ABSTRACT
Human immunodeficiency virus (HIV) infection and the psychostimulant drug cocaine are known to induce epigenetic changes in DNA methylation that are linked with the severity of viral replication and disease progression, which impair neuronal functions. Increasing evidence suggests that changes in DNA methylation and hydroxymethylation occur in mitochondrial DNA (mtDNA) and represent mitochondrial genome epigenetic modifications (mitoepigenetic modifications). These modifications likely regulate both mtDNA replication and gene expression. However, mtDNA methylation has not been studied extensively in the contexts of cocaine abuse and HIV-1 infection. In the present study, epigenetic factors changed the levels of the DNA methyltransferases (DNMTs) DNMT1, DNMT3a, and DNMT3b, the Ten-eleven translocation (TET) enzymes 1, 2, and 3, and mitochondrial DNMTs (mtDNMTs) both in vitro and in vivo. These changes resulted in alterations in mtDNA methylation levels at CpG and non-CpG sites in human primary astrocytes as measured using targeted next-generation bisulphite sequencing (TNGBS). Moreover, mitochondrial methylation levels in the MT-RNR1, MT-ND5, MT-ND1, D-loop and MT-CYB regions of mtDNA were lower in the HIV-1 Tat and cocaine treatment groups than in the control group. In summary, the present findings suggest that mitoepigenetic modification in the human brain causes the mitochondrial dysfunction that gives rise to neuro-AIDS.
Introduction
Epigenetic changes are known as heritable changes in gene expression without underlying changes in the encoding DNA sequence and are influenced by external environmental factors[Citation1]. Epigenetic changes include DNA methylation and histone modifications [Citation2]. DNA methylation is a covalent modification that occurs on cytosine (C) residues, mostly those located in CG dinucleotides (CpGs), by means of a reaction catalysed by DNA methyltransferases (DNMTs). DNMTs are responsible for maintaining methylation, in which a methyl group is transferred from S-adenyl methionine (SAM) to the fifth carbon of a C residue to form 5-methylcytosine (5mC) [Citation3]. However, the nervous system is a complex biological system in which cell differentiation into diverse lineages occurs as a result of tissue-specific methylation patterns [Citation4]. Moreover, ten eleven translocation (TET) enzymes oxidize 5mCs and promote locus-specific reversal of DNA methylation. An estimated ∼2–8% of the total cytosines in human genomic DNA are 5mCs, suggesting that epigenetic modification of the 5mC level may impact a broad range of biological functions including neuronal differentiation, neural plasticity, and brain functions [Citation5].
Human immunodeficiency virus (HIV) is known for its high evolutionary potential, mutation rate and rapid turnover upon infection in a human host [Citation6]. HIV targets immune cells, and the viral genome assumes control over the cellular machinery and ultimately affects the central nervous system (CNS) [Citation7]. HIV infection alters redox changes, mitochondrial biogenesis and epigenetic modifications, including DNA methylation, which regulates gene expression cascades in humans [Citation8–10]. Trans-activator of transcription protein (Tat), which has a length of 86–102 amino acids (aa), is a viral protein secreted from HIV-infected cells and is known to control HIV transcription [Citation11]. HIV-1 Tat is known to disrupt cellular homoeostasis and induce oxidative stress that leads to reactive oxygen species (ROS) accumulation, which can further alter CNS functions [Citation12,Citation13]. Studies have shown that HIV-1 Tat activates epigenetic modification in the animal brain [Citation14] as well as mouse primary microglia [Citation15]. HIV-1 Tat affects energy deficits in astrocytes and impairs mitochondrial functions, leading to cell death [Citation16]. Mouse model studies have shown that exposure to HIV-1 Tat not only affects the membrane potential [Citation17] but also alters mitochondrial functions [Citation18]. Moreover, HIV-1 Tat affects mitochondrial dynamics by disrupting the functions of fission and fusion proteins, ultimately causing damage to mitochondrial DNA (mtDNA) and proteins [Citation19,Citation20]. HIV-1 Tat exposure-mediated mtDNA damage induces genetic changes in immune cells [Citation21,Citation22]. Additionally, studies have documented that mtDNA mutations are associated with various CNS-related disorders, including ataxia, seizures and dementia [Citation23]. These effects are commonly associated with biochemical defects in oxidative phosphorylation (OXPHOS) related to seven subunits of complex I (NADH dehydrogenases 1–6 [ND1–6] and ND4L), three subunits of complex IV (cytochrome c oxidase subunits I–III [COX1–3]), two subunits of complex V (ATPase 6 and ATPase 8), and one subunit of complex III (cytochrome b) [Citation24].
The psychostimulant drug cocaine has been shown to affect the CNS in a number of manners, such as by significantly impairing cellular functions, in both in vitro and in vivo studies [Citation25,Citation26]. Clinical studies have also demonstrated that cocaine increases oxidative stress, inflammation and mitochondrial biogenesis, which can cause brain damage [Citation27,Citation28]. Alterations in mitochondrial function are known to impact fission, fusion and oxidation, which are mediated by mtDNA [Citation29,Citation30]. Recent in vitro, in vivo and clinical studies have shown that cocaine alters several cellular functions, including mitochondrial biogenesis and epigenetic modification of DNA methylation [Citation31,Citation32]. Notably, these activities are accelerated by the presence of HIV infection [Citation9]. Moreover, studies have shown that cocaine abuse and addiction can compromise judgement and decision-making power, which may increase the risk of HIV infection [Citation33]. Furthermore, cocaine abuse induces oxidative stress and ROS production, which may affect mtDNA and thus impact mitochondrial functions, including oxidative phosphorylation (OXPHOS).
Extensive studies have consistently demonstrated that cocaine abuse and HIV infections lead to neuronal dysfunction and are involved in HIV-associated neurocognitive disorder (HAND) [Citation16,Citation34]. HIV-positive cocaine users have shown synergistic potentiation of viral replication and disease progression compared with HIV-positive subjects who do not use cocaine or HIV-negative cocaine users [Citation35,Citation36]. Studies have shown that HIV infection potentiates alterations in DNA methylation machinery, particularly induction of DNMT1 expression [Citation37]. In addition, cocaine exposure elevates DNA methylation, DNMT3a and DNMT3b levels and MeCP2 binding, which decreases gene expression patterns in the nucleus accumbens [Citation38]. A study in which rats were treated with cocaine revealed that inhibition of DNMTs decreases DNA hypermethylation and attenuates the drug-induced downregulation of gene expression in the nucleus accumbens [Citation39]. However, the mechanisms underlying the link between HIV-1 Tat and cocaine-mediated mtDNA methylation-induced neurodegeneration are not clearly understood.
In the present study, we investigated whether HIV-1 Tat and cocaine, either alone or in combination, induce modification of mtDNA methylation by altering DNMTs and mitochondrial DNMT (mtDNMT) activity. We also performed expression studies. We found that HIV-1 Tat and cocaine exposure reduced mtDNA methylation and altered the expression of DNMTs, mtDNMTs and TET proteins. We also analysed the methylation patterns in the genomic regions of the mtDNA D-loop and ND1–ND6 via targeted next-generation bisulphite sequencing (TNGBS) in human primary astrocytes exposed to HIV-1 Tat and cocaine.
Materials and methods
Cell culture and reagents
Cell culture reagents were purchased from ScienCell (Carlsbad, CA, USA). DNMT1, DNMT3a, DNMT3b, TET1, TET2, and TET3 antibodies; goat anti-rabbit IgG and goat anti-mouse IgG antibodies; and kits for analysis of global DNA methylation of 5mC were purchased from Epigentek Group Inc. (Farmingdale, NY, USA). Mitochondrial isolation kits were purchased from Abcam (CA). Electrophoresis reagents were purchased from Bio-Rad (Richmond, CA, USA), and nitrocellulose membranes were purchased from Amersham Scientific (Piscataway, NJ).
HIV-1 Tat proteins
HIV-1 Tat (Catalogue number #2222) was obtained from the NIH AIDS Research and Reference Reagent Program. The recombinant Tat proteins were tested and revealed to be of >95% purity.
Chemicals and reagents
All chemicals and reagents were obtained from Sigma-Aldrich (St. Louis, MO). Doxycycline (Dox) was dissolved in 0.9% saline before injection.
Human primary astrocytes
In this study, we used commercially obtained human primary astrocytes isolated from normal human brain cerebellum tissue. The astrocytes were maintained in astrocyte basal medium supplemented with foetal bovine serum to a final concentration of 10% and 1% antibiotic/antimycotic solution (ScienCell, Carlsbad, CA, USA).
Animals and housing
Adult male Tat-inducible transgenic mice (iTat, formerly known as GT-tg bigenic mice) [Citation40] were bred from a colony started by progenitors generously donated by Dr. Johnny He. All mice (8–10 weeks of age) were maintained at the University of Florida’s animal facilities and used in experiments according to protocols approved by the Institutional Animal Care and Use Committee, University of Florida, Gainesville, Florida. Procedures for creating HIV-1 iTat mice and confirming the genotype of the inducible and brain-targeted HIV-1 Tat protein are described in detail elsewhere [Citation40,Citation41].
