ABSTRACT
Imprinted genes are differentially expressed in a parent-of-origin-specific manner. Parental origin of the alleles is discriminated by intragenic DNA polymorphisms. Comparisons of parental allelic expression have been analysed by semiquantitative RT-PCR. Here, we developed a novel quantitative method for allelic expression of the imprinted gene Ube3a, which inactivation and mutations cause Angelman syndrome and predominantly expressed by the maternal allele in neuronal tissues. In this method, cDNA was amplified by droplet digital PCR (ddPCR) coupled with allele-specific locked nucleic acid (LNA) TaqMan probes, which labelled by FAM and HEX were designed to detect the SNPs in the target regions. ddPCR assay demonstrated that the sense transcript of Ube3a was equally expressed from both parental alleles in adult tissues except neuronal tissues, where Ube3a expression from the paternal allele was about 10 to 14% of total Ube3a expression in adult brain, and 20% in spinal cord. The antisense transcript of Ube3a was expressed at 60% to 70% of the sense transcript of Ube3a in adult brain. Changes in the Ube3a transcripts during postnatal brain development were also evaluated by ddPCR. The ddPCR method is far more reliable and simpler to use than semiquantitative PCR to analyse skewed or faint allelic expression of imprinted genes.
Introduction
Genomic imprinting is a form of regulation causing nonequivalence between maternal and paternal mammalian genomes. Most genes are equally expressed in both parental alleles. In contrast, imprinted genes are expressed exclusively or preferentially by one or the other parental allele [Citation1,Citation2]. Imprinted genes may be specific to a tissue, developmental stage, species, or promoter. Moreover, the imprinting may be partial. Imprinted genes play important roles in numerous congenital human syndromes [Citation3]. The imprinted human UBE3A gene is implicated in Angelman syndrome (AS) (MIM 105830) which is characterized by neurobehavioral anomalies. UBE3A encodes an E3 ubiquitin ligase which ubiquitinates other proteins to be degraded in the ubiquitin proteasome pathway [Citation4]. UBE3A is ubiquitously expressed at different expression levels. Nearly all of the phenotypic abnormalities observed in AS are attributed to abnormal brain development and function. Thus, UBE3A may have brain-specific imprinting. Direct evidence for brain – specific UBE3A imprinting was demonstrated in human foetal – and AS deletion brains [Citation5,Citation6]. RT-PCR revealed that UBE3A is imprinted mainly in the brain (but not other tissues) and is preferentially expressed by the maternal allele.
Imprinted Ube3a expression has been measured in mouse brains [Citation7–9]. Ube3a maternal-deficient mice have been reported by this group [Citation8] and others [Citation9]. These animals presented with markedly reduced Ube3a expression in the hippocampus and dentate gyrus relative to those of Ube3a paternal – deficient mice. Judson et al extensively mapped allele-specific Ube3a protein expression by an immunofluorescent staining protocol. They disclosed that paternal Ube3a expression is characteristic of immature neurons in the postnatal brain whereas the more mature neurons lacked paternal Ube3a expression [Citation10]. Neuron-specific imprinting was also demonstrated for in vitro neuron culture systems [Citation11].
In mature neurons, antisense transcripts of Ube3a (Ube3a-ATS) ()) contribute to the 3′ end of a paternal large noncoding RNA transcript (lncRNA) including exons upstream of Snrpn (U exons), Snrpn, and snoRNAs (Snord115 and Snord116) [Citation12]. Ube3a-ATS is a large and complex transcript identified by northern blot as a hybridization smear [Citation13]. Ube3a-ATS may interfere with paternal Ube3a transcription from the opposite DNA strand by an antisense-mediated mechanism [Citation14–16]. Inhibiting lncRNA transcript extension by inserting a transcriptional termination sequence [Citation16], administering topoisomerase inhibitor drugs [Citation17], or using antisense oligonucleotides [Citation18] partially unsilences paternal Ube3a. These inhibitors of Ube3a-ATS transcript are applicable for AS therapy, however effective technique to quantitatively analyse the expression of Ube3a-ATS remains unknown. Since the discovery of Ube3a-ATS in the brain was first reported [Citation19], Ube3a-ATS expression was evaluated by northern blot [Citation13] and quantitative PCR (qPCR) [Citation11–18]. Skewed allelic Ube3a expression described for postnatal brains and various brain regions was not consistently biased towards maternal expression because of the inherent limitations of semiquantitative PCR [Citation7–9,Citation11,Citation13]. To elucidate the balance of expression between Ube3a and Ube3a-ATS, including the allelic contributions of the Ube3a transcripts, we analysed the absolute allelic expressions of Ube3a and Ube3a-ATS by droplet digital PCR (ddPCR) coupled with locked nucleic acid (LNA) TaqMan probes.
