ABSTRACT
Studies have suggested that abrogated expression of detoxification enzymes, UGT2B15 and UGT2B17, are associated with prostate tumour risk and progression. We investigated the role of EGF on the expression of these enzymes since it interacts with signalling pathways to also affect prostate tumour progression and is additionally associated with decreased DNA methylation. The expression of UGT2B15, UGT2B17, de novo methyltransferases, DNMT3A and DNMT3B was assessed in prostate cancer cells (LNCaP) treated with EGF, an EGFR inhibitor PD16893, and the methyltransferase inhibitor, 5-azacytidine, respectively. The results showed that EGF treatment decreased levels of expression of all four genes and that their expression was reversed by PD16893. Treatment with 5-azacytidine, markedly decreased expression of UGT2B15 and UGT2B17 over 85% as well as significantly decreased expression of DNMT3B, but not the expression of DNMT3A. DNMT3B siRNA treated LNCaP cells had decreased expression of UGT2B15 and UGT2B17, while DNMT3A siRNA treated cells had only moderately decreased UGT2B15 expression. Treatment with DNMT methyltransferase inhibitor, RG108, significantly decreased UGT2B17 expression. Additionally, methylation differences between prostate cancer samples and benign prostate samples from an Illumina 450K Methylation Array study were assessed. The results taken together suggest that hypomethylation of the UGT2B15 and UGT2B17 genes contributes to increased risk of prostate cancer and may provide a putative biomarker or epigenetic target for chemotherapeutics. Mechanistic studies are warranted to determine the role of the methylation marks in prostate cancer.
Introduction
In the U.S., cancer is the second leading cause of death and prostate cancer is the second highest occurring cancer in men after lung cancer. The number of new prostate cancer cases for 2020 was estimated to be 191,930[Citation1]. For over 10 years, the DNA sequence variants of the enzymes of the human UDP-glucuronosyltransferases 2B (UGT2B) subfamily have been investigated and shown to be associated with increased risk for prostate cancer [Citation2,Citation3]. The UGT2B subfamily of enzymes are part of the UGT (UGT, EC 2.4.1.17) superfamily of drug-metabolizing enzymes that are bound to the endoplasmic reticulum in hepatic and extrahepatic cells to facilitate the clearance of exogenous and endogenous toxins[Citation4]. The UGT superfamily consists of two main gene subfamilies, UGT1, located on chromosome 2 which contains nine unique first exons that are spliced to common exons 2–5; and UGT2, located on chromosome 4 which contains nine distinct genes. The UGT2 subfamily is further divided into UGT2A and UGT2B[Citation5].
The UGT2B subfamily is comprised of UGT2B4, UGT2B7, UGT2B11, UGT2B15, UGT2B17, and UGT2B28 genes along with six pseudogenes that are interspersed among the genes in the region[Citation6]. Enzymes UGT2B15 and UGT2B17 exhibit substrate specificity for androgens such as testosterone, dihydrotestosterone (DHT), DHT metabolites, androsterone, and 5α-androstane-3α, 17β-diol [Citation7–9]. One of the tissues where the UGT2B15 and UGT2B17 enzymes typically catalyse the clearance of these chemicals is the prostate. Results from a recent study were suggestive of an association of overexpression of UGT2B17 with biochemical recurrence of prostate cancer in a Veterans’ Medical Centre (VA) population including both African American and Caucasian patients[Citation10]. Other lines of evidence showed that UGT2B15 genes are associated with risk for prostate cancer in African American men [Citation3,Citation11]. These associations suggest that the development of prostate cancer disease has UGT2B genetic underpinnings. However, the impact on gene expression remains unclear.
Other lines of evidence suggest that epigenetic alterations may play a role in the regulation of UGT2B15 and UGT2B17. For instance, hyper and hypomethylation changes in the TSS1500 interval of UGT1A and UGT2 genes have been differentially associated with transcription in individuals with non-alcoholic fatty liver disease[Citation12]. On the other hand, DNA hypermethylation and histone hypoacetylation has been shown to abrogate transcriptional regulation of UGT1A1 in human kidney cells[Citation13]. The investigators in that study further showed that tissue-specific expression of UGT1A10 was mediated by transcription factors whose efficacy was affected by promoter methylation of that gene[Citation14].
An early study of UGT2B15 and UGT2B17 gene expression suggested a regulatory role for epidermal growth factor (EGF)[Citation15]. That study and others showed that UGT2B17 expression and glucuronidation function was downregulated in prostate cancer cells treated with EGF [Citation16,Citation17]. Aberrant expression resulting from EGF/EGFR signalling has been observed in androgen-independent and metastatic prostate cancer; associated with epithelial–mesenchymal transition and reduced survival rate[Citation18]. This signalling impacts the efficacy of UGT glucuronidation and subsequently its antiproliferative effects on prostate cancer cells in androgen-dependent cancer[Citation19]. Moreover, EGF was later found to alter global DNA methylation in gastric cancer cell lines when used as treatment[Citation20]. In addition, another study suggests that EGF may impact methylation status by modulating DNA methyltransferase activities[Citation21]. In that study, long-term EGF treatment or constitutively active EGFR expression increased DNA methyltransferase activity in ovarian cancer cells. These changes are widely believed to be caused by de novo methylation by DNA methyltransferases, DNMT3A and DNMT3B; which target DNA sequences in a complex with DNA methyltransferase, DNMT3L, that subsequently becomes associated with histone 3 unmethylated at lysine 4 (H3K4)[Citation22].