Drug treatment
Brain-targeted Tat was induced with Dox treatment with or without cocaine exposure. To induce expression of HIV-1 Tat [1-86], iTat mice were administered Dox via intraperitoneal injection of a single daily dose of 100 mg/kg dissolved in 0.9% saline in a volume of 0.3 ml/30 g body weight for 14 days (iTat-Dox; n = 3). iTat mice treated with saline served as controls (n = 3), as indicated and characterized previously [Citation41]. An additional set of mice were treated with cocaine administered subcutaneously at a dose of 10 mg/kg/d in 0.9% saline for 14 days (n = 3). Finally, a final set of iTat mice (n = 3) were treated with Dox (100 mg/kg/d, i.p.) followed by cocaine (10 mg/kg/d, s.c.) 2 min later.
Isolation of brain specimens for gene analysis, immunoblotting or quantitative image analysis
After the 14-day treatment period, mice were anesthetized with isoflurane (4%) before being euthanized and then subjected to transcardial perfusion with cold saline (0.9%), and tissues were harvested. The frontal lobe region of the brain was extracted from three euthanized mice for each treatment set, flash-frozen in liquid nitrogen, and stored at −80°C for further analysis of gene and protein modifications, as detailed below.
Human primary astrocyte treatment
Briefly, astrocytes (1 x 106) were separately treated with HIV-1 Tat (50 ng/ml) and cocaine (0.5 µM) for 24 hr. At the end of the incubation period, the cells were digested with 0.05% trypsin-EDTA and washed with PBS. The cells were then pelleted by centrifugation (1000 × g for 10 min). The total and mitochondrial fractions were then resuspended in lysis buffer for further experimental procedures.
Mitochondrial isolation
A mitochondrial isolation kit (Abcam, CA) was used to isolate mitochondria from astrocytes. For the in vitro model, human primary astrocytes (3 × 10Citation8) were grown and treated with HIV-1 Tat (50 ng/ml) and cocaine (0.5 µM) for 24 hr. At the end of the incubation, the cells were lysed with 0.05% trypsin-EDTA and washed with PBS. The lysed cells were centrifuged (1000 × g for 10 min) at room temperature, and the pellets formed were resuspended in mitochondrial assay buffer. The cells were incubated in the solution on ice for 2 min and homogenized with a glass homogenizer using 30 up-and-down strokes, and cellular disruption was confirmed by microscopy. For the in vivo experiment, samples of brain tissues (frontal lobe) were collected from mice subjected to different treatments: saline, cocaine, Dox to induce HIV-1 iTat expression and cocaine + iTat induction with Dox (as detailed in ‘Drug treatment’ and ‘Isolation’ above). Fifty milligrams of brain tissue was homogenized, and then the extract was centrifuged at 800 x g for 10 min. The pellet formed was the nuclear fraction and was discarded. The supernatant was collected and centrifuged again at 12,000 × g for 15 min at 4°C. The pellet formed was the mitochondrial fraction, which was then resuspended in lysis buffer for further analyses, including western blot analysis.
Western blot analysis
Whole-cell protein lysates and mitochondrial fractions were extracted from HIV-1 Tat- and cocaine-exposed primary astrocytes as well as from the brains of cocaine- and/or Tat-exposed iTat mice using ice-cold RIPA buffer supplemented with protease inhibitors. Equal amounts of primary astrocyte proteins and mouse brain tissue total lysate, cytoplasmic fraction and mitochondrial fraction proteins were resolved by 4–15% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated with the appropriate primary antibodies. Primary antibodies against DNMT3a (A-1003-050, 1:500) and DNMT3b (A-1004-050, 1:500) were obtained from Abcam. Monoclonal antibodies against DNMT1 (A-1001-050, 1:1000), TET1 (A-1020-050, 1:1000), TET2 (A-1701-050, 1:1000), TET3 (A50520-020, 1:1000) and β-actin (A-0800-100, 1:1000) were obtained from Epigentek (Brooklyn, NY, USA). The immunoreactive bands were visualized using a chemiluminescence western blotting system according to the manufacturer’s instructions (Bio-Rad). The relative protein levels were quantified with ImageJ software. The relative intensity was calculated for each DNMT band with normalization to the relative intensity of actin in each sample.
DNMT activity assay
Cultured human primary astrocytes were harvested after 24 hr of treatment with cocaine, HIV-1 Tat, or both. Nuclear extracts were prepared with nuclear extraction reagent (Pierce, Rockford, IL). Overall DNMT activity was determined using an EpiQuik DNA Methyltransferase Activity Assay Kit (Epigentek) according to the manufacturer’s protocol. This analysis provided the levels of overall DNMT activity and was not specific to any one DNMT. The data are presented as units calculated according to the manufacturer’s manual.
Enzyme-linked immunosorbent assay (ELISA) of DNA 5mC content
The 5mC and 5hmC levels of DNA were measured with a MethylFlash Methylated DNA 5-mC Quantification Kit and a MethylFlash Hydroxy methylated DNA 5-hmC Quantification Kit (Colorimetric) (Epigentek, Brooklyn, NY, USA), respectively. Briefly, 100 ng of the appropriate DNA was bound to a 96-well plate. The methylated fraction of DNA was detected using the corresponding capture and detection antibodies and quantified colorimetrically by reading the absorbance at 450 nm in a microplate spectrophotometer (Bio-Rad, Model 550, Hercules, CA, USA). The results are expressed in units that were calculated according to the manufacturer’s manual.
DNA bisulphite conversion
A total of 250–500 ng of extracted DNA was bisulphite-modified using a Zymo EZ-96 DNA Methylation™ Kit (Zymo Research, CA) according to the manufacturer’s protocol with minor modifications. The bisulphite-modified DNA samples were eluted using M-elution buffer in a volume of 46 µl.
Pyrosequencing methylation analysis
A Line-1 Pyrosequencing methylation assay was used for quality control (QC) of the bisulphite-modified DNA samples. ADS2888-FS1re (D-loop), ADS2886-FS2 (CYB), ADS9500-FS1 (RNR1), and ADS9515-FS2 pyrosequencing assays were performed on mtDNA and cell line DNA. Pyrosequencing was performed using a PSQ 96HS system or a PSQ 96HSA system. The systems are equivalent to the PyroMark MD system. Briefly, the PCR product was bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences), after which the immobilized PCR products were purified, washed, denatured with a 0.2 µM NaOH solution, and rewashed using Pyrosequencing Vacuum Prep Tool (Qiagen) according to the manufacturer’s protocol. The sequencing and data analysis were performed using Q96 software.
Multiplex PCR, library preparation and sequencing for TNGBS
All bisulphite-modified DNA samples were amplified using separate multiplex or simplex PCRs, as listed in Supplementary Table 1. The assay designed using Assay Design Service (ADS) software for genes or regions is described in Supplementary Table 2. The PCRs included 0.5 units of Qiagen HotStarTaq, 0.2 µM primers, and 3 µl of bisulphite-treated DNA in a 20 µl reaction. All PCR products were verified and quantified using the QIAxcel Advanced System. Prior to library preparation, PCR products from the same sample were pooled and purified using QIAquick PCR Purification Kit columns (Qiagen). Libraries were prepared using a KAPA Library Preparation Kit for Ion Torrent Platforms (cat# KK8310) and Ion Xpress™ Barcode Adapters (Thermo Fisher). Next, the libraries were purified using Agencourt AMPure XP beads (Beckman Coulter) and quantified using the Qiagen QIAxcel Advanced System. The barcoded samples were pooled in an equimolar fashion before template preparation, and enrichment was performed on an Ion Chef™ system (Thermo Fisher) using Ion 520™ & Ion 530™ Chef reagents. Next, the enriched, template-positive libraries were sequenced on an Ion S5™ sequencer using Ion 530™ sequencing chips (Thermo Fisher).
Bioinformatics – data filtering and alignment
For all TNGBS runs, FASTQ files were downloaded from the Ion Torrent S5 server. All reads were then further assessed through FastQC v0.72 for standard QC. The reads were trimmed with Trim Galore version 0.6.3 with a minimum quality cut-off of 20 bp. Following adapter trimming, all reads with scores below a QC threshold of 20 were discarded. Since we used a Pico Methyl-Seq Kit, we trimmed off at least the first 10 bp from both the 5ʹ and 3ʹ ends of each read to remove biases seen in the base composition and M-bias plots. The FASTQ files from the Ion Torrent S5 server were aligned to the NC_012920.1 human mitochondrial genome using open-source Bismark Bisulfite Read Mapper software with the Bowtie2 alignment algorithm. The methylation levels were calculated in Bismark by dividing the number of methylated reads by the total number of reads [Citation42]. As per the Bismark reference guide, we used directional libraries to obtain the two different bisulphite strands (OT, OB) present in the library. We also used the Bismark Deduplicate tool to remove alignments to the same position in the genome, which can arise by excessive PCR amplification, from the Bismark mapping output for single-end SAM files. Bismark Pretty Report was used to generate a graphical HTML report page from the report outputs of Bismark deduplicate and the Bismark mapping and Bismark methylation extractor tools. A summary of all data reports generated using the Bismark Pretty Report is shown in Supplementary Table 3.