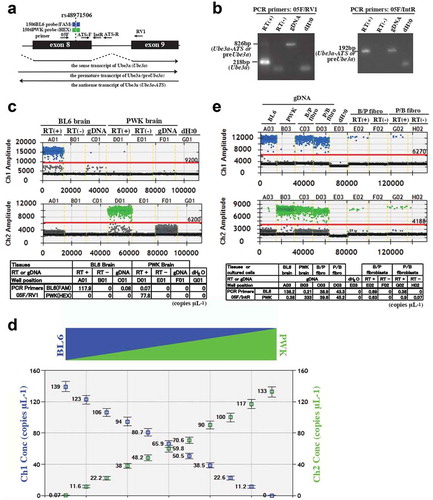
Figure 1. Ube3a and Ube3a-ATS detection by RT-PCR and ddPCR. (a) Schematic of Ube3a exons 8 and 9 with primers (arrows), LNA TaqMan probes, and the transcripts Ube3a, preUbe3a, and Ube3a- ATS. (b) RT-PCR products in agarose gel. Brain cDNA and genomic DNA (gDNA) were amplified by the primers 05 F and RV1 (left panel) and by the primers 05 F and IntR (right panel). Agarose gel electrophoresis showed PCR products as Ube3a, preUbe3a, or Ube3a-ATS. (c) ddPCR analysis of allele-specific Ube3a detection in cDNA amplified by primers 05 F and RV1. The cDNA and gDNA used were homozygous for the nucleotide of interest. (d) ddPCR analysis of the allelic ratios of Ube3a transcripts in mixed cDNA amplified by primers 05 F and RV1. The upper panel shows an illustrated panel of mixed cDNAs from the BL6 (blue) and PWK (green) brains at different ratios. The lower panel is a concentration plot for each positive droplet shown in channel 1 and channel 2 concentrations (Ch1 Conc and Ch2 Conc) as concentrations of FAM- and HEX-positive droplets, selectively hybridized to BL6 and PWK alleles, respectively. Poisson error bars were calculated in QuantaSoft™ as 95% confidence intervals. (e) ddPCR analysis of allele-specific detection of Ube3a-ATS length in gDNA and cDNA amplified by primers 05 F and IntR. The gDNA from BL6, PWK, and F1 hybrids were amplified using primers 05 F and IntR. The cDNAs primed by random hexamers in embryonic fibroblasts of F1 hybrids (B/P- or P/B fibroblasts) were amplified. (c, e) The upper- and middle panels are one- dimensional plots of droplets whose fluorescence signals (amplitudes indicated on y-axis) emitted from FAM- and HEX-labelled LNA TaqMan probes were measured and selectively hybridized to the BL6 and PWK alleles, respectively. Red horizontal lines are thresholds that were manually determined to exclude false positive droplets based on the results of the control gDNA or cDNA. FAM- and HEX-positive droplets are shown in blue and green, respectively. Negative droplets are shown in black. Tissues were analysed as RT (+) and RT (-) samples meaning with- and without RT treatment, respectively. Lower tables show concentrations (copies μL−1) of the transcripts or genomic copy numbers at the PCR target sites processed in QuantaSoft™
The ddPCR is a new digital PCR method performed using droplets of water-oil emulsion and microfluidic techniques. It can be adapted to a wide range of biomedical applications. It provides absolute quantification of nucleic acid target sequences with high degrees of sensitivity and accuracy [Citation20]. The ddPCR has several advantages over qPCR such as absolute quantification without standard assays, reduced sensitivity to inhibitors, and reproducibility between interassays and intraassays. To distinguish and determine the parental origins of the transcribed allele, we used LNA TaqMan probes which enhance hybridization affinity for complementary sequences [Citation21,Citation22]. As LNA bases significantly increase melting temperatures (Tm), they produce probes that are shorter than the standard DNA type. A short TaqMan probe with LNA has relatively better quenching, higher signal-to-noise ratio, greater sensitive, and an improved ability to distinguish mutations or single nucleotide polymorphisms (SNPs) [Citation23,Citation24].
The ddPCR assay was first applied to screen for genomic imprinting in chicken [Citation25]. It was then used in the expression analysis of the murine Gnas locus. This gene is predominantly expressed from the maternal allele in brown fat tissue [Citation26]. Here, we described a simple ddPCR assay using LNA TaqMan probes, attempted the absolute quantitation of the allelic expression at 3 loci of the imprinted Ube3a gene, and determined the allelic ratios of Ube3a transcripts during brain development.
Material and methods
Animals and tissues
All experiments were designed to minimize animal pain and were conducted with the approval of the Institutional Animal Care and Use Committee (Project/File 1503111200-2) of Nagasaki University, Nagasaki, Japan. Female C57BL/6 J (BL6) mice were crossed with male PWK mice (B/P: BL6 x PWK), and vice-versa (P/B: PWK x BL6). The mice were euthanized by cervical dislocation, and their tissues were excised from the mice at P26 and classified as adult tissues. The brains were carefully dissected using a stereomicroscope to isolate the brain regions of interest for this experiment. Cerebra was prepared from P0, P9, P16, and P26. Embryonic fibroblasts were prepared from E15.5 embryos which had been removed from the uteri of timed pregnant mice and placed in Petri dishes containing ice-cold HEPES. The tissues were used for RNA extraction and the embryonic tissues were used in primary cultures.
cDNA synthesis
Total RNA was isolated with ISOGEN II (NIPPON GENE, cat.#317-07363) according to the manufacturer’s protocol. RNA was treated with amplification-grade DNase I (Invitrogen, cat.# 18068015) to degrade any genomic DNA present. The cDNA was generated from total RNA with SUPERSCRIPT III reverse transcriptase (Invitrogen, cat.# 18080093) primed with random hexamer primers, specific primers, or nothing (no priming). First-strand cDNAs primed with or without random hexamers were synthesized at 25°C for 5 min followed by 50°C for 50 min. First-strand cDNAs primed with or without strand-specific primers were synthesized at 55°C for 50 min. The mRNA-cDNA chains were denatured and reverse transcriptase activity was arrested by heating at 70°C for 15 min. An identical reaction was carried out without reverse transcriptase as a negative control (RT (-)).