The regulation of UGT expression by DNA methylation and its impact on UGT2B15 and UGT2B17 expression in prostate cancer cells is unknown. Thus, we sought to determine the relationship of EGF/EGFR regulation and DNA methylation on UGT2B gene expression in prostate cancer cells. We hypothesized that EGF downregulation of UGT2B gene expression in prostate cancer cells is associated with changes in DNA methyltransferase activity. Here, we show that changes in UGT2B17 and UGT2B15 expression were altered in the prostate cancer cell line, LNCaP, treated with DNMT3A or DNMT3B siRNAs. In addition, we present data that shows specific UGT2B17 and UGT2B15 CpG sites are associated with prostate cancer as determined by data from the Illumina Infinium HumanMethylation 450K Beadchip array.
Results
EGF downregulated gene expression in prostate cancer cells is reversed by EGFR inhibitor, PD168393
Using RT-PCR, expression of UGT2B15, UGT2B17, DMNT3A, and DNMT3B was measured in LNCaP cells treated with EGF and the EGFR inhibitor, PD68393 [Citation23]. Treatment with 10 ng/ml EGF decreases UGT2B15 mRNA expression by nearly 50% when compared to control (p < 0.0001) and that the inhibition is completely reversed by treatment with 10 μM of PD168393 ()). UGT2B17 mRNA levels are also reduced by more than 50% with EGF treatment (p < 0.0001) when compared to the control while treatment with 1 and 10 μM of PD168393 increases mRNA levels significantly when compared to EGF treatment alone (p < 0.0001), ()). Similarly, mRNA levels of DMNT3A are decreased by 50% upon EGF treatment when compared to the control while treatment with 1 and 10 μM PD168393 significantly increases mRNA levels when compared to EGF treatment alone (p < 0.001), ()). Finally, EGF treated mRNA levels of DNMT3B are decreased approximately by 50% when compared to the control while treatment with 1 and 10 μM PD168393 significantly increases mRNA levels when compared to EGF treatment alone (p < 0.0001), ()).
Figure 1. EGF mediated down regulation of UGT2B15, UGT2B17, DNMT3A and DNMT3B expression is reversed by EGFR inhibitor, PD168393 (PD). LNCaP cells were treated for 24 h with vehicle (DMSO), EGF and EGF+PD at concentrations indicated in the figure. mRNA was extracted and quantitated using RT-PCR as described in materials and methods. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc test. The mRNA levels are reported as the mean ± SEM of 3 independent experiments, each performed in triplicate. Means that do not share letters are significantly different from each other. (a) Effect of EGF and PD168393 on UGT2B15 expression. (b) Effect of EGF and PD168393 on UGT2B17 expression (c) Effect of EGF and PD168393 on DNMT3A expression. (d) Effect of EGF and PD168393 on DNMT3B expression. Vehicle = Control
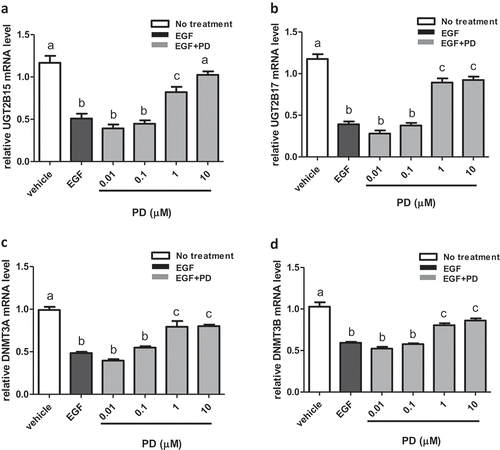
In summary, these results show that EGF treatment inhibits UGT2B15, UGT2B17, DMNT3A, and DNMT3B expression which is then reversed by the EGFR inhibitor, PD168393.
Demethylation agent, 5-azacytidine, alters gene expression in LNCaP cells in a manner similar to EGF effect
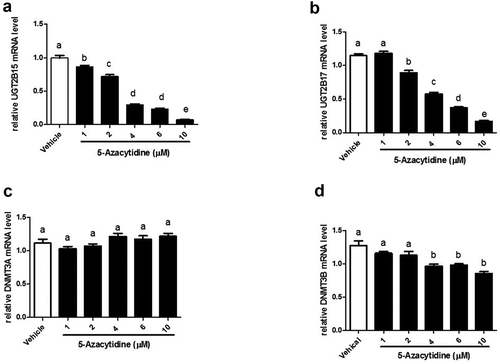
To assess the effect of 5-azacytidine on the expression of UGT2B15, UGT2B17, DMNT3A, and DNMT3B, cells were treated with 1–10 μM 5-azacytidine and mRNA was measured using RT-PCR. The results show that 5-azacytidine decreases expression of both UGT2B15 and UGT2B17 in a dose-response manner ()). At 10 μM, 5-azacytidine inhibited UGT2B15 and UGT2B17expression by 93.2% and 85.4%, respectively (p < 0.0001), and inhibited DNMT3B expression by 33% (p < 0.0001). In contrast, the mRNA levels of DNMT3A remained unchanged ()).