Methylation calculations
The methylation levels were calculated in Bismark by dividing the number of methylated reads by the total number of reads. The following formulas were used to determine the percentage of methylation across the mitochondrial genome for TNGBS data:
% methylation (context) = 100 * methylated Cs (context)/(methylated Cs (context) + unmethylated Cs (context)).
For pyrosequencing data analysis, the percentage of methylation was calculated as % methylation = methylated C (C)/C + unmethylated C (T).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6. In order to analyse main effect, differences between control and treatment groups, including in vitro HIV-1 Tat, cocaine and HIV-1 Tat combined with cocaine and in vivo HIV-1 iTat, cocaine and HIV-1 iTat combined with cocaine, were calculated using two-way ANOVAs followed by Tukey’s post hoc tests. In order to analyse interaction effect between HIV-1 Tat and cocaine treatments, the effect of HIV-1 Tat alone and cocaine alone treatments were compared with combined treatment of cocaine with HIV-1 Tat were calculated using two-way ANOVAs followed by Tukey’s post hoc tests. In the mitochondrial methylation data analysis, differentially methylated regions and CpG sites were analysed by performing a Student’s t-test of each CpG site and each amplicon. The values are expressed as the means ± standard error, and a significance level of p < 0.05 was used.
Results
HIV infection and cocaine abuse are known to cause epigenome-wide differential DNA methylation [Citation8,Citation38]. Epigenetic factors can target the mitochondrial genome and alter mtDNA methylation patterns, leading to disruption of neuronal function [Citation43]. Multiple DNMTs have previously been shown to be present in mitochondria at different levels and to play roles in regulating methylation patterns at mitochondrial CpG sites [Citation44–46]. Recently, a study reported that the non-CpG sites of mtDNA are extensively and predominantly methylated. In that study, methylation quantification was carried out using whole-genome bisulphite sequencing (WGBS) at single-base pair resolution [Citation47]. However, HIV-1 Tat- and cocaine-induced epigenetic changes in mtDNA have not yet been studied from any perspective relevant to the CNS. Therefore, in the present study, we studied whether HIV-1 Tat and cocaine affect TET, DNMT and mtDNMT levels and thus potentially alter mitochondrial genome methylation patterns in astrocytes via similar or distinct mechanisms.
HIV-1 Tat and cocaine exposure impact DNMT and TET protein levels in human primary astrocytes
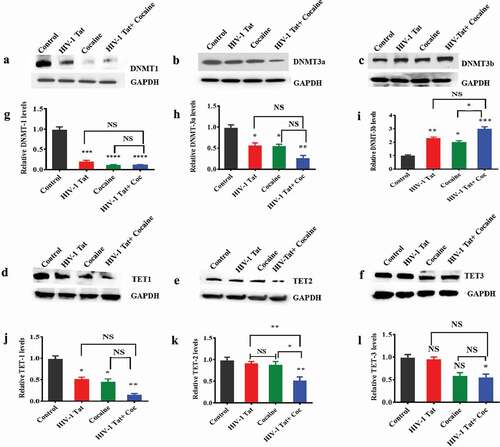
DNMTs play a major role in the methylation of DNA in humans [Citation48]. To understand the effects of HIV-1 Tat and cocaine exposure on DNMT protein levels in vitro, human primary astrocytes were treated with cocaine, HIV-1 Tat, or HIV-1 Tat combined with cocaine. The control group was treated with medium only (without drug exposure). Our findings demonstrated that HIV-1 Tat alone exposure (F (3, 6) = 60.68, P = 0.0002), cocaine alone (F (3, 6) = 60.68, P = 0.0001) and HIV-1 Tat and cocaine coexposure (F (3, 6) = 60.68, P = 0.0001) significantly decreased the DNMT1 protein level compared to control treatment, as shown in ). Cocaine exposure significantly (F (3, 6) = 60.68, P = 0.0001) downregulated DNMT1 protein to a negligible level ()). Moreover, we also compared the effects of cocaine and HIV-1 Tat coexposure and cocaine or HIV-1 Tat alone on DNMT1 protein levels, and the results were non-significant, as shown in ). Similarly, we investigated the effects of cocaine, HIV-1 Tat and HIV-1 Tat combined with cocaine on DNMT3a in human primary astrocytes. We observed that the DNMT3a level was significantly reduced with exposure to HIV-1 Tat (F (3, 6) = 12.34, P = 0.0475) and cocaine (F (3, 6) = 12.34, P = 0.0405) ()). Moreover, HIV-1 Tat and cocaine coexposure significantly (F (3, 6) = 12.34, P = 0.0038) decreased the DNMT3a level, as shown in ). The effects of HIV-1 Tat exposure alone and cocaine exposure alone on DNMT3a levels were not statistically different from those of HIV-1 Tat and cocaine coexposure, as shown in ). Furthermore, the DNMT3b level was significantly increased (F (3, 6) = 27.88, P = 0.0045) with HIV-1 Tat and cocaine treatment (F (3, 6) = 27.88, P = 0.0161) ()). HIV-1 Tat and cocaine coexposure (F (3, 6) = 27.88, P = 0.0004) accelerated these effects on DNMT3b compared to control treatment ()). We compared the effects of cocaine and HIV-1 Tat coexposure and cocaine or HIV-1 Tat alone on DNMT3b protein levels, and the results were statistically significant for cocaine (F (3, 6) = 27.88, P = 0.0161) interaction as shown in ). DNMT level changes have been found to be epigenetically linked with TET enzymes, which oxidize 5mCs and promote locus-specific DNA demethylation [Citation49]. Therefore, we examined the effects of treatment on TET protein levels. TET protein showed an interesting pattern with exposure to cocaine, HIV-1 Tat and cocaine with HIV-1 Tat. When primary astrocytes were exposed to cocaine or HIV-1 Tat, the TET1 protein level was significantly downregulated (F (3, 6) = 19.08, P = 0.0121 and F (3, 6) = 19.08, P = 0.0224 respectively). Cocaine and HIV-1 Tat coexposure also significantly downregulated (F (3, 6) = 19.08, P = 0.0012) the TET1 protein level compared to control treatment, as shown in ). The effects of HIV-1 Tat and cocaine coexposure on TET1 protein levels showed no statistically significant difference compared to those of cocaine alone and HIV-1 Tat alone exposure in astrocytes as shown in ). Human primary astrocytes showed no significant changes in TET2 protein levels when exposed to cocaine or HIV-1 Tat, while HIV-1 Tat and cocaine coexposure significantly downregulated (F (3, 6) = 15.32, P = 0.0034) TET2 protein levels compared with control treatment, as shown in ). The effects of HIV-1 Tat and cocaine coexposure on TET2 protein levels showed statistically significant difference compared to those of cocaine exposure alone (F (3, 6) = 15.32, P = 0.0114) as well as the effects of HIV-1 Tat exposure alone showed a significant difference (F (3, 6) = 15.32, P = 0.0075) ()). Moreover, we observed that the TET3 protein level did not change when astrocytes were exposed to HIV-1 Tat and cocaine alone and HIV-1 Tat and cocaine coexposure significantly (F (3, 6) = 7.342, P = 0.0435) downregulated TET3 protein levels compared with control treatment ()). Moreover, the effects of HIV-1 Tat and cocaine coexposure on TET3 protein levels showed no statistically significant difference compared to those of cocaine and HIV-1 Tat exposure alone as shown in ). ) show the densitometric values for DNMT1, DNMT3a, DNMT3b, TET1, TET2 and TET3 protein levels (fold change compared to the control), respectively. The data are expressed as the mean of the control level ± SE for 3 independent experiments.