PCR
Ube3a expression was analysed with primers 05 F, IntR, and RV1 in exon 8, intron 8, and exon 9, respectively ()). Each PCR product was electrophoresed in 1–2% agarose gel. All of the primers used for PCR, qPCR, and ddPCR are listed in Supplementary Table.
ddPCR assay
Genomic DNA (gDNA) or cDNA was added to a 20-μL PCR mixture containing 10 μL of 2x ddPCR Supermix for probes (no dUTP) (Bio-Rad Laboratories, cat.# 1863023), 900 nM of target-specific PCR primers, and 250 nM of LNA-BL6 and LNA-PWK TaqMan probes. The LNA TaqMan probes were purchased from and designed in consultation with Integrated DNA Technologies, Inc. (IDT, Coralville, IA, USA) and labelled with 6-carboxyfluorescein (FAM) or hexachloro-6-carboxyfluorescein (HEX) on the 5‘ end and Iowa Black FQ quencher at the 3‘ end. Twenty microlitres of PCR mixture and 70 μL of droplet generation oil for probes (Bio-Rad Laboratories, cat.#1863005) were mixed. Droplets were produced with a QX200 Droplet Generator (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s manual. The droplet emulsion was thermally cycled as follows: denaturing at 95°C for 10 min; 43 cycles of PCR at 94°C for 30 s; 58–60°C for 1 min, depending on the probes (Supplementary Table) and a final extension at 98°C for 10 min.
The products were scanned on a QX200 Droplet Reader (Bio-Rad Laboratories) and the data were analysed in QuantaSoft™ ver. 1.7.4.0917 software (Bio-Rad Laboratories). The thresholds between the positive and negative droplet signals were manually set to exclude false positive droplets.
qPCR assay
RT-PCR was performed on the cDNAs using SYBR Premix Ex Taq II (Takara Bio Inc., cat.# RR820S) on QuantStudio 12 K Flex (Life Technologies Inc.). The relative standard curve method was adopted for relative quantification of gene expression. PCR was performed at least in triplicate to control for reaction variations. To standardize each experiment, data were represented as a percentage of expression calculated by dividing the average expression of the target gene by that of an internal control gene Gapdh. Each experiment was repeated twice or thrice on independent RNAs. Data are means ± SEM of five biological replicates.
Results
Allele-specific detection of Ube3a transcripts
The Ube3a gene is expressed as sense (Ube3a) and antisense (Ube3a-ATS) transcripts ()). Ube3a is biallelically expressed in most tissues except neuronal tissues. In the latter, the maternal allele is predominantly expressed. Ube3a-ATS expression by the paternal allele is restricted in neuronal tissues. RT-PCR analysis of brain cDNA primed by random hexamers revealed that Ube3a was amplified as a 281-bp product by primer 05 F in exon 8 and by primer RV1 in exon 9. Ube3a-ATS was detected as an amplified product 192 bp in size by primer 05 F in exon 8 and IntR in intron 8. It was also found as a faint 826-bp product by primers 05 F and RV1 ()). In brain cDNA, the large 826-bp PCR product amplified from Ube3a-ATS was not distinguishable from the premature transcript of unspliced Ube3a (preUbe3a) ()). To differentiate allelic Ube3a and Ube3a-ATS transcripts, we crossed the C57BL/6 J (BL6) and PWK strains. The F1 hybrids (B/P; BL6 female x PWK male, P/B; PWK female x BL6 male) were applied in the expression analysis by using the Ube3a DNA polymorphism between the BL6 and PWK strains. The allelic concentrations of the Ube3a transcripts were estimated by ddPCR as copies per μL (copies μL−1) using allele-specific LNA TaqMan probes (1506BL6 and 1506PWK). The probes were designed to detect either thymine (T) in the BL6 allele or cytosine (C) in the PWK allele, namely, the SNP (dbSNP:rs48971506) in Ube3a exon 8 labelled by FAM and HEX, respectively ()). ddPCR data can be assessed as a one-dimensional (1-D) plot with each droplet from a sample plotted on a graph of fluorescence intensity vs. droplet number. It was confirmed that the 1506PWK probe could not hybridize to the transcripts in the brain cDNA from the BL6 strain and formed no positive drops with HEX fluorescent and vise-versa ()). In the embryonic fibroblasts cultured from F1 hybrids (B/P and P/B), the allelic Ube3a transcript concentrations from the BL6 and PWK alleles were nearly equal (Supplementary Figure S1A).
Allele-specific detection of Ube3a transcripts in the mixed cDNA from BL6 and PWK brains
To determine if the allelic concentrations of Ube3a transcripts estimated by ddPCR reflect the allelic ratio of transcripts, cDNAs from the BL6 and PWK brains were mixed and analysed by ddPCR ()). cDNA from the BL6 brain with 139 copies μL−1 of the Ube3a transcript was mixed with that from the PWK brain with 133 copies μL−1. The ratio of the BL6 and PWK cDNA volumes was changed in a gradient-manner for every 10%. The threshold was set manually in the 1-D plot to remove the false positive droplets based on the amplitude signal observed in genomic DNA samples for negative controls (Supplementary Figure S2A). Competing duplex reactions, in which one primer pair binds the same region with two probes can also be viewed in a two-dimensional (2-D) plot in which the channel 1 fluorescence (FAM) is plotted against the channel 2 fluorescence (HEX) for each droplet (Supplementary Figure S2B). The thresholds were set in the 1-D plot for each probe channel, and then, only a few double-positive droplets were detected. The concentration plot in each sample revealed a nearly 10% decrease in allelic transcripts from BL6, which reflected the ratio of BL6 transcripts in the mixed cDNA ().
