we could not adjust for heterogeneity in cell-type composition.
Conclusions
The existence of mtDNA methylation has been a topic of debate. Our study highlights that, while previously overestimated, significant methylation is present on specific CpG sites in the mitochondrial genome. We also demonstrate a significant association between placental mtDNA methylation levels and prenatal exposure to ambient air pollutants and tobacco smoke.
Supplemental Material
Download Zip (578.6 KB)Acknowledgments
The authors thank the participating women and neonates, as well as the staff of the maternity ward, midwives, and the staff of the clinical laboratory of East-Limburg Hospital in Genk. This research is supported by the Flemish Scientific Fund N1518119 and G082317N. Bram Janssen and Dries Martens are postdoctoral fellows funded by the Research Foundation Flanders (FWO) (12W3218N and 12X9620N respectively).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Sahin E, Colla S, Liesa M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365.
- Soberanes S, Urich D, Baker CM, et al. Mitochondrial Complex III-generated oxidants activate ASK1 and JNK to induce alveolar epithelial cell death following exposure to particulate matter air pollution. J Biol Chem. 2009;284(4):2176–2186.
- Waterhouse NJ, Goldstein JC, von Ahsen O, et al. Cytochrome C maintains mitochondrial transmembrane potential and Atp generation after outer mitochondrial membrane permeabilization during the apoptotic process. J Cell Biol. 2001;153(2):319.
- Hiura TS, Li N, Kaplan R, et al. The role of a mitochondrial pathway in the induction of apoptosis by chemicals extracted from diesel exhaust particles. J Immunol. 2000;165(5):2703.
- Janssen BG, Byun H-M, Roels HA, et al. Regulating role of fetal thyroid hormones on placental mitochondrial DNA methylation: epidemiological evidence from the ENVIRONAGE birth cohort study. Clin Epigenetics. 2017;9:66.
- Sun X, Vaghjiani V, Jayasekara WSN, et al. The degree of mitochondrial DNA methylation in tumor models of glioblastoma and osteosarcoma. Clin Epigenetics. 2018;10(1):157.
- Feng S, Xiong L, Ji Z, et al. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol Med Rep. 2012;6(1):125–130.
- Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging. 2012;33(12):2881–2891.
- Pirola CJ, Gianotti TF, Burgueno AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363.
- Shock LS, Thakkar PV, Peterson EJ, et al. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108(9):3630–3635.
- Liu B, Du Q, Chen L, et al. CpG methylation patterns of human mitochondrial DNA. Sci Rep. 2016;6:23421.
- Mechta M, Ingerslev LR, Fabre O, et al. Evidence suggesting absence of mitochondrial DNA methylation. Front Genet. 2017;8:166.
- Hong EE, Okitsu CY, Smith AD, et al. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol. 2013;33(14):2683–2690.
- van der Wijst MG, van Tilburg AY, Ruiters MH, et al. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci Rep. 2017;7(1):177.
- Janssen BG, Byun H-M, Gyselaers W, et al. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: an ENVIR ON AGE birth cohort study. Epigenetics. 2015;10(6):536–544.
- Janssen BG, Gyselaers W, Byun HM, et al. Placental mitochondrial DNA and CYP1A1 gene methylation as molecular signatures for tobacco smoke exposure in pregnant women and the relevance for birth weight. J Transl Med. 2017;15(1):5.
- Manokhina I, Del Gobbo GF, Konwar C, et al. Review: placental biomarkers for assessing fetal health. Hum Mol Genet. 2017;26(R2):R237–r45.
- Wakefield SL, Lane M, Mitchell M. Impaired mitochondrial function in the preimplantation embryo perturbs fetal and placental development in the mouse. Biol Reprod. 2011;84(3):572–580.
- Mayeur S, Lancel S, Theys N, et al. Maternal calorie restriction modulates placental mitochondrial biogenesis and bioenergetic efficiency: putative involvement in fetoplacental growth defects in rats. Am J Physiol Endocrinol Metab. 2012;304(1):E14–E22.