Figure 1. Effects of HIV-1 Tat and cocaine on DNMT and TET protein levels in human primary astrocytes. Human primary astrocytes (1x106 cells/ml) were treated with HIV-1 Tat (100 ng), cocaine (0.5 µM) or a combination of HIV-1 Tat and cocaine for 24 hr. Total cell lysates were resolved by SDS-PAGE and analysed by western blotting for DNMT1 (a), DNMT3a (b), DNMT3b (c), TET-1 (d), TET-2 (e), TET-3 (f) and total protein GAPDH. (g), (h), (i), (j), (k) and (l) show the densitometric values for DNMT1, DNMT3a, DNMT3b, TET1, TET2 and TET3 protein levels (fold change compared to the control), respectively. The data are expressed as the mean ± SE of three independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, NS – Nonsignificant
HIV-1 Tat and cocaine impact DNMT enzyme activity and 5mC levels in human primary astrocytes
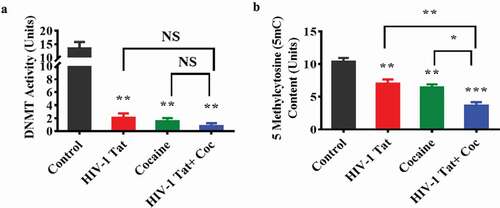
To further explore the changes in DNMT and TET levels, we investigated the effects of HIV-1 Tat and cocaine on DNMT enzyme activity and global DNA 5mC and 5hmC levels in primary astrocytes. ) shows the effects of HIV-Tat, cocaine, and the combination of HIV-1 Tat with cocaine on total DNMT enzyme activity. The results confirmed that overall DNMT enzyme activity significantly decreased with HIV-1 Tat (F (3, 6) = 26.63, P = 0.0019), cocaine (F (3, 6) = 26.63, P = 0.0015) and combined cocaine and HIV-1 Tat exposure (F (3, 6) = 26.63, P = 0.0010) compared to the control. The effects of HIV-1 Tat and cocaine coexposure on DNMT enzyme activity showed no statistically significant difference compared to those of cocaine treatment alone and HIV-1 Tat treatment alone, as shown in ). To further strengthen this study, we also analysed whether HIV-1 Tat and cocaine exposure affect 5mC and 5hmC content in primary astrocytes. We observed similar results as those found for DNMT enzyme activity, namely, significant decreases in overall 5mC levels in the treatment groups (cocaine F (3, 6) = 35.84, P = 0.0040, HIV-1 Tat F (3, 6) = 35.84, P = 0.0088 and HIV-1 Tat with cocaine F (3, 6) = 35.84, P = 0.0002) compared to the control group, as shown in ). Moreover, the effects of HIV-1 Tat and cocaine coexposure on the overall 5mC content showed statistically significant difference compared to those of cocaine exposure alone (F (3, 6) = 35.84, P = 0.0188), while HIV-1 Tat exposure alone showed a significant difference (F (3, 6) = 35.84, P = 0.0080), as shown in ). Interestingly, 5hmC was not detectable in the HIV-1 Tat and cocaine exposure group.
Figure 2. HIV-1 Tat and cocaine affect global DNA methylation (5mC) content and DNMT enzyme activity in human primary astrocytes. Total cellular DNA and nuclear extracts were obtained from human primary astrocytes treated with HIV-Tat alone, cocaine alone, or HIV-1 Tat and cocaine for 24 hr. DNA was isolated, and equal amounts were used to determine the overall DNMT activity (a) and global DNA methylation (5mC) by ELISA (b). The data are expressed as fold changes compared to the control level ± SE for 3 independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, NS – Nonsignificant
HIV-1 Tat and cocaine impact DNMT and TET protein levels in a transgenic mouse model
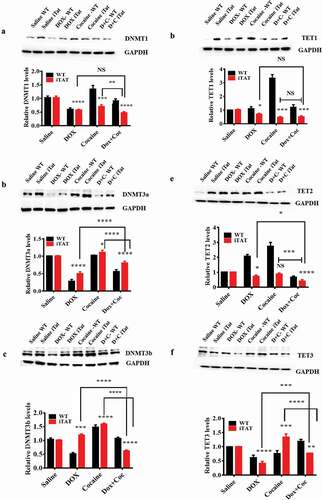
Our in vitro study revealed that HIV-Tat and cocaine exposure significantly changed DNMT and TET protein levels. Furthermore, we performed a validation study and found that cocaine- and Dox-exposed [inducible HIV-1 Tat (iTat)] mouse brains demonstrated alterations in the levels of DNMT and TET proteins. Specifically, we found that DNMT1 levels were significantly decreased in mice expressing Tat protein when treated with cocaine (F (3, 16) = 64.16, P = 0.0001), HIV-iTat (Dox) (F (3, 16) = 64.16, P = 0.0001) or both (F (3, 16) = 64.16, P = 0.0001) ()). Moreover, we also compared the effects of cocaine and HIV-1 iTat coexposure and cocaine or HIV-1 iTat exposure alone on DNMT1 protein levels. We observed that the comparisons of effects on DNMT1 levels between HIV-1 iTat and cocaine coexposure and HIV-1 iTat exposure alone did not reveal significant differences, while cocaine exposure alone produced a significant difference (F (3, 16) = 64.16, P = 0.0020), as shown in ). Furthermore, we studied the effects cocaine, HIV-1 iTat and HIV-1 iTat with cocaine on DNMT3a protein levels in iTat mice and observed that HIV-1 iTat (F (3, 16) = 309.1, P = 0.0001), cocaine (F (3, 16) = 309.1, P = 0.0345) and cocaine with HIV-1 iTat (F (3, 16) = 309.1, P = 0.0001) caused significant changes in DNMT3a protein levels compared to control treatment. We compared the effects of HIV-1 iTat with cocaine and HIV-1 iTat alone or cocaine alone on DNMT3a levels and observed significant differences (F (3, 16) = 309.1, P = 0.0001), as shown in ). We also observed that cocaine (F (3, 16) = 287.8, P = 0.0001), HIV-1 iTat (F (3, 16) = 287.8, P = 0.0006) and HIV-1 iTat with cocaine (F (3, 16) = 287.8, P = 0.0001) showed significant effects on protein level changes in DNMT3b in iTat mice compared to control treatment. We observed that the effects of HIV-1 iTat with cocaine and HIV-1 iTat alone or cocaine alone on DNMT3b levels were significantly different (F (3, 16) = 287.8, P = 0.0001), as shown in ). Moreover, we observed that TET1 levels were decreased in mice expressing HIV-1 iTat (F (3, 16) = 78.95, P = 0.0275) exposed to cocaine (F (3, 16) = 78.95, P = 0.0003) or to a combination of cocaine and HIV-1 iTat protein (F (3, 16) = 78.95, P = 0.0007), as shown in ). We also observed that cocaine (NS), HIV-1 iTat (F (3, 16) = 125.1, P = 0.0238) and HIV-1 iTat with cocaine (F (3, 16) = 125.1, P = 0.0001) showed effects on protein level changes in TET2 in iTat mice compared to control treatment, as shown in ). We observed that the effects of HIV-1 iTat with cocaine and HIV-1 iTat alone (F (3, 16) = 125.1, P = 0.0126) or cocaine alone (F (3, 16) = 125.1, P = 0.0005) on TET2 levels were significantly different as shown in ). Cocaine (F (3, 16) = 61.62, P = 0.0002), HIV-1 iTat (F (3, 16) = 61.62, P = 0.0001) and HIV-1 iTat with cocaine (F (3, 16) = 61.62, P = 0.0069) showed significant effects on protein level changes in TET3 in iTat mice compared to control treatment, as shown in ). We observed that the effects of HIV-1 iTat with cocaine and HIV-1 iTat alone (F (3, 16) = 61.62, P = 0.0001) or cocaine alone (F (3, 16) = 61.62, P = 0.0001) on TET3 levels were significantly different as shown in ) show the densitometric values for DNMT1, DNMT3a, DNMT3b, TET1, TET2 and TET3 protein levels (fold change compared to the control), respectively. The data are expressed as the mean of the control level ± SE for 3 independent experiments.