Distinction between Ube3a-ATS and preUbe3a by ddPCR
LNA TaqMan probe selectivity in Ube3a-ATS was verified by ddPCR using primers 05 F and IntR. The allelic concentrations of the genomic copy numbers were analysed by ddPCR using primers 05 F and IntR ()). The 1506BL6 and 1506PWK probes hybridized to the BL6 and PWK alleles, respectively. In the genomic DNA of the F1 hybrids, the allelic concentrations were almost equal: 38.9 copies μL−1 and 39.6 copies μL−1 from the BL6 and PWK allelles, respectively, were detected in the B/P fibroblasts, and 43.3 copies μL−1 and 45.2 copies μL−1 from the BL6 and PWK alleles, respectively, were detected in the P/B fibroblasts ()). The cDNA from the embryonic fibroblasts of the F1 hybrids was then analysed. The RT (+) samples had low allelic concentrations (0.89 copies μL−1 and 0.63 copies μL−1 from the BL6 and PWK alleles, respectively, in the B/P fibroblasts, and 0.38 copies μL−1 and 0.9 copies μL−1 from the BL6 and PWK alleles, respectively, in the P/B fibroblasts). In contrast, no transcripts were detected in the RT (–) samples ()). Strand-specific cDNA was primed by 05 F or IntR to determine the origins of these transcripts. RNA primed by the sense-strand 05 F was reverse-transcribed to antisense-strand cDNA (Ube3a-ATS). RNA primed by the antisense-strand IntR was reverse- transcribed to sense-strand cDNA (preUbe3a). Low concentrations of both alleles were detected in the cDNA primed by IntR whereas none was detected in the cDNA primed by 05 F (Supplementary Figure S1B).
Allele-specific detection of Ube3a and Ube3a-ATS in adult tissues
Ube3a is biallelically expressed in most tissues except neuronal tissues. In the latter, the maternal allele is predominantly expressed whereas Ube3a-ATS expression is restricted to the neuronal tissues derived from the paternal allele. We analysed the allelic expression of these transcripts in the adult tissues of the F1 hybrid (B/P) at postnatal day 26 (P26). We used cDNA primed by random hexamers (random primed cDNA). The absolute Ube3a concentrations were measured by ddPCR using the primers 05 F and RV1. In the liver, kidney, spleen, heart, testis, and lung, the Ube3a levels were almost equal in both parental alleles ()). In the neuronal tissues, the paternal contributions to the total Ube3a concentrations were 9.7%, 14%, 11%, and 20% in the cerebrum, cerebellum, hippocampus, and spinal cord, respectively ()).
Figure 2. Allele-specific Ube3a-ATS and Ube3a detection by ddPCR. (a)-(c) One-dimensional plots of droplets measured for FAM- and HEX fluorescence signals in adult tissues (upper and middle panels). Data tables (lower panels) are also shown. Transcript concentrations (copies μL−1) were processed in QuantaSoft™. (a) cDNA primed by random hexamers from adult F1 (B/P) tissues at P26 and amplified by primers 05 F and RV1. Proportions of the paternal transcripts in the total Ube3a (pat/total) were calculated (lower tables). (b) The same volume of cDNA as in (a) was amplified by primers 05 F and IntR. Proportions of the maternal transcript in the total Ube3a (mat/total) were calculated (lower tables). (c) Strand-specific cDNA amplified by primers 05 F or IntR for detection of antisense- (Ube3a-ATS) or sense (preUbe3a) transcripts, respectively. Liv: liver; Kid: kidney; Spl: spleen; Hr: heart; Tes: testis; Lun: lung; Cerebr: cerebrum; Cerebel: cerebellum; Hip: hippocampus; SpC: spinal cord. Transcripts originating from paternal and maternal alleles are indicated by pat and mat, respectively. (d) Summary of ddPCR assay in adult neuronal tissues. cDNA primed by random hexamers was used. Transcripts amplified by primers 05 F and RV1 as in (a) are described as Ube3a and those amplified by primers 05 F and IntR as in (b) are described as preUbe3a or Ube3a-ATS, depending on the parental alleles
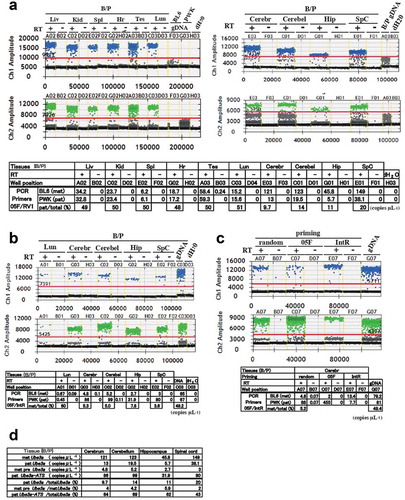
The Ube3a-ATS and/or preUbe3a concentrations in the adult neural tissues were also analysed with the primers 05 F and IntR (). The ddPCR assays showed that the ratios of maternal to total allelic concentrations were 5.3%, 5.0%, 7.8%, and 3.6% in the cerebrum, cerebellum, hippocampus, and spinal cord, respectively ()). The allelic expression of these transcripts in the adult neuronal tissues was also confirmed in the F1 hybrid (P/B) of the reciprocal cross (Supplementary Figure S1C- E).
To distinguish the Ube3a-ATS from the preUbe3a detected in the random primed cDNA, the allelic concentration in the strand-specific cDNA primed by 05 F or IntR was compared with that in the random primed cDNA. The few transcripts from the maternal BL6 allele detected in the random primed cDNA originated mainly from preUbe3a detected in the IntR-primed cDNA. Most transcripts from the paternal PWK allele were derived from Ube3a-ATS ()).