- Martens DS, Cox B, Janssen BG, et al. Prenatal air pollution and newborns’ predisposition to accelerated biological aging. JAMA Pediatr. 2017;171(12):1160–1167.
- Janssen BG, Munters E, Pieters N, et al. Placental mitochondrial DNA content and particulate air pollution during in utero life. Environ Health Perspect. 2012;120(9):1346–1352.
- Bouhours-Nouet N, May-Panloup P, Coutant R, et al. Maternal smoking is associated with mitochondrial DNA depletion and respiratory chain complex III deficiency in placenta. Am J Physiol Endocrinol Metab. 2005;288(1):E171–7.
- Wadhwa PD, Buss C, Entringer S, et al. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27(5):358–368.
- Janssen S, Dumont G, Fierens F, et al. Spatial interpolation of air pollution measurements using CORINE land cover data. Atmos Environ. 2008;42(20):4884–4903. doi:10.1016/j.atmosenv.2008.02.043.
- Lefebvre W, Degrawe B, Beckx C, et al. Presentation and evaluation of an integrated model chain to respond to traffic- and health-related policy questions. Environ Modell Software. 2013;40:160–170.
- Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;1(1):1–48.
- Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest package: tests in linear mixed effects models. J Stat Softw. 2017;1(13):1–26.
- Shmookler Reis RJ, Goldstein S. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J Biol Chem. 1983;258(15):9078–9085.
- Pollack Y, Kasir J, Shemer R, et al. Methylation pattern of mouse mitochondrial DNA. Nucleic Acids Res. 1984;12(12):4811–4824.
- Infantino V, Castegna A, Iacobazzi F, et al. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down’s syndrome. Mol Genet Metab. 2011;102(3):378–382.
- Bellizzi D, D’Aquila P, Scafone T, et al. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013;20(6):537–547.
- Patil V, Cuenin C, Chung F, et al. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 2019;47(19):10072–10085.
- Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9(6):823–828.
- Morris MJ, Hesson LB, Youngson NA. Non-CpG methylation biases bisulphite PCR towards low or unmethylated mitochondrial DNA: recommendations for the field. Environ Epigenet. 2020;6(1):dvaa001–dvaa.
- Neven KY, Saenen ND, Tarantini L, et al. Placental promoter methylation of DNA repair genes and prenatal exposure to particulate air pollution: an ENVIRONAGE cohort study. Lancet Planet Health. 2018;2(4):e174–e83.
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51(5):440–450.
- Truban D, Hou X, Caulfield TR, et al. PINK1, Parkin, and mitochondrial quality control: what can we learn about Parkinson’s disease pathobiology? J Parkinsons Dis. 2017;7(1):13–29.
- Baccarelli AA, Byun H-M. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clin Epigenetics. 2015;7(1):1–9.
- Corsi S, Iodice S, Vigna L, et al. Platelet mitochondrial DNA methylation predicts future cardiovascular outcome in adults with overweight and obesity. Clin Epigenetics. 2020;12(1):29.
- Lawlor DA, Ronalds G, Clark H, et al. Birth weight is inversely associated with incident coronary heart disease and stroke among individuals born in the 1950s: findings from the Aberdeen Children of the 1950s prospective cohort study. Circulation. 2005;112(10):1414–1418.
- Gennser G, Rymark P, Isberg PE. Low birth weight and risk of high blood pressure in adulthood. Br Med J (Clin Res Ed). 1988;296(6635):1498–1500.
- Crump C, Winkleby MA, Sundquist K, et al. Preterm birth and psychiatric medication prescription in young adulthood: a Swedish national cohort study. Int J Epidemiol. 2010;39(6):1522–1530.
- Jayaprakash AD, Benson EK, Gone S, et al. Stable heteroplasmy at the single-cell level is facilitated by intercellular exchange of mtDNA. Nucleic Acids Res. 2015;43(4):2177–2187.
- Baccarelli AA, Byun H-M. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clin Epigenetics. 2015;7(1):44.