Figure 3. Effects of HIV-1 Tat and cocaine on DNMTs and TET proteins. HIV-1 inducible Tat transgenic (iTat) or saline-treated (control) mice received daily intraperitoneal injections of saline or Dox (100 mg/kg/d) over 14 days either with or without s.c. cocaine (10 mg/kg/d). Following treatment, equal amounts of protein lysates from harvested brains were resolved, and we analysed protein expression using western blot analysis. The results show the levels of DNMT1 (a), DNMT3a (b), DNMT3b (c), TET1 (d), TET2 (e) and TET3 (f) in HIV-Tat (GT-tg) mouse brains and the levels of total GAPDH protein. (g), (h), (i), (j), (k) and (l) show the densitometric values for DNMT1, DNMT3a, DNMT3b, TET1, TET2 and TET3 protein levels (fold change compared to the control), respectively. The data are expressed as the mean ± SE of three independent experiments. The data represent 3 independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, NS – Nonsignificant
HIV-1 Tat and cocaine impact mitochondrial DNMT protein levels in human primary astrocytes and in an HIV-1 iTat mouse model
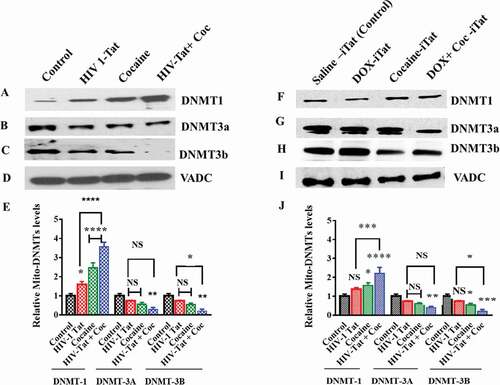
Furthermore, we studied the effects of HIV-1 Tat and cocaine exposure on mtDNMTs in both human primary astrocytes and a iTat mouse model. We isolated mitochondria from human primary astrocytes after 24 hr of exposure to cocaine, HIV-1 Tat, or both HIV-1 Tat and cocaine. Similarly, we isolated mitochondria from the brains of HIV-iTat mice with and without 14 days of exposure to cocaine. Furthermore, we resolved the mitochondrial fractions by SDS-PAGE and analysed them by western blot analysis for DNMT1, DNMT3a and DNMT3b levels both in vitro and in vivo, as shown in . We observed that the DNMT1 levels in the mitochondrial extracts of human primary astrocytes were significantly higher in the HIV-1 Tat (F (2, 24) = 165.7, P = 0.0133), cocaine (F (2, 24) = 165.7, P = 0.0001) and combined HIV-1 Tat/cocaine (F (2, 24) = 165.7, P = 0.0001) exposure groups than in the control group ()). Moreover, we also check the differences between HIV-1 Tat alone or cocaine alone exposure and coexposure of cocaine with HIV-1 Tat. The results revealed that both cocaine alone and HIV-1 Tat alone (F (2, 24) = 165.7, P = 0.0001) exposure were significantly different than coexposure of cocaine with HIV-1 Tat. On the other hand, we observed that DNMT3a and DNMT3b levels were not significantly changed due to exposure to HIV-1 Tat and cocaine. The HIV-1 Tat and cocaine coexposure group showed significant decreases in DNMT3a (F (2, 24) = 165.7, P = 0.0031) and DNMT3b (F (2, 24) = 165.7, P = 0.0011) compared to the control group, as shown in ), respectively. ) shows the results for total VDAC protein as a loading control. Additionally, we observed similar results in HIV-iTat mouse brain mitochondrial extracts: DNMT1 levels ()) were significantly higher with cocaine exposure (F (2, 24) = 63.04, P = 0.0129) and coexposure with HIV-1 Tat and cocaine (F (2, 24) = 63.04, P = 0.0001) compared to those in the control group. DNMT3a levels were not significantly different ()) in HIV-iTat- and cocaine-exposed mice. However, HIV-iTat mice with cocaine exposure showed a significant (F (2, 24) = 63.04, P = 0.0083) reduction in DNMT3a levels compared to saline-treated mice. Furthermore, we also measured DNMT3b levels in the in vivo samples. We observed that DNMT3b levels in iTat mouse brain mitochondrial extracts were significantly lower in the cocaine (F (2, 24) = 63.04, P = 0.0443) and coexposure with HIV-1 Tat and cocaine (F (2, 24) = 63.04, P = 0.0006) treatment groups than in the control group, as shown in ). ) shows the results for total VDAC protein as a loading control. ) shows the densitometric values for DNMT1, DNMT3a, and DNMT3b protein levels collected in vitro from human primary astrocyte mitochondrial fractions (fold change compared to the control), whereas ) shows the densitometric values for DNMT1, DNMT3a, and DNMT3b protein levels from in vivo mouse brain mitochondrial fractions (fold change compared to the control). The data are representative of 3 independent experiments.
Figure 4. Effects of HIV-1 Tat and cocaine on in vitro and in vivo mtDNMTs. For the in vitro experiment, human primary astrocytes (3 × 108 cells/ml) were treated with HIV-Tat (100 ng) or cocaine (0.5 µM) for 24 hr. For the in vivo experiment, HIV-1 inducible Tat transgenic (iTat) or saline-treated (control) mice received daily intraperitoneal injections of saline or Dox (100 mg/kg/d) over 14 days either with or without s.c. cocaine (10 mg/kg/d). At the end of the experiment, mitochondria were isolated from human primary astrocytes and HIV-1 iTat mice, and equal amounts of mitochondrial fraction were resolved by SDS-PAGE and analysed by western blot analysis. DNMT1 (a), DNMT3a (b) and DNMT3b (c) levels in human primary astrocytes; DNMT1 (f), DNMT3a (g) and DNMT3b (h) levels in HIV-iTat mouse brains; and total protein VDAC levels were analysed in vitro (d) and in vivo (i). (e) shows the densitometric values for DNMT1, DNMT3a, and DNMT3b protein levels (fold change compared to the control) in human primary astrocyte mitochondrial fractions in vitro, and (j) shows the densitometric values for DNMT1, DNMT3a, and DNMT3b protein levels (fold change compared to the control) in mouse brain mitochondrial fractions in vivo. The data represent 3 independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, NS – Nonsignificant
HIV-1 Tat and cocaine impact human primary astrocyte mtDNA methylation
DNA methylation is a significant component of epigenetic modification. Studies have recently reported that mtDNA methylation plays an essential role in mitochondrial gene regulation [Citation50]. In human cells, methylation has been observed at CpG, CHG and CHH sites (where H is A, C or T). The mtDNA genome is a double-stranded, negatively supercoiled circular genome 16,569 base pairs in length that encodes 37 genes. These genes include two rRNA molecules, 22 tRNA molecules, and 13 protein subunits of the electron transport chain (ETC)/OXPHOS system [Citation51]. Our findings clearly showed that cocaine and HIV-1 Tat exposure reduced total DNMT enzyme activity and 5mC levels in astrocytes. To further investigate whether changes in DNMT and TET protein levels due to exposure to HIV-1 Tat and cocaine affect mtDNA methylation, we performed mtDNA bisulphite modification followed by pyrosequencing methylation analysis to detect C methylation at CpG islands of the mitochondrial genome. Pyrosequencing relies on light emitted during an enzymatic reaction that is set in motion by the release of pyrophosphate upon addition of a base during the sequencing process. We studied specific regions of methylation via sequencing on a Pyromark Q24 system. To perform the assay, we used Assay Design Service (ADS) software to investigate specific CpG regions in the mitochondrial genome. We analysed the percentage of methylation in mitochondrial regions such as ADS2886-FS2 (MT-CYB), ADS9500-FS1 (MT-RNR1), ADS2888-FS1re (D-loop), and ADS9515-FS2 (MT-ND1) using pyrosequencing assays for mtDNA, as shown in ), respectively. We found that the percentage of mtDNA methylation in HIV-1 Tat-treated astrocyte mitochondria was significantly lower at ChrM:14,957
Figure 5. Impacts of HIV-1 Tat and cocaine on mtDNA methylation, as analysed by pyrosequencing. mtDNA methylation profiles were determined by pyrosequencing in order to compare the levels of methylation among primary astrocytes treated with HIV-1 Tat/cocaine. The levels of methylation (% methylation) at CpG sites across the A) MT-CYB and MT-RNR1 and B) MT-ND1 and D-loop regions are shown. All of the values are expressed as the mean±SD of each group. The data represent 3 independent experiments. *P < 0.05
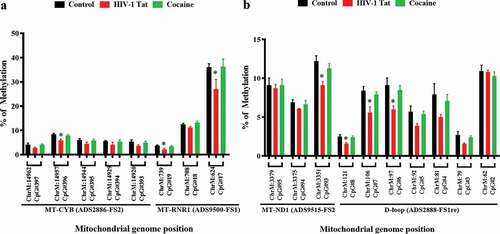
CpG#396 (MT-CYB), ChrM:739 CpG#19 (MT- RNR1), ChrM:624 CpG#17 (MT- RNR1), ChrM:3351 CpG#93 (MT- ND1), ChrM:121 CpG #8 (D-loop), ChrM:106 CpG #7 (D-loop) and ChrM:97 CpG #6 (D-loop) than that in untreated human primary astrocyte mitochondria, as shown in ). Cocaine-exposed human primary astrocytes showed slightly lower percentages of mtDNA methylation than control astrocytes, as shown in ). Furthermore, to validate our pyrosequencing findings, we analysed CpG and non-CpG methylation sites in the mitochondrial genome using TNGBS, which is a more sensitive technique that provides 1000-times more coverage than pyrosequencing and can be used to measure the percent methylation of multiple target regions (PCR amplicons) and multiple samples in a single run. TNGBS can also be used for DNA methylation analysis covering many areas of one gene or a diverse set of regions across multiple genes [Citation52]. ) shows 64 in silico-designed bisulphite PCR primers that were used for amplification in 8 multiplex PCRs across the human mitochondrial genome. We observed that the genomic regions of MT-RNR1, MT-ND5, and MT-CYB were differentially methylated (p < 0.02) in cocaine- and HIV-1 Tat-exposed astrocyte samples compared with control samples, as shown in ). ) shows the human mitochondrial genome regions that were screened for methylation levels in various regulatory regions, such as the MT-RNR1, MT-ND5, MT-ND1, D-loop and MT-CYB regions. All bisulphite-modified DNA samples were amplified using 8 separate multiplex PCRs as listed in Supplementary Table 1.