Allele-specific detection of preUbe3a and Ube3a-ATS in other SNPs
The ddPCR assay of strand-specific cDNAs in rs48971506 detected 2 copies μL−1 of transcripts from the maternal allele in the 05 F-primed cDNA and 13.4 and 7.7 copies μL−1of transcripts from both parental alleles in the IntR-primed cDNA ()) despite the low paternal Ube3a contribution (9.7%) in the adult brain ()). Are these transcripts originated from Ube3a-ATS or preUbe3a? To establish whether the allelic-specific detection of these transcripts depended on SNP location, ddPCR assays were run on other SNPs in introns (dbSNP: rs248854698, and rs49512826) () and Supplementary Figure S3A). Probe selectivity was confirmed for each SNP (Supplementary Figure S4B), and the transcripts were analysed in each strand-specific cDNA. The transcripts from the maternal allele in forward primer-primed cDNA was detected at concentrations of 12.3 and 16.3 copies μL−1 of transcripts in rs248854698 and at 2.4 and 3.6 copies μL−1 of transcripts in rs49512862 in F1hybrid brains (B/P and P/B), respectively () and Supplementary Figure S3C and 3D). The transcripts in the reverse primer-primed cDNA was biallelically detected in 15 ~ 60 copies μL−1 in rs248854698 and rs49512862. These results indicate that the transcript from the maternal allele in the forward primer- primed cDNA and those from both parental alleles in the reverse primer-primed cDNA exist, independent of SNP location.
Figure 3. Allele-specific expression of Ube3a-ATS and preUbe3a in SNPs. (a) Schematic of Ube3a with exons, primers (arrowheads), and SNP locations. (b) Summary of ddPCR assay in SNPs. One microgram of adult brain RNA (P26) was used per cDNAs synthesis primed by random hexamers (random), specific primers, or no primers (no) at 50°C or at 55°C. Details of the LNA probes, one-dimensional plots including the RT (–) samples are shown in Supplementary Figure S3 and S4. The copy numbers with one asterisk (*) are considered to be noises in RT reaction, compared to the value of copy numbers in non-specific cDNAs by Poisson error bars in supplementary Figure S3E and S4C. The copy numbers with two asterisk (**) are also considered to be noises because of the location of primers, used for priming for RT
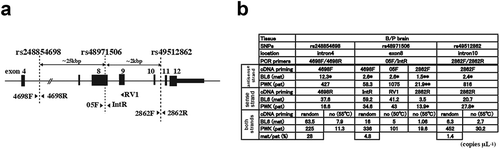
Allele-specific detection of preUbe3a and Ube3a-ATS in strand-specific cDNAs in rs48971506
The allelic-specific transcripts in cDNAs primed by different primers was analysed in rs48971506. The concentrations of the transcript from the paternal allele varied in 2 ~ 3 copies μL−1 with the cDNAs primed by different forward primers (4698 F, 05 F, and 2866 F). These were 25 kbp and 60 bp upstream and 2kbp downstream of the SNP rs48971506, respectively ()). In the cDNA primed by 2866 F, there should be no transcripts amplified by primers 05 F and IntR, upstream of primer 2866 F, however 0.6–1.5 copy μL−1 of the transcript from the maternal allele and 10–20 copies μL−1 of the transcript from the paternal allele were detected () and supplementary Figure S4B). Varying degrees of skewed biallelic preUbe3a expression were also detected in strand-specific cDNAs primed by IntR, RV1, and 2862 R located at 100 bp, 700 bp, and 2 kbp downstream of the SNP, respectively ()).
False positive detection in strand-specific cDNAs
The result that the transcripts were detected by ddPCR using the primers 05 F and IntR in strand-specific cDNAs primed by 2862 F ()) suggested that non-specific cDNA syntheses, which could occur concomitantly during strand-specific cDNA synthesis might affect the ddPCR results, because a ddPCR system has maximum amplification and detection. The cDNA synthesized without exogenous primers during RT was used as non-specific cDNA in ddPCR. We first considered allelic expression in the strand- specific- and non-specific cDNAs in rs48971506. In the non-specific cDNA, the transcript was detected at the rates of 1 and 0.4 copy μL−1 from the maternal and 20 and 14 copies μL−1 from the paternal alleles in F1 hybrids,B/P and P/B, respectively () and Supplementary Figure S4B).
In other SNPs, the non-specific cDNA showed positive detection of the maternal and paternal transcripts () and Supplementary Figure S3C-D). The low concentrations of the transcript from the maternal allele detected in non-specific cDNA did not significantly differ from those detected in strand-specific cDNAs by Poisson statistics. Most of the lower error bars for the measured concentrations in strand-specific cDNAs overlapped with the upper error bars for the measured concentrations in non-specific cDNAs (Supplementary Figure S3E and S4C). False positive Ube3a- ATS detection was higher in non-specific cDNA synthesis at a reverse transcription temperature of 50°C than at the RT temperature of 55°C () and Supplementary Figure S4A). These results mean that in the cDNAs synthesized by strand-specific primers, false positive detection of both strand- transcripts from the non-specific cDNA inevitably exists as a noise in the process of RT synthesis. As for Ube3a-ATS, significant numbers of the transcript were not detected from the maternal allele in the cDNAs primed by forward primers, compared to the those in non-specific cDNA.