Figure 2. 5-Azacytidine treatment of LNCaP cells downregulates expression of UGT2B15, UGT2B17 and DNMT3B mRNA in a dose-response manner. LNCaP cells were treated with vehicle (DMSO) and increasing amounts (1–10 µM) of 5-Azacytidine for 24 h. mRNA was quantitated and statistically analysed as described in legend. The mRNA levels are reported as the mean ± SEM of 3 independent experiments, each performed in triplicate. Means that do not share letters are significantly different from each other. (a) Effect of 5-Azacytidine on UGT2B15 expression. (b) Effect of 5-Azacytidine on UGT2B17 expression. (c) Effect of 5-Azacytidine on DNMT3A expression and (d) Effect of 5-Azacytidine on DNMT3B expression. Vehicle = Control
Increased expression of both p53 and EGFR have been previously determined in prostate cancer[Citation24]. To demonstrate the gene-specific response of UGT2B and DNMT3 genes to 5-azacytidine, the impact of treatment with 10 ng/ml EGF, its inhibitor, and 5-azacytdine on p53 expression in LNCaP cells was determined (Figure S1). The results show that treatment with 1–10 μM 5-azacytidine increases p53 gene expression in a dose-response manner. At 10 μM, 5-azacytidine increased p53 gene expression (by 46.7%, p < 0.0001), (Figure S1A). Treatment with 10 ng/ml EGF significantly increases levels of p53 mRNA which is then reversed by treatment with EGFR inhibitor, PD168393 (Figure S1B).
In summary, these results show that the demethylation agent, 5-azacytidine decreases UGT2B15, UGT2B17, and DNMT3B expression in a gene-specific manner that mimics the EGF effect. In contrast 5-azacytidine and EGF increases p53 gene expression.
DNMT3A and DNMT3B silencing downregulates gene expression in LNCaP cells
Because treatment with 5-azacytidine inhibited UGT2B15, UGT2B17 and DNMT3B expression, we used DNMT3A or DNMT3B siRNA to confirm that DNMTs drive this effect.
Transfection with DNMT3B siRNA decreased DNMT3B expression by 72% when compared to cells transfected with non-targeting control (negative control) siRNAs (p < 0.0001) ()). DNMT3B silencing significantly downregulated UGT2B17 and UGT2B15 expression by 45% and 47% (p < 0.0001), respectively, when compared to negative control ()). Unexpectedly, however, p53 mRNA levels were also significantly decreased by 40% (p < 0.0001) ()) after having observed increased expression when hypomethylated by 5-azacytidine.
Figure 3. DNMT3B and DNMT3A siRNA knockdown in LNCaP cells showing knockdown expression of UGT2B17, UGT2B15, and P53. The mRNA was quantitated and statistically analysed as described in legend. The mRNA levels are reported as the mean ± SEM of 3 independent experiments, each performed in triplicate. Means that do not share letters are significantly different from each other. (a) Knockdown expression of DNMT3B; (b) UGT2B17 in DNMT3B knockdown LNCaP cells; (c) decreased UGT2B15 in DNMT3B knockdown LNCaP cells at p < 0.0001; and (d) decreased p53 in DNMT3B knockdown LNCaP cells at p < 0.0001. DNMT3A siRNA knockdown in LNCaP cells showing (e) knockdown expression of DNMT3A; (f) decreased UGT2B17 in DNMT3A knockdown LNCaP cells at p < 0.0001; (g) decreased UGT2B15 in DNMT3A knockdown LNCaP cells at p < 0.02; and (h) p53 in DNMT3A knockdown LNCaP cells. Negative control = negative control siRNA; DNMT3A or DNMT3B siRNA = NeoFX Transfection Agent 0.8 ul (30 nM) siRNA
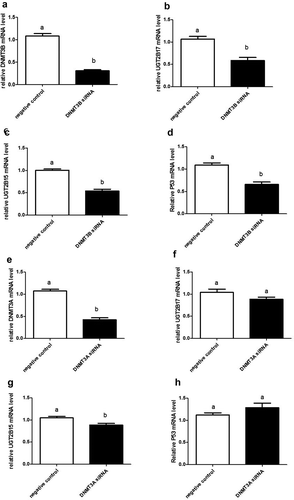
Transfection with DNMT3A siRNA reduced DNMT3A by 60% (p < 0.0001) when compared to cells transfected with nonsense siRNAs ()). DNMT3A silencing slightly decreased expression of UGT2B17 and p53 genes ()), but this reduction was not significant. However, levels of UGT2B15 mRNA were significantly decreased by 16% (p < 0.02) ()) when compared to nonsense controls.
In summary, DNMT3B silencing in prostate cancer cells effectively and significantly decreases expression of UGT2B15, UGT2B17, and p53; whereas DNMT3A silencing significantly reduces only UGT2B15 expression.