Figure 6. Mitochondrial methylation is shown with reference to the human mitochondrial genome. A) Sixty-four bisulphite PCR primers were designed using Qiagen ADSW to investigate 326 CpG sites in the mitochondrial genome. mtDNA methylation was analysed by TNGBS of the mitochondrial genome in primary astrocytes. B) The mtDNA methylation shown at different CpG sites is representative of three independent experiments, and the results were aligned to the human mitochondrial genome (NCBI reference sequence: NC_012920.1, length: 16,569 bp). *P < 0.05
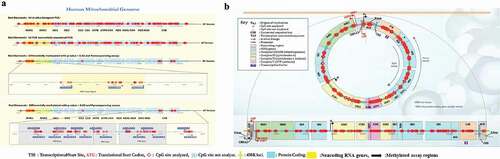
mtDNA methylation analysis using TNGBS
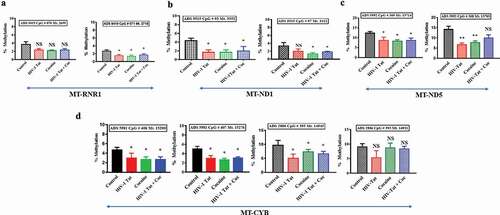
To determine the baseline methylation patterns within the mitochondrial genome, human primary astrocytes were first assessed. We found that a total of 31.29% of all Cs of the mitochondrial genome were methylated in human primary astrocytes ()). We then used bismark_methylation_extractor to summarize the number of reads supporting methylated Cs and the number of reads supporting unmethylated Cs for every C in the reference genome. The methylation calls for every single C analysed depended on the context CpG, CHG or CHH (where H can be A, T or C). The percentages of methylation detected within the CpG, CHC and CHH dinucleotide contexts were 8.63%, 8.76% and 13.9%, respectively ()). Interestingly, HIV-1 Tat-exposed astrocyte mitochondria showed a total of ~28% methylation (CpG 7.9%, CHG 7.8% and CHH 12.8%), which was slightly lower (by almost ~3%) than that of the control astrocyte mitochondria ()). In addition, we found a total of 27.82% (CpG 7.46%, CHG 7.46% and CHH 12.9%) methylation in cocaine-exposed astrocyte mitochondria, which was slightly lower (by almost ~3.5%) than that in control astrocyte mitochondria, as shown in ). A striking difference in methylation was observed across the RNR1-, ND5-, ND1- and CYB-encoding regions of the mitochondrial genome in cocaine- and HIV-1 Tat-treated astrocytes compared with control astrocytes, as shown in . The total percentage of mitochondrial methylation in HIV-1 Tat- and cocaine-treated human primary astrocytes was not significantly different from that in the control group, as shown in . However, the TNGBS results showed that at specific mitochondrial regions, mitochondrial methylation levels with HIV-1 Tat treatment were significantly different from those in the control group (). The TNGBS results showed lower CpG island methylation at ADS9419 in the MT-RNR1 gene in the cocaine (p < 0.05), HIV-1 Tat (p < 0.05) and HIV-1 Tat with cocaine (p < 0.04) treatment groups than in the control group ()). Similarly, we observed significantly lower methylation levels at ADS9515 in the MT-ND1 gene in the cocaine (p < 0.05), HIV-1 Tat (p < 0.05) and HIV-1 Tat with cocaine (p < 0.05) treatment groups than in the control group ()). Moreover, we found a similar trend in the MT-ND5 gene (ADS5992): significantly lower methylation levels were observed in the cocaine (p < 0.05), HIV-1 Tat (p < 0.05) and HIV-1 Tat with cocaine (p < 0.05) groups than in the control group ()). In the case of the MT-CYB gene (ADS5981 and ADS2886), we again observed significantly lower methylation levels in the cocaine (p < 0.05), HIV-1 Tat (p < 0.05) and HIV-1 Tat with cocaine (p < 0.05) groups than in the control group ()).
Figure 7. Summary statistics of the percentages of mitochondrial methylation in CpG and non-CpG sites in primary astrocytes. The mtDNA methylation (methylation %) patterns in primary astrocytes differed among the control (a), HIV-1 Tat (b) and cocaine (c) groups. The data represent 3 independent experiments
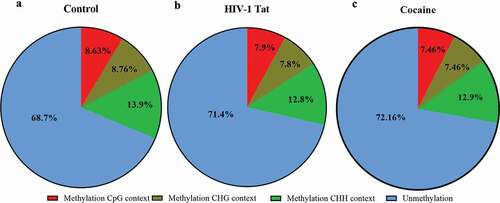
Figure 8. Impacts of HIV-1 Tat and cocaine on mtDNA methylation, as analysed by TNGBS. The mtDNA methylation profiles were determined by mitochondrial genome bisulphite sequencing to compare the levels of methylation among primary astrocytes treated with HIV-1 Tat/cocaine. The levels of methylation (% methylation) at CpG sites across the A) RNR1 gene B) ND1 gene C) ND5 gene D) CYB genes are shown. All of the values are expressed as the mean±SD of each group. The data represent 3 independent experiments. **P < 0.01, *P < 0.05, NS – Nonsignificant
Discussion
HAND is characterized by dementia, which involves varying degrees of cognitive impairment, asymptomatic neuropsychological impairment and behavioural changes [Citation53]. Studies have shown the roles of HIV-1 Tat proteins [Citation54] in HIV-positive patients presenting with HAND who show neuropathological conditions, and astrocytes are major contributors to the development of HAND in the CNS [Citation55,Citation56], triggering oxidative stress that leads to neuronal impairment [Citation57]. Considerable evidence suggests that mitochondrial dysfunction occurs early in neurodegenerative diseases [Citation58]. HIV infection [Citation8] and the psychostimulant drug cocaine [Citation59] are known to alter cellular homoeostasis, including epigenetic changes such as DNA methylation, and to affect different parts of the brain [Citation60,Citation61]. The impacts of HIV infection and cocaine on mitochondrial dysfunction have been studied previously [Citation62,Citation63], but the effects of exposure to HIV-1 Tat and cocaine on mitochondrial methylation patterns in astrocytes have not yet been investigated. In the present work, we examined the effects of cocaine and/or HIV-1 Tat on the mitochondrial epigenome in astrocytes for the first time.
Astrocytes play major roles in the regulation of oxidative stress, energy storage, mitochondrial biogenesis, neurogenesis and synapse modulation in the CNS [Citation64]. Astrocytes are modulated by HIV infection as well as drug abuse, both of which lead to neurodegeneration [Citation16,Citation65]. Earlier studies have also shown that mtDNA methylation patterns are directly linked to DNMT enzyme levels [Citation44,Citation47]. Furthermore, HIV infection [Citation66] and cocaine [Citation67] have independent effects on DNMT levels and expression. Therefore, we investigated whether HIV-1 Tat and cocaine exert similar or different effects on DNMT levels in human primary astrocytes. We observed that HIV-1 Tat treatment decreased the protein levels of DNMT1, and this effect was accelerated by cocaine in vitro. Since DNMT1 is responsible for maintaining the methylation levels of DNA, the reductions in DNMT1 protein levels show that HIV-1 Tat and cocaine exposure may reduce DNA methylation levels. We also observed significant changes in the levels of the de novo proteins DNMT3a and DNMT3b that may have reflected changes in DNA methylation due to treatment with HIV-1 Tat and cocaine. Moreover, we found that TET protein levels were decreased in vitro in astrocytes exposed to cocaine and HIV-1 Tat. Similarly, we observed reduced levels of TET proteins in the HIV-iTat and combined cocaine/HIV-iTat treatment groups compared to the control group. TET1 has been shown to be able to modify methylcytosines and potentially erase DNA methylation [Citation68]. The TET family includes three proteins – TET1, TET2, and TET3 – that catalyse the successive oxidation of 5mC into 5–5hmC, 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) [Citation69,Citation70]. C modifications are forms of epigenetic modifications that are associated with compaction and transcriptional silencing [Citation71]. Cs in CpG dinucleotides can be modified via methylation of methyl (5mC) or hydroxymethyl (5hmC) groups [Citation71] or by demethylation [Citation72] of these respective regions.
Among astrocytes, those exposed to HIV-1 Tat alone, cocaine alone, and HIV-1 Tat with cocaine showed significantly lower DNMT enzyme activity than the controls. Recent studies have also indicated that DNMTs are potential predictive diagnostic biomarkers and therapeutic targets for several diseases, including neuronal dysfunction-related diseases [Citation73–75]. Since DNMT enzymes mainly act on the 5-carbon positions of the pyrimidine rings of Cs within CpG sites, the levels of methylation at the 5-carbon positions must be measured [Citation76]. Previous research has shown that brain cells show overall decreases in the total genomic 5mC content during DNA methylation drift [Citation77]. 5mC oxidation products have been implicated as intermediates in the conversion from 5mC to unmodified C, potentially representing the first steps in a pathway of active DNA demethylation; thus, DNA methylation patterns may not be as static as previously assumed. These results point towards decreases in methylation at C5 residues at both CpG and non-CpG sites of DNA. Furthermore, we performed an in vivo study to investigate the effects of HIV-Tat alone, cocaine alone and HIV-iTat with cocaine on the levels of DNMT and TET proteins in iTat mice. iTat mice express HIV-Tat protein when treated with doxycycline, facilitating controlled expression of the protein in the CNS of mice [Citation40]. We observed significantly lower DNMT and TET protein levels in the combined HIV-Tat/cocaine treatment group compared to the control group. This in vivo study showed a trend similar to that in the in vitro study, thus confirming the changes in DNMT and TET protein levels that occur after exposure to cocaine and HIV-1 Tat.
Recently, researchers have discovered human mitochondria-specific DNMT enzymes and their roles in mitochondrial methylation [Citation78]. Therefore, we investigated the effects of HIV-1 Tat and cocaine on mtDNMTs. We observed significantly lower DNMT3a and DNMT3b protein levels and significantly higher DNMT1 protein levels after HIV-1 Tat and cocaine exposure than after control treatment in both in vitro and in vivo experiments. Our observations provide new insights into the alterations in mitochondrial genome methylation profiles and the expression of enzymes and proteins associated with mitochondrial methylation, such as DNMTs and TETs, caused by HIV-1 Tat and cocaine exposure. These alterations may affect mitochondrial biogenesis and serve as a potential sign of neurodegeneration.