Allelic expression analysis of Ube3a and Ube3a-ATS during postnatal brain development
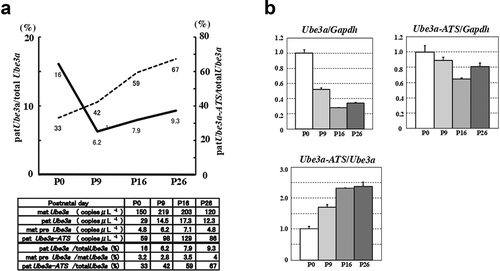
To determine whether allelic expression of Ube3a and Ube3a-ATS is age-dependent, we measured the absolute Ube3a concentrations in random primed cDNAs from the cerebra at various ages via ddPCR using the primers 05 F and RV1 (Supplementary Figure S5A). In the neonatal cerebrum, the paternal contribution to total Ube3a concentration was 16%. In cerebra at P9 and later, however, it was 6–10% ()). Using identical cDNA samples, the absolute Ube3a-ATS concentrations were measured with the primers 05 F and IntR (Supplementary Figure S5B). The ratio of Ube3a-ATS to total Ube3a increased from 33% to 67% in an age-dependent manner ()). Total Ube3a expression was evaluated by quantitative PCR (qPCR) using the same primers as those for ddPCR. Total Ube3a expression was high in the neonatal brain but decreased to ~20% in the adult brain. The Ube3a-ATS expression in P9–P26 was ~60–80% that of P0 ()). The qPCR showed that the ratio of Ube3a- ATS to total Ube3a at P26 was nearly 2× greater than that at P0 ()). This ratio was almost the same as that calculated by ddPCR (33% at P0 and 67% at P26; ().)
Figure 4. Developmental changes in Ube3a-ATS and Ube3a in the cerebrum. (a) Line chart of the proportion of pat Ube3a in total Ube3a (solid line) and the proportion of pat Ube3a-ATS in total Ube3a (dotted line). Line chart is based on ddPCR of allele-specific detection of Ube3a and Ube3a-ATS in random primed cDNA (lower panel) (Supplementary Figure S4). (b) Total expression of Ube3a and Ube3a-ATS determined by qPCR. Expression levels of Ube3a and Ube3a-ATS were evaluated with primers 05 F and RV1 and primers 05 F and IntR, respectively. Experiments were normalized to Gapdh (Ube3a/Gapdh and Ube3a-ATS/Gapdh) or Ube3a controls (Ube3a-ATS/Ube3a)
Discussion
Ube3a is imprinted in the brain. It is expressed there as well as in other neuronal tissues mainly by the maternal allele. However, skewed expression between parental alleles in neuronal tissues has seldom been investigated. Conventional analysis of allelic gene expression is restricted to enzymatic digestion or RT-PCR product sequencing, which spanned polymorphic sites and distinguished alleles. The extent of skewness in allelic expression may vary with the number of PCR cycles and PCR efficiency. Here, we analysed allelic gene expression by ddPCR which determines the absolute nucleic acid concentrations at the target sites with 3 SNPs. The ddPCR system partitions nucleic acid samples into thousands of nanoliter-sized water-oil emulsion droplets. PCR amplification is conducted on each droplet. The proportions of PCR-positive droplets are evaluated by Poisson statistics. The concentrations of the target DNA templates are measured in copies μL−1. The accuracy of allelic gene expression analysis depends on ddPCR sensitivity and TaqMan probe allele specificity. The ddPCR assay is very susceptible to nucleic acid contamination because of maximum target site amplification in the droplets. No positive droplets were detected in most RT (-) samples. Thus, DNase pretreatment before RT completely digested any residual DNA in the RNA samples. To differentiate the allelic origins of the transcript, we adapted locked nucleic acid (LNA) TaqMan probes incorporating several LNA residues. LNA nucleotides contain methylene bridges connecting the 2‘-O of ribose with the 4‘-C and forming a locked 3‘-endo conformation that reduces the conformational flexibility of the ribose. LNA oligonucleotides improve hybridization affinity for complementary sequences [Citation23,Citation24] and enhance reaction specificity. Our LNA probes were designed to recognize SNP sites (C/T) and specifically targeted the complementary sequences of the parental alleles. In ddPCR, the concentration of the amplified region in the sample is calculated from the proportions of positive and negative droplets and by Poisson statistics. For droplets not clearly defined as negative or positive, the threshold was manually determined based on control genomic DNA or cDNA. For Ube3a measurement in random primed cDNA, amplification by primers 05 F and RV1 in exons 8 and 9 created two groups of positive droplets with different fluorescence intensities ()). Droplets with low fluorescence intensity were also detected in genomic DNA samples. Thus, the 826-bp fragments would be incompletely amplified in cDNA ()). To determine the correct Ube3a concentration, the threshold in the cDNA samples was manually set to exclude the droplets which may have originated from Ube3a-ATS or preUbe3a. Allele-specific detection of transcripts is specified by the signal from the bound probe in a competing duplex reaction, in that, one primer pair binds the same region with two probes, which selectively hybridize each SNP. A competing duplex reaction is typically performed for quantifying localized variants in the abundant wild-type. In the present study, the ratios of variants were correctly detected in control samples with various abundances of variants ()). Although it is sometimes difficult to position thresholds to partition the double-positive droplets in a 2-D plot [Citation27], in this study, the thresholds were manually set in a 1-D plot to exclude false positive droplets based on the results of the control gDNA or cDNA, resulting in few double-positive droplets in the 2-D plot.