Methyltransferase inhibitor, RG108, alters UGT2B17 expression in LNCaP cells
To assess the effect of directly inhibiting DNA methyltransferase activity, LNCaP cells were treated with methyltransferase inhibitor, RG108[Citation25]. This inhibitor was identified through a screen using methyltransferase, DNMT1; however, its highly conserved catalytic domains are shared with methyltransferases, DNMT3A and DNMT3B. The results show that mRNA levels were unchanged between vehicle and RG108 treatment for DNMT3A, DNMT3B, and p53 (, respectively). Notably, the expression of UGT2B17 was decreased by 32% (p < 0.0001) ()).
Figure 4. DNA methyltransferase inhibitor RG108 treatment of LNCaP cells downregulates UGT2B17 expression. LNCaP cells were treated with vehicle (DMSO) and RG108 (100 µM) for 48 h. mRNA was quantitated as described in material and methods. Data was analysed using Nonparametric Student t test. mRNA levels are reported as the mean ± SEM of experiments done in triplicate. (a) Effect of RG108 treatment on DNMT3A expression. (b) Effect of RG108 treatment on DNMT3B expression. (c) Effect of RG108 treatment on UGT2B17 expression . (d) Effect of RG108 on p53 expression. Vehicle = Control
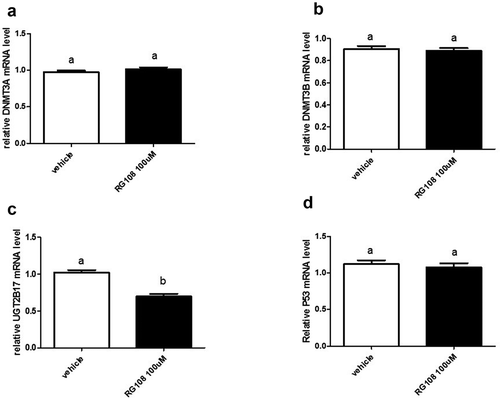
In summary, the inhibition of methyltransferase activity with RG108 unlike 5-azacytidine only altered UGT2B17 gene expression.
Discussion
Previous studies have suggested an association between EGF modulated gene expression in cancer cells and DNA methylation status. As hypothesized we show EGF treatment decreases mRNA expression in DNMT3s and UGT2Bs. Moreover, the treatment of LNCaP cells with DNA methyltransferase inhibitor 5-azacytidine decreased mRNA expression of UGT2B15 and UGT2B17 as well as DNMT3B while having lesser effect on DNMT3A mRNA expression. While the decrease in DNMT3B expression was moderate in LNCaP cells treated with 5-azacytidine when compared to that of UGT2B15 and UGT2B17, the decrease in global methyl acceptor activity in the presence of EGF observed in Figure S2 suggests a role for methyltransferases. The results from the silencing of DNMT3B suggest an unknown interaction between the methyltransferase in decreasing UGT2B gene expression since p53 expression was decreased by DNMT3B siRNA treatment but had the expected increase in expression under conditions created by 5-azacytidine. Results from a study of 73 curated clinically annotated fresh-frozen prostate cancer tissues and 63 benign prostate tissues using the Illumina Infinium HumanMethylation 450K Beadchip array showed significant differences in methylation in the regulatory region of UGT2B15 and UGT2B17 that were indicative of hypomethylation (; cg20840634; 11.64% (p = 1.9 x 10 −12))[Citation26]. Taken together these results suggest that DNA methyltransferases play a role in UGT2B gene regulation ().
Table 1. Analysis of study GSE76938 of 73 prostate cancers vs. 63 benign tissue with Illumina HumanMethylation 450 K Beadship array
Figure 5. Downregulation of UGT2B17 and UGT2B15 gene expression induced by increased EGFR activity and decreased DNMT3B expression. Specific CpG dinucleotides within UGT2B regulatory regions are hypomethylated at the proximal promoter and coding region and are associated with prostate cancer risk. EGF/EGFR signalling may mediate the downregulation of DNMT3B methyltransferase activity resulting in decreased expression of UGTB217 and UGT2B15 genes from the hypomethylation of regulatory CpG dinucleotides in the promotor and first exon
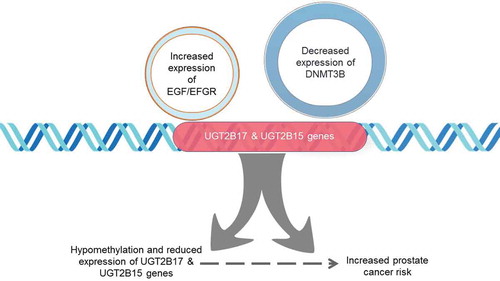
Research nearly twenty years ago first demonstrated that LNCaP cells treated with EGF had decreased expression of UGT2B17 mRNA but not UGT2B15 [Citation15–17]. The results in this study showed that the decrease in expression was observed in both UGT2B15 and UGT2B17 when measured with gene-specific custom PCR primers[Citation27]. Reversal of the downregulation by EGFR inhibitor, PD168393, demonstrates that EGF acts through that receptor in prostate cancer cells to decrease UGT2B15 and UGT2B17 expression. The inhibitor has been used in synergy with other chemotherapeutic drugs in prostate cancer patients to abrogate the proliferative effect that has been observed because of EGFR activity in cancers [Citation28–30]. In addition, reduced levels of UGT2B15 and UGT2B17 from UGT2B15/B17 siRNA treated cells resulted in lower levels of glucuronidation of UGT2B15/B17 androgen metabolites leading to increased levels of dihydrotestosterone (DHT)[Citation31]. The inverse relationship between UGT2B15/B17 expression and levels of DHT may be linked to activated androgen receptor and EGFR interactions associated with progression to castration-resistant prostate cancer (CRPC) or resistance to androgen-derived therapies [Citation32,Citation33]. Thus, decreased expression of UGT2B15 and UGT2B17 may be markers for the development of CRPC.