For the first time, our present study used an integrated experimental approach incorporating exposure of human primary astrocytes to cocaine and HIV-1 Tat and quantitative assessment of mtDNA methylation at both CpG and non-CpG methylation sites at single-base pair resolution. The results showed that methylation in human primary astrocytes occurred predominantly in non-CpG and CpG contexts and was more prevalent on the H-strand than on the L-strand. Analysis of our TNGBS results showed that mtDNA methylation levels were lower in the HIV-1 Tat- and cocaine-exposed group than in the control group. Moreover, we also showed that HIV-1 Tat with cocaine exposure changes DNMT protein levels. These changes in mitochondrial DNMT1, DNMT3a and DNMT3b protein levels may have associations with changes in the level of mitochondrial methylation. Our results revealed that mitochondrial DNMT1 protein expression increased while DNMT3a and DNMT3b protein expression decreased when astrocytes were exposed to HIV-1 Tat and cocaine. These findings suggest that DNMT3a and DNMT3b rather than DNMT1 may be major players in the regulation of methylation levels in mitochondria. These changes may have resulted in mitochondrial gene expression and protein modifications since mitochondria are known players in cellular energy homoeostasis. Therefore, we hypothesize that alterations in mtDNA methylation may play important roles in regulating specific genes responsible for transient changes in cellular energy demands caused by exposure to HIV-1 Tat and cocaine. In support of this notion, our previous report demonstrated that HIV infection alters energy deficits, and that the effects are accelerated by cocaine [Citation79]. Epigenetic changes in DNA, such as hypomethylation and hypermethylation, have been identified as major causes of various human neurodegenerative diseases [Citation80,Citation81]. Studies have also shown that HIV-1 Tat is responsible for DNA methylation at specific CpG sites of genes [Citation82]. mtDNA methylation has been reported at both CpG sites and non-CpG sites [Citation47], and changes in mtDNA methylation patterns are associated with neurovegetative diseases [Citation83].
Interestingly, our analysis of mtDNA methylation showed significant alterations in the methylation levels of Cs present in the genomic regions of the D-loop, MT-RNR1, MT-ND5, MT-ND1 and MT-CYB genes. These genes are involved in various biological pathways; for example, the MT-RNR1 gene regulates metabolic homoeostasis through activation of 5ʹ-AMP-activated protein kinase (AMPK), while MT-ND5, MT-ND1 and MT-CYB are involved in proper functioning of the mitochondrial respiratory chain [Citation84,Citation86]. Therefore, hypomethylation at regions of the MT-RNR1, MT-ND5, MT-ND1 and MT-CYB genes may result in perturbation of mitochondrial function in human primary astrocytes. In particular, reduced MT-ND1 methylation levels have been observed in the brains of patients with Alzheimer’s disease (AD)-related pathology compared with healthy control brains [Citation87]. Defects in MT-ND5 and MT-RNR1 expression are associated with defects in brain energy homoeostasis and with neurodegenerative disease, supporting the idea that changes in expression may be linked to changes in methylation levels in the mitochondrial genome [Citation85,Citation88].
Conclusions
In summary, the present study revealed via a next-generation sequencing (NGS)-based bisulphite sequencing method, for the first time, that HIV-1 Tat and cocaine exposure alters mtDNA methylation patterns and that the alterations may be functionally linked to improper mitochondrial gene function. We also found that mtDNA methylation changes were linked to changes in the levels of TETs, DNMTs and mtDNMTs both in vitro and in vivo. This study provides insight into mitochondrial methylation at both CpG and non-CpG sites in human primary astrocytes exposed to external stimuli such as viral protein (HIV-1 Tat) and a psychostimulant drug (cocaine). We found that the MT-RNR1, MT-ND5, MT-ND1 and MT-CYB regions were significantly hypomethylated due to exposure to cocaine and HIV-1 Tat, providing a direction for further investigation of epigenetic regulation of mitochondrial function. mtDNA methylation may be a useful biomarker for the detection and diagnosis of mitochondrial disease progression related to HAND and cocaine abuse. In the future, our findings will aid in elucidation of mtDNA methylation-oriented signalling pathways and interactions and of communications among mtDNA sequence regions and between mtDNA and nuclear DNA (nDNA). Future efforts aimed at deciphering the functional and biological consequences of alterations in mtDNA methylation will be highly useful in elucidating the various biological mechanisms involved in neuro-AIDS.
Disclosure statement
No potential conflict of interest is reported by the authors.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials.
Additional information
Funding
References
- Hu XL, Wang Y, Shen Q. Epigenetic control on cell fate choice in neural stem cells. Protein Cell. 2012;3:278–290.
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128(4):635–638.
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38.
- Singh RP, Shiue K, Schomberg D, et al. Cellular epigeneti epigenetic modifications of neural stem cell differentiation. Cell Transplant. 2009;18:1197–1211.
- Jiang D, Zhang Y, Hart RP, et al. Alteration in 5-hydroxymethylcytosine-mediated epigenetic regulation leads to Purkinje cell vulnerability in ATM deficiency. Brain. [Internet] 2015; 138:3520–3536. [cited 2020 Jul 24]. Available from: https://academic.oup.com/brain/article-abstract/138/12/3520/413106
- Troyer RM, Collins KR, Abraha A, et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. [Internet] 2005 ; 79: 9006–9018. [cited 2020 Apr 6]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15994794
- Valcour V, Sithinamsuwan P, Letendre S, et al. Pathogenesis of HIV in the central nervous system. Curr HIV/AIDS Rep. 2011;8:54–61.
- Zhang X, Justice AC, Hu Y, et al. Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics. 2016;11:750–760.
- Samikkannu T, Rao KVK, Ding H, et al. Immunopathogenesis of HIV infection in cocaine users: role of arachidonic acid. PLoS One. [Internet] 2014 ; 9:e106348. [cited 2020 Feb 12]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25171226
- Minagar A, Shapshak P, Fujimura R, et al., The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: HIV-associated dementia, Alzheimer disease, and multiple sclerosis [Internet]. J Neurol Sci 2002;202:13–23. cited 2020 Feb 19. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12220687
- Dayton AI, Sodroski JG, Rosen CA, et al. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986;44:941–947.
- Fields J, Dumaop W, Crews L, et al. Mechanisms of HIV-1 Tat neurotoxicity via CDK5 translocation and hyper-activation: role in HIV-associated neurocognitive disorders. Curr HIV Res. 2015;13(1):43–54.
- Ivanov AV, Valuev-Elliston VT, Ivanova ON, et al. Oxidative stress during HIV infection: mechanisms and Consequences. Oxid Med Cell Longev. 2016;2016:8910396.
- Zhao X, Fan Y, Vann PH, et al. 1 Tat expression in the brain led to neurobehavioral, pathological, and epigenetic changes reminiscent of accelerated aging. 2019; 11:1–15.
- Periyasamy P, Thangaraj A, Guo ML, et al. Epigenetic promoter DNA methylation of mir-124 promotes HIV-1 tat-mediated microglial activation via MECP2-STAT3 axis. J Neurosci. 2018;38:5367–5383.
- Natarajaseenivasan K, Cotto B, Shanmughapriya S, et al. Astrocytic metabolic switch is a novel etiology for Cocaine and HIV-1 Tat-mediated neurotoxicity article. Cell Death Dis. 2018;9(4):415.
- Lecoeur H, Borgne-Sanchez A, Chaloin O, et al. HIV-1 Tat protein directly induces mitochondrial membrane permeabilization and inactivates cytochrome c oxidase. Cell Death Dis. 2012;3(3): e282.
- Norman JP, Perry SW, Reynolds HM, et al. HIV-1 Tat activates neuronal ryanodine receptors with rapid induction of the unfolded protein response and mitochondrial hyperpolarization. PLoS One. 2008;3(11):e3731.
- Pernas L, Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol. 2016;78:505–531.
- Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770.
- El-Amine R, Germini D, Zakharova VV, et al. HIV-1 Tat protein induces DNA damage in human peripheral blood B-lymphocytes via mitochondrial ROS production. Redox Biol. 2018;15:97–108.
- Rodríguez-Mora S, Mateos E, Moran M, et al. Intracellular expression of Tat alters mitochondrial functions in T cells: A potential mechanism to understand mitochondrial damage during HIV-1 replication. Retrovirology. 2015;12; 12:78.
- Turnbull HE, Lax NZ, Diodato D, et al. The mitochondrial brain: from mitochondrial genome to neurodegeneration [Internet]. Biochim Biophys Acta - Mol Basis Dis. 2010;1802:111–121. [cited 2020 Jul 24]. Available from: /pmc/articles/PMC2795853/?report=abstract
- Jaeger VK, Lebrecht D, Nicholson AG, et al. Mitochondrial DNA mutations and respiratory chain dysfunction in idiopathic and connective tissue disease-related lung fibrosis. Sci Rep. 2019;9:1–8.