A limitation of the ddPCR assay for Ube3-ATS analysis is that it cannot distinguish preUbe3a from Ube3-ATS in random primed cDNA. Strand-specific cDNAs primed by sense- or antisense-strand primer can clearly differentiate them. However, these specific cDNAs cannot directly compare preUbe3a and Ube3a-ATS concentrations because of primer specificity in cDNA synthesis. Random primed cDNA was used for Ube3a-ATS analysis. Nevertheless, the transcripts originating from preUbe3a were inevitably contaminated in the concentrations detected for random primed cDNA. Contamination of maternal preUbe3a transcript in the concentrations of maternal Ube3a (mat preUbe3a/mat Ube3a) was 4–6% ()). Therefore, the concentration of pat preUbe3a is low. Concentration from the paternal allele detected by primers 05 F and IntR in random primed cDNA was considered to be the same as that of patUbe3a-ATS ()).
Allelic discrimination of transcripts with low expression and analysis of the skewness in allelic expression are difficult to perform by conventional densitometry, enzymatic digestions, or chromatographic sequencing of RT-PCR products. We recently used the ddPCR assay with LNA TaqMan probes for imprinting analysis of tissues with faint Ndn expression [Citation28]. Here, we applied it to analyse tissue- and cell-specific Ube3a imprinting. The ddPCR assay was reliable for the assessment of allele-specific Ube3a expression, especially in tissues with low Ube3a levels such as the spleen and the lung ()). We demonstrated that Ube3a was equally expressed by both parental alleles in adult tissues even in the lung with low expression [Citation29,Citation30]. Northern blot hybridization disclosed a strong bias (> 10:1) towards maternal expression in the brain [Citation13]. Ube3a was predominantly expressed by the maternal allele in the neuronal tissues. The paternal contributions to the total Ube3a transcripts were 9.7–20% (). The differences in paternal contribution may depend on the ratios of glial or unknown neuronal cells with biallelic Ube3a expression. Biallelic Ube3a expression was reported for bichiasmatic neurons and ganglia [Citation31,Citation32]. Ube3a-ATS is expressed only in neurons [Citation11]. Therefore, the relatively lower ratio of pat Ube3a-ATS to total Ube3a (patUbe3a-ATS/totalUbe3a) (43%) in the spinal cord may indicate comparatively lower neuron densities in the spinal cord or the presence of unidentified spinal neurons with biallelic Ube3a expression.
Several results of the analysis of strand-specific cDNAs were inconsistent with those of previous observations that Ube3a-ATS is expressed exclusively from the paternal allele. In rs48971506 in exon 8, ddPCR using primers 05 F and IntR detected a small amount of transcript from the maternal allele in 05 F-primed cDNA and biallelically equal expression in IntR-primed cDNA despite low paternal Ube3a expression (1/10 of the total) ()). In addition, the maternal transcripts were detected in forward primer-specific cDNAs of other SNPs in introns ()), whereas no transcripts were detected in the RT (–) samples. To resolve these inconsistencies, we used non-specific cDNAs in ddPCR. These were synthesized by RT without exogenous primers. In the non-specific cDNAs, ddPCR using the primers in introns detected copy numbers of the maternal transcript were close to those in the forward primer-primed cDNA (Supplementary Figure S3E and S4C), while copy numbers of the paternal transcript were over one-half of those in the reverse primer-primed cDNA (Supplementary Figure S3C- D and S4A-B). It indicates that ddPCR inevitably detects false positive transcripts from non-specific cDNA as noises in the strand-specific cDNA. In human RNAs, RT initiation by self-priming occurs without exogenous primers [Citation33]. Whenever strand-specific cDNAs are used to detect antisense transcripts, especially for intronless imprinted genes, the data must be compared with those obtained for non-specific cDNAs. As far as in our ddPCR assay, it is conceivable that Ube3a-ATS was not expressed from the maternal allele and most of preUbe3a was originated from the maternal allele.
The ddPCR analysis of the postnatal brain revealed that the paternal contributions to the total Ube3a transcripts (patUbe3a/totalUbe3a) markedly decreased during the neonatal period but the proportion of pat Ube3a-ATS in the total Ube3a (patUbe3a-ATS/totalUbe3a) gradually increased ()). The dramatic decrease in the patUbe3a/totalUbe3a ratio by P9 was consistent with immunohistochemical observations that neocortical neurons stop expressing detectable paternal Ube3a by P7 [Citation10]. Why does the patUbe3a-ATS/totalUbe3a ratio increase during postnatal brain development? The qPCR indicated that the total Ube3a expression at P0 was ~3× higher than it was at P26 whereas total Ube3a-ATS expression in P9–P26 was ~60–80% of that at P0 ()). The qPCR showed that the increase in Ube3a-ATS/Ube3a ratio depends mainly on drastic decreases in total Ube3a expression and a less severe decrease in Ube3a-ATS expression. Neural progenitor cells in the neonatal brain might express Ube3a-ATS which does not suffice to repress paternal Ube3a in a dose-dependent manner [Citation15], that is also suggested in neural cells derived from induced human pluripotent stem cells (iPSCs) [Citation34,Citation35]. Ube3a-ATS transcription was unexpectedly high and reached 60–70% of the total Ube3a expression in the adult brain. In a previous study, subcellular localization analysis of RNA extracted from primary neuronal cultures revealed that Ube3a has a cytoplasm-to-nucleus (c/n) ratio of 0.61 whereas that for Ube3a-ATS is 0.01–0.03. Thus, most of the Ube3a-ATS is localized in the nucleus [Citation15]. In addition, the half-life of Ube3a-ATS is much shorter (~4 h) than that of Ube3a (~48 h) [Citation15]. Therefore, Ube3a-ATS may be persistently and frequently transcribed from the paternal allele and much more abundant than Ube3a in mature neuron nucleus.