To date, only two studies have shown an association between EGF/EGFR activation and DNA methylation. In one study, 10 nM of EGF was shown to induce the demethylation of DNA in treated gastric cancer cell lines [Citation20] while in another study by Samudio-Ruiz and Hudson [Citation21], EGF treatment of ovarian cancer cells significantly increased DNMT activity and subsequently global DNA methylation. In that same study, the treatment of prostate cancer cells with the same concentration of EGF resulted in decreased global methyltransferase activity consistent with decreased expression of UGT2B15, UGT2B17, DNMT3A, and DNMT3B observed in prostate cancer cells in our study. Only one study has shown a link between methylation at the UGT2B15 locus and gene expression[Citation34]. In that study, differences in levels of methylation were found between Asians and Caucasians. Other studies focused on regulation of expression of other enzymes in the UGT1 and UGT2 subfamilies by methylation. [Citation12,Citation35] Because there is evidence that EGF increases DNMT methyltransferase activity in ovarian cancer cells, it is highly likely that expression of DNMT3A and DNMT3B is affected by EGF[Citation21]. That study measured global methyltransferase activity and did not distinguish between DNMT1, DNMT3A, or DNMT3B. Our study represents the first measurement of specific DNMT3A/3B expression in prostate cancer cells treated with EGF and 5-azacytidine. In our study, the silencing of the DNMT3B gene demonstrated decreased expression of UGT2B15 and UGT2B17 in a hypomethylated cellular environment. In addition, DNMT3B and not DNMT3A expression in prostate cancer cells was shown to be affected by treatment with a plant-derived carbazole alkaloid, which resulted in the restoration of expression a tumour suppressor gene[Citation36]. These data suggest that DNMT3A and DNMT3B methyltransferases may have differential roles in regulation of UGT2B15 and UGT2B17 expression even though they have very similar responses to EGF treatment. In a study, bicalutamide, used in antiandrogenic therapy, increased the expression of DNMT3B but not DNMT3A in 22Rv1 cells suggestive of a relationship of DNMT mediated hypermethylation and prostate cancer progression to the castration-resistant phenotype[Citation37]. In that same study that expression was attenuated by treatment with 5-azacytidine. These results are consistent with the observations from another study that showed increased expression of UGT2B15 and UGT2B17 in LNCaP cells treated with bicalutamide[Citation33].
Several studies have examined epigenetic mechanisms of gene regulation of UGT enzymes including the methylation of distinct CpG sites [Citation12–14,Citation38-43]. The current literature indicates that only one study has examined the regulation of UGT2B15 and UGT2B17 by site-specific DNA methylation in human normal hepatic tissue[Citation34]. In that study, UGT2B15 was found to be differentially methylated at four CpG loci, cg09189601, cg07973162, cg10632656, and cg07952421, in hepatic samples from Caucasians and Asians. Site cg20840634 () was hypomethylated in the transcriptional start site regions of UGT2B15 and UGT2B17 in prostate cancer tissue when compared to normal tissue. However, that same loci showed no difference in methylation patterns in hepatic tissue[Citation34]. Further, loci cg22161115, located in the first exon of UGT2B15, was also shown to be associated with hypomethylation in prostate cancer tissue when compared to normal tissue but not showing differential methylation in hepatic tissue. These observations are evidence that methylation patterns may differ by gene location and tissue specificity and that specific sites may be an important mechanism in DMNT3A/3B mediated gene regulation of UGT2B15 and UGT2B17. While changes in global methylation have been observed in EGF treated gastric cancer cells [Citation20], specific CpG dinucleotides may be responsible for downregulation of UGT2B15 and UGT2B17 genes in prostate cancer. Notably, two CpG dinucleotide loci associated with hypermethylation of prostate cancer tissue were located in the body and intergenetic region of UGT2B15 and UGT2B15/UGT2B17, respectively ().
Future studies that implement more sensitive, genome-wide methods will be needed to determine the relationship between UGT2B region methylation and prostate cancer progression including to CRPC to establish the biomarker potential of such patterns. Such approaches have been explored with other genes associated with prostate cancer risk and progression[Citation44].