In the present study, we used ddPCR with LNA TaqMan probes to measure the absolute concentrations of allelic Ube3a sense- and antisense (Ube3a-ATS) transcripts at 3 loci and to evaluate the relationship between them during brain development and in various brain regions. The expression level of Ube3a- ATS was unexpectedly high despite its short half-life and nuclear localization. The ddPCR assay with LNA TaqMan probes is far more reliable and simpler than semiquantitative PCR for the measurement of skewed or faint allelic expression of imprinted genes. This method can also be applied to other experiments that require allelic discrimination. In the chromatin immunoprecipitation assay (ChIP), allelic precipitation ratio can be calculated in SNPs in 5ʹUTR.
Author contributions
M.M. and T.K. carried out cDNA synthesis, ddPCR and qPCR assays, and S.H. maintained the mouse strains. T.K designed the study, analysed results and drafted the manuscript. All authors reviewed the manuscript.
Supplemental Material
Download PDF (3.5 MB)Supplemental Material
Download PDF (58.8 KB)Acknowledgments
The authors thank Ayumi Nakagaki for technical support of ddPCR assay and Katsunori Hironaka (Bio- Rad Laboratories, Tokyo, Japan) for helpful suggestions regarding the ddPCR analysis and we would like to thank Editage (www.editage.com) for English language editing.
Disclosure statement
The authors declare that they have no competing interests.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu Rev Genet. 1988;22(1):127–146.
- Surani MA. Genomic imprinting: developmental significance and molecular mechanism. Curr Opin Genet Dev. 1991;1(2):241–246.
- Soellner L, Begemann M, Mackay DJ, et al. Recent advances in imprinting disorders. Clin Genet. 2017;91(1):3–13.
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83.
- Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17(1):12–13.
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17(1):14–15.
- Albrecht U, Sutcliffe JS, Cattanach BM, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17(1):75–78.
- Miura K, Kishino T, Li E, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9(2):149–159.
- Jiang YH, Armstrong D, Albrecht U, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811.
- Judson MC, Sosa-Pagan JO, Del Cid WA, et al. Allelic specificity of Ube3a expression in the mouse brain during postnatal development. J Comp Neurol. 2014;52(8):1874–1896.
- Yamasaki K, Joh K, Ohta T, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12(8):837–847.
- Landers M, Bancescu DL, Le Meur E, et al. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32(11):3480–3492.
- Landers M, Calciano MA, Colosi D, et al. Maternal disruption of Ube3a leads to increased expression of Ube3a-ATS in trans. Nucleic Acids Res. 2005;33(13):3976–3984.
- Chamberlain SJ, Lalande M. Angelman syndrome, a genomic imprinting disorder of the brain. J Neurosci. 2010;30(30):9958–9963.
- Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet. 2012;21(13):3001–3012.
- Meng L, Person RE, Huang W, et al. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013;9(12):e1004039.
- Huang HS, Allen JA, Mabb AM, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2011;481(7380):185–189. .
- Meng L, Ward AJ, Chun S, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412.
- Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73(3):316–322.
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610.
- Petersen M, Nielsen CB, Nielsen KE, et al. The conformations of locked nucleic acids (LNA). J Mol Recognit. 2000;13(1):44–53.
- Jensen GA, Singh SK, Kumar R, et al. A comparison of the solution structures of an LNA:DNA duplex and the unmodified DNA:DNA duplex. J Chem Soc Perkin Trans. 2001;2(7):1224–1232.
- Johnson MP, Haupt LM, Griffiths LR. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32(6):e55.
- Ugozzoli LA, Latorra D, Puckett R, et al. Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal Biochem. 2004;324(1):143–152.
- Frésard L, Leroux S, Servin B, et al. Transcriptome-wide investigation of genomic imprinting in chicken. Nucleic Acids Res. 2014;42(6):3768–3782. .
- Tafaj O, Hann S, Ayturk U, et al. Mice maintain predominantly maternal Gαs expression throughout life in brown fat tissue (BAT), but not other tissues. Bone. 2017;103:177–187.
- Whale AS, Huggett JF, Tzonev S. Fundamentals of multiplexing with digital PCR. Biomol Detect Quantif. 2016;10:15–23.
- Nakagaki A, Hirano S, Urakawa A, et al. Transgenic mice with a tandem duplication of the Necdin gene overexpress Necdin. Mamm Genome. 2018;29(9–10):680–689.
- Sutcliffe JS, Jiang YH, Galijaard RJ, et al. The E6-Ap ubiquitin-proteinLigase (UBE3A) gene is localized within a narrowed angelman syndrome critical region. Genome Res. 1997;7(4):368–377.
- Kishino T, Wagstaff J. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 1998;47(1):101–107.
- Jones KA, Han JE, DeBruyne JP, et al. Persistent neuronal Ube3a expression in the suprachiasmatic nucleus of Angelman syndrome model mice. Sci Rep. 2016;6(1):28238.
- McCoy ES, Taylor-Blake B, Aita M, et al. Enhanced nociception in angelman syndrome model mice. J Neurosci. 2017;37(42):10230–10239.
- Moison C, Arimondo PB, Guieysse-Peugeot AL. Commercial reverse transcriptase as source of false-positive strand-specific RNA detection in human cells. Biochimie. 2011;93(10):1731–1737.
- Chamberlain SJ, Chen PF, Ng KY, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader- Willi syndromes. Proc Natl Acad Sci U S A. 2010;107(41):17668–17673.
- Hsiao JS, Germain ND, Wilderman A, et al. A bipartite boundary element restricts UBE3A imprinting to mature neurons. Proc Natl Acad Sci U S A. 2019;116(6):2181–2186.