Materials and methods
Cell culture
LNCaP cells were purchased from American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI (Gibco, ThermoFisher Scientific, USA), 1% Penicillin-Streptomycin (Gibco), and 10% Heat-Inactivated Foetal Bovine Serum (FBS) (Gibco). Cells were grown in 25 or 75 cm2 flask (Corning) for siRNA experiments and protein isolation for Western Blots, respectively. Cells were grown to 80% confluency, trypsinized (0.25% Trypsin-EDTA, Gibco ThermoFisher Scientific, USA), collected in a 15-mL tube (Fisher Scientific), and centrifuged (125 x g) for 5 min. The cell pellet was collected and reconstituted in antibiotic-free RPMI supplemented with 2% Charcoal-stripped FBS. Cells were seeded (72 h) in a 96-well plate (Corning, Tweksbury, MA) at 20,000 cells/well, in triplicates, for 24 hr treatments.
Cell culture treatments
For EGF (Gibco, ThermoFisher Scientific, USA) treatment, 100 μl of complete RPMI 1640 media (10% charcoal stripped serum, 1% penicillin/streptomycin, 1% sodium bicarbonate) was added to wells in a 96 well-dish and pre-incubated at 37°C to adjust for pH and temperature. Cells were then added to each well at 200 cells/ μl in a 50 ul volume then incubated at 37°C and 5% CO2 for 72 hr. The media in the wells was replaced with fresh complete RPMI 1640 media containing 10 ng/ml EGF and 0.01, 0.1,1, and 10 ng/ml EGFR inhibitor. In treatments with PD168393, (Calbiochem Corp., San Diego, CA) DMSO was used as vehicle for PD168393 and the control consisted of a DMSO vehicle control. For samples with dual treatments of EGF and PD168393, the controls consisted of the DMSO vehicle control. For cells treated with 5-azacytidine (MP Biomedicals, Santa Ana, CA) the pre-incubation procedure was followed but the media in the wells was replaced with fresh complete RPMI 1640 media containing 1, 2, 4, 6 and 10 μM 5-azacytidine. DMSO vehicle control was included in control and 5-azacytidine treated cells. These treatments were incubated for 24 h. For cells treated with 100 μM DNA methyltransferase inhibitor, RG108 [Citation45], DMSO vehicle control was included in control and RG108 treated cells. The experiments were performed in triplicate. All treatments were incubated for 48 hr at standard culture conditions.
Methyltransferase activity measurement
The methyltransferase activity was determined by use of a commercially prepared kit from Fermentas (Fermentas-ThermoFisher Scientific). LNCaP cells were treated with EGF and PD168393 as described above in Cell Culture Treatments in 75 ml culture flasks. The cells were incubated 48 hours and nuclear protein isolated quantitated using Pierce protein isolation kit (Thermo Scientific). Universal DNMT substrates are stably coated onto microplate wells and the activity in nuclear extracts transfer methyl groups to cytosine from Adomet to methylate DNA substrate. The wells are sequentially incubated with different antibodies, washed and a colour developing solution added developed and absorbance measured at 450 nm.
siRNA transfection
Short interfering RNAs (siRNA) specific for human Deoxyribonucleic Acid (DNA) (cytosine-5-) methyltransferase 3 alpha (DNMT3A) (s200426), Deoxyribonucleic Acid (DNA) (cytosine-5-) methyltransferase 3 beta (DNMT3B) (s4223) and Negative Control siRNA were purchased from Applied Biosystems (Foster City, CA). siPORTTM NeoFXTM Transfection Agent (Ambion) and siRNA were individually diluted in Opti-Mem I medium (Gibco), incubated for 10 mins at room temperature (RT), then combined 1:1 to form a siRNA/siPORTTM NeoFXTM complex. The final concentration of siRNA is 30 nM. This complex was incubated for 10 min at RT and added to the cells which were incubated under standard cell culture conditions for 48 hr.
Quantitative real-time PCR
Expression was measured in siRNA treated cells after 48 hr using quantitative-RT PCR (Q-RTPCR) using TaqMan® Gene Expression Cells-to-CTTM (Thermo Fisher-Applied Biosystems, AM1728, Foster City, CA) technology was used to lyse the cells and prepare cDNA. Briefly, the cells were washed with cold 1X Phosphate Buffered Solution (PBS). Next, 50 μl of lysis solution is added, mixed five times and incubated 5 min at room temperature. After 5 μl of stop solution is added and mixed five times the solution is incubated for 2 min at room temperature. Reverse transcription was accomplished by incubating ten microlitres of crude cell lysate with RT Master Mix and running samples in RT thermal cycler program to convert RNA to cDNA. Standard PCR was accomplished using TaqMan PCR Master Mix and 5–10 μl cDNA template using commercially available primers and 7900HT Fast Real-Time PCR Systems with 96 well block (ThermoFisher-Applied Biosystems, AM1728, Foster City, CA). Commercial primers used as follows: TaqMan® Gene Expression AssaysTM for DNMT3B (Assay ID. Hs00171876_m1) and DNMT3A (Assay ID. Hs00602456_m1) were purchased from Applied Biosystems (ThermoFisher-Applied Biosystems, AM1728, Foster City, CA). The DNMT3B assay includes six Interrogated Reference Sequences: NM_001204055.1, NM_001207056.1, NM_006892.3, NM_175848.1, NM_175849.1 and NM_175850.2. The DNMT3A assay includes four Interrogated Reference Sequences: NM_001320892.1, NM_022552.4, NM_175629.2 and NM_175630.1. Knockdown percentage was quantified by using the comparative ΔΔCt with 18S as an internal control. Relative abundance of subsequent genes, UDP glucuronosyltransferase family 2 member B17 (UGT2B17) (Assay ID. Hs00854486_sH), UGT2B15 (Assay ID. EX25) and Tumour Protein P53 (TP53) (Assay ID. Hs00153349_m1) were also analysed.
Statistical analysis
Statistical calculations were performed using Prism Version 5.0 (GraphPad Software, Inc., San Diego, CA). For RT-PCR experiments, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used to determine statistical differences among controls and treatments (EGF, PD168393, RG108 and 5-azacytidine). Data are reported as the mean ± SEM of three independent experiments, each performed in triplicate. Nonparametric Student t test was performed to analyse statistical differences between siRNA transfected groups, and comparison of subsequent gene expression. Data are reported as the mean ± SEM of experiments done in triplicate. p < 0.05 was considered a significant difference.
GEO analysis
Regularized logistic regression was performed (rlm), and betas determined for ~ Tumour/Normal, between tumour and normal samples for UGT genes from study GSE76938 which examined 73 clinically annotated fresh-frozen prostate tissues and 63 benign prostate tissues using the Illumina Infinium HumanMethylation 450K Beadchip array (Kirby et al., 2017 BMC Cancer April 17;17(1):273. PMID: 28,412,973). Negative BETA values represent hypomethylation and positive represents hypermethylation. The significance is based on multiple-corrected p-value using Benjamine Hockereg multiple correction (padj).
Supplemental Material
Download Zip (81.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
- Habibi M, Mirfakhraie R, Khani M, et al. Genetic variations in UGT2B28, UGT2B17, UGT2B15 genes and the risk of prostate cancer: A case-control study. Gene. 2017;634:47–52.
- Hu DG, Mackenzie PI, McKinnon RA, et al. Genetic polymorphisms of human UDP-glucuronosyltransferase (UGT) genes and cancer risk. Drug Metab Rev. 2016;48:47–69.
- Fujiwara R, Yokoi T, Nakajima M. Structure and protein-protein interactions of human UDP-Glucuronosyltransferases. Front Pharmacol. 2016;7:388.
- Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45:1121–1132.
- Guillemette C, Levesque E, Harvey M, et al. UGT genomic diversity: beyond gene duplication. Drug Metab Rev. 2010;42:24–44.
- Turgeon D, Carrier JS, Levesque E, et al. Isolation and characterization of the human UGT2B15 gene, localized within a cluster of UGT2B genes and pseudogenes on chromosome 4. J Mol Biol. 2000;295:489–504.
- Turgeon D, Carrier JS, Levesque E, et al. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787.
- Migita T, Takayama KI, Urano T, et al. ACSL3 promotes intratumoral steroidogenesis in prostate cancer cells. Cancer Sci. 2017;108:2011–2021.
- Grant DJ, Chen Z, Howard LE, et al. UDP-glucuronosyltransferases and biochemical recurrence in prostate cancer progression. BMC Cancer. 2017;17:463.
- Vidal AC, Tucker C, Schildkraut JM, et al. Novel associations of UDP-glucuronosyltransferase 2B gene variants with prostate cancer risk in a multiethnic study. BMC Cancer. 2013;13:556.
- Schioth HB, Bostrom A, Murphy SK, et al. A targeted analysis reveals relevant shifts in the methylation and transcription of genes responsible for bile acid homeostasis and drug metabolism in non-alcoholic fatty liver disease. BMC Genomics. 2016;17:462.
- Oda S, Fukami T, Yokoi T, et al. Epigenetic regulation is a crucial factor in the repression of UGT1A1 expression in the human kidney. Drug Metab Dispos. 2013;41:1738–1743.
- Oda S, Fukami T, Yokoi T, et al. Epigenetic regulation of the tissue-specific expression of human UDP-glucuronosyltransferase (UGT) 1A10. Biochem Pharmacol. 2014;87:660–667.
- Belanger A, Hum DW, Beaulieu M, et al. Characterization and regulation of UDP-glucuronosyltransferases in steroid target tissues. J Steroid Biochem Mol Biol. 1998;65:301–310.
- Chouinard S, Pelletier G, Belanger A, et al. Isoform-specific regulation of uridine diphosphate-glucuronosyltransferase 2B enzymes in the human prostate: differential consequences for androgen and bioactive lipid inactivation. Endocrinology. 2006;147:5431–5442.
- Guillemette C, Levesque E, Beaulieu M, et al. Differential regulation of two uridine diphospho-glucuronosyltransferases, UGT2B15 and UGT2B17, in human prostate LNCaP cells. Endocrinology. 1997;138:2998–3005.
- Montanari M, Rossetti S, Cavaliere C, et al. Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget. 2017;8:35376–35389.
- Gauthier-Landry L, Belanger A, Barbier O. Multiple roles for UDP-glucuronosyltransferase (UGT)2B15 and UGT2B17 enzymes in androgen metabolism and prostate cancer evolution. J Steroid Biochem Mol Biol. 2015;145:187–192.
- Saikawa Y, Kubota T, Otani Y, et al. Alteration of DNA methylation status induced by epidermal growth factor in gastric cancer cell line, MKN-74. Anticancer Res. 2003;23:143–148.
- Samudio-Ruiz SL, Hudson LG. Increased DNA methyltransferase activity and DNA methylation following Epidermal Growth Factor stimulation in ovarian cancer cells. Epigenetics. 2012;7:216–224.
- Edwards JR, Yarychkivska O, Boulard M, et al. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23.
- Pu YS, Hsieh MW, Wang CW, et al. Epidermal growth factor receptor inhibitor (PD168393) potentiates cytotoxic effects of paclitaxel against androgen-independent prostate cancer cells. Biochem Pharmacol. 2006;71:751–760.
- Sauer L, Gitenay D, Vo C, et al. Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene. 2010;29:2628–2637.
- Savickiene J, Treigyte G, Jazdauskaite A, et al. DNA methyltransferase inhibitor RG108 and histone deacetylase inhibitors cooperate to enhance NB4 cell differentiation and E-cadherin re-expression by chromatin remodelling. Cell Biol Int. 2012;36:1067–1078.
- Kirby MK, Ramaker RC, Roberts BS, et al. Genome-wide DNA methylation measurements in prostate tissues uncovers novel prostate cancer diagnostic biomarkers and transcription factor binding patterns. BMC Cancer. 2017;17:273.
- Wilson W 3rd, Pardo-manuel de Villena F, Lyn-Cook BD, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714.
- Mitchell RA, Luwor RB, Burgess AW. Epidermal growth factor receptor: structure-function informing the design of anticancer therapeutics. Exp Cell Res. 2018;371:1–19.
- Xu MJ, Johnson DE, Grandis JR. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017;36:463–473.
- Resnick MB, Routhier J, Konkin T, et al. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res off J Am Assoc Cancer Res. 2004;10:3069–3075.
- Chouinard S, Barbier O, Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J Biol Chem. 2007;282:33466–33474.
- Mandel A, Larsson P, Sarwar M, et al. The interplay between AR, EGF receptor and MMP-9 signaling pathways in invasive prostate cancer. Mol Med. 2018;24:34.
- Grosse L, Paquet S, Caron P, et al. Androgen glucuronidation: an unexpected target for androgen deprivation therapy, with prognosis and diagnostic implications. Cancer Res. 2013;73:6963–6971.
- Oeser SG, Bingham JP, Collier AC. Regulation of hepatic UGT2B15 by methylation in adults of asian descent. Pharmaceutics. 2018;10:6.
- Yasar U, Greenblatt DJ, Guillemette C, et al. Evidence for regulation of UDP-glucuronosyltransferase (UGT) 1A1 protein expression and activity via DNA methylation in healthy human livers. J Pharm Pharmacol. 2013;65:874–883.
- Agarwal S, Amin KS, Jagadeesh S, et al. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol Cancer. 2013;12:99.
- Gravina GL, Marampon F, Piccolella M, et al. Hormonal therapy promotes hormone-resistant phenotype by increasing DNMT activity and expression in prostate cancer models. Endocrinology. 2011;152:4550–4561.
- Kalthoff S, Winkler A, Freiberg N, et al. Gender matters: estrogen receptor alpha (ERalpha) and histone deacetylase (HDAC) 1 and 2 control the gender-specific transcriptional regulation of human uridine diphosphate glucuronosyltransferases genes (UGT1A). J Hepatol. 2013;59:797–804.
- Dluzen DF, Sun D, Salzberg AC, et al. Regulation of UDP-glucuronosyltransferase 1A1 expression and activity by microRNA 491-3p. J Pharmacol Exp Ther. 2014;348:465–477.
- Hu DG, Meech R, McKinnon RA, et al. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug Metab Rev. 2014;46:421–458.
- Dluzen DF, Sutliff AK, Chen G, et al. Regulation of UGT2B expression and activity by miR-216b-5p in liver cancer cell lines. J Pharmacol Exp Ther. 2016;359:182–193.
- Papageorgiou I, Freytsis M, Court MH. Transcriptome association analysis identifies miR-375 as a major determinant of variable acetaminophen glucuronidation by human liver. Biochem Pharmacol. 2016;117:78–87.
- Papageorgiou I, Court MH. Identification and validation of the microRNA response elements in the 3ʹ-untranslated region of the UDP glucuronosyltransferase (UGT) 2B7 and 2B15 genes by a functional genomics approach. Biochem Pharmacol. 2017;146:199–213.
- Massie CE, Mills IG, Lynch AG. The importance of DNA methylation in prostate cancer development. J Steroid Biochem Mol Biol. 2017;166:1–15.
- Ou Y, Zhang Q, Tang Y, et al. DNA methylation enzyme inhibitor RG108 suppresses the radioresistance of esophageal cancer. Oncol Rep. 2018;39:993–1002.