- Dhillon NK, Peng F, Bokhari S, et al. Cocaine-mediated alteration in tight junction protein expression and modulation of CCL2/CCR2 axis across the blood-brain barrier: implications for HIV-dementia. J NeuroImmune Pharmacol. [Internet] 2008 ; 3 52–56. [cited 2020 Apr 6]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18046654
- Gandhi N, Saiyed ZM, Napuri J, et al. Interactive role of human immunodeficiency virus type 1 (HIV-1) clade-specific Tat protein and cocaine in blood-brain barrier dysfunction: implications for HIV-1associated neurocognitive disorder. J Neurovirol. [Internet] 2010 ; 16: 295–306. [cited 2020 Apr 6]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20624003
- Ersche KD, Jones PS, Williams GB, et al. Cocaine dependence: A fast-track for brain ageing. Mol Psychiatry. 2013;18:134–135.
- Vonmoos M, Hulka LM, Preller KH, et al. Cognitive impairment in cocaine users is drug-induced but partially reversible: evidence from a longitudinal study. Neuropsychopharmacology. 2014;39:2200–2210.
- Elachouri G, Vidoni S, Zanna C, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20.
- Rouzier C, Bannwarth S, Chaussenot A, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain [Internet]. 2011;135:23–34.
- Samikkannu T, Atluri VSR, Nair MPN. HIV and cocaine impact glial metabolism: energy sensor AMP-activated protein kinase role in mitochondrial biogenesis and epigenetic remodeling. Sci Rep. 2016;6:1–11.
- Cunha-Oliveira T, Silva L, Silva AM, et al. Mitochondrial complex I dysfunction induced by cocaine and cocaine plus morphine in brain and liver mitochondria. Toxicol Lett. [Internet] 2013 ; 219: 298–306. [cited 2020 Apr 6]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23542814
- US NIH. How does drug abuse affect the HIV epidemic? | National Institute on Drug Abuse (NIDA) [Internet]. 2012. [cited 2020 Apr 6]. Available from: https://www.drugabuse.gov/publications/research-reports/hivaids/how-does-drug-abuse-affect-hiv-epidemic
- Samikkannu T, Agudelo M, Gandhi N, et al. Human immunodeficiency virus type 1 clade B and C gp120 differentially induce neurotoxin arachidonic acid in human astrocytes: implications for neuroAIDS. J Neurovirol. 2011;17:230–238.
- Baum MK, Rafie C, Lai S, et al. Crack-cocaine use accelerates HIV disease progression in a cohort of HIV-positive drug users. J Acquir Immune Defic Syndr. 2009;50:93–99.
- Samikkannu T, Rao KVK, Arias AY, et al. HIV infection and drugs of abuse: role of acute phase proteins. J Neuroinflammation. 2013;10:113.
- Desplats P, Dumaop W, Cronin P, et al. Epigenetic alterations in the brain associated with HIV-1 infection and methamphetamine dependence. PLoS One. 2014;9(7):e102555.
- Anier K, Malinovskaja K, Aonurm-Helm A, et al. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35:2450–2461.
- Carouge D, Host L, Aunis D, et al. CDKL5 is a brain MeCP2 target gene regulated by DNA methylation. Neurobiol Dis. 2010;38:414–424.
- Kim BO, Liu Y, Ruan Y, et al. Neuropathologies in transgenic mice expressing human immunodeficiency virus type 1 Tat protein under the regulation of the astrocyte-specific glial fibrillary acidic protein promoter and doxycycline. Am J Pathol. 2003;162:1693–1707.
- Paris JJ, Singh HD, Ganno ML, et al. Anxiety-like behavior of mice produced by conditional central expression of the HIV-1 regulatory protein, Tat. Psychopharmacology (Berl). 2014;231:2349–2360.
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma Appl Note. [Internet] 2011 ; 27 1571–1572. [cited 2020 Apr 13]. Available from: www.bioinformatics.bbsrc.ac.uk/projects/bismark/
- Coppedè F, Stoccoro A. Mitoepigenetics and neurodegenerative diseases. Front Endocrinol (Lausanne). 2019;10:86.
- Shock LS, Thakkar PV, Peterson EJ, et al. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. [Internet] 2011. ; 108 3630–3635. [cited 2020 Mar 4]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21321201
- Wong M, Gertz B, Chestnut BA, et al. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci. [Internet] 2013 ; 7:279. [cited 2020 Mar 4]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24399935
- Bellizzi D, D’aquila P, Scafone T, et al. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. [Internet] 2013 ; 20 537–547. [cited 2020 Mar 4]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23804556
- Patil V, Cuenin C, Chung F, et al. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. [Internet] 2019 ; 47:10072–10085. [cited 2020 Mar 4]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/
- Jin B, Robertson KD DNA methyltransferases, DNA damage repair, and cancer. In: Advances in experimental medicine and biology. NIH Public Access; 2013. 3–29.
- Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750.
- Sun H, Wang Xeditors. Mitochondrial DNA and diseases [Internet]. Singapore: Springer Singapore. 2017. cited 2020 Apr 16. Available from: http://link.springer.com/https://doi.org/10.1007/978–981–10–6674–0
- Brandon MC, Lott MT, Nguyen KC, et al. MITOMAP: a human mitochondrial genome database–2004 update. Nucleic Acids Res. [Internet] 2005 ; 33:D611–3. [cited 2020 Apr 13]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15608272
- Masser DR, Stanford DR, Freeman WM. Targeted DNA methylation analysis by next-generation sequencing. J Vis Exp. 2015.
- Gray F, Adle-Biassette H, Chretien F, et al. Neuropathology and neurodegeneration in human immunodeficiency virus infection. Pathogenesis of HIV-induced lesions of the brain, correlations with HIV-associated disorders and modifications according to treatments. Clin Neuropathol [Internet] ; 20(4):146–155. [cited 2020 May 13]. Available from:http://www.ncbi.nlm.nih.gov/pubmed/11495003
- Churchill MJ, Cowley DJ, Wesselingh SL, et al. HIV-1 transcriptional regulation in the central nervous system and implications for HIV cure research. J Neurovirol. [Internet] 2015 ; 21: 290–300. [cited 2020 May 13]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25060300
- Gray LR, Turville SG, HItchen TL, et al. HIV-1 entry and trans-infection of astrocytes involves CD81 vesicles. PLoS One. [Internet] 2014 ; 9:e90620. [cited 2020 May 17]. Available from: https://dx.plos.org/https://doi.org/10.1371/journal.pone.0090620
- Churchill MJ, Wesselingh SL, Cowley D, et al. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann Neurol. [Internet] 2009 ; 66 253–258. [cited 2020 May 17]. Available from: http://doi.wiley.com/https://doi.org/10.1002/ana.21697
- Vignoli AL, Martini I, Haglid KG, et al. Neuronal glycolytic pathway impairment induced by HIV envelope glycoprotein gp120. Mol Cell Biochem. [Internet] 2000 ; 215:73–80. [cited 2020 May 13]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11204458
- Lunnon K, Ibrahima Z, Proitsi P, et al. Mitochondrial dysfunction and immune activation are detectable in early alzheimer’s disease blood. J Alzheimer’s Dis. [Internet] 2012 ; 30 685–710. [cited 2020 May 5]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22466004
- Sadri-Vakili G. Cocaine triggers epigenetic alterations in the corticostriatal circuit. Brain Res. 2015;1628:50–59.
- Breiter HC, Gollub RL, Weisskoff RM, et al. Acute effects of cocaine on human brain activity and emotion. Neuron. 1997;19:591–611.
- Hategan A, Bianchet MA, Steiner J, et al. HIV Tat protein and amyloid-β peptide form multifibrillar structures that cause neurotoxicity. Nat Struct Mol Biol. [Internet] 2017 ; 24:379–386. [cited 2020 May 13]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28218748
- Miro O, Lopez S, Martinez E, et al. Mitochondrial effects of HIV infection on the peripheral blood mononuclear cells of hiv-infected patients who were never treated with antiretrovirals. Clin Infect Dis. [Internet] 2004 ; 39:710–716. [cited 2020 May 11]. Available from: https://academic.oup.com/cid/article-lookup/doi/https://doi.org/10.1086/423176
- Graziani M, Sarti P, Arese M, et al. Cardiovascular mitochondrial dysfunction induced by cocaine: biomarkers and possible beneficial effects of modulators of oxidative stress. 2017 [cited 2020 May 11]. DOI: https://doi.org/10.1155/2017/3034245
- Siracusa R, Fusco R, Cuzzocrea S. Astrocytes: role and functions in brain pathologies. Front Pharmacol. 2019;10:1114.
- Buch S, Yao H, Guo M, et al. Cocaine and HIV-1 interplay in CNS: cellular and molecular mechanisms. Curr HIV Res. 2012;10:425–428.
- Zhao X, Fan Y, Vann PH, et al. Long-term HIV-1 Tat expression in the brain led to neurobehavioral, pathological, and epigenetic changes reminiscent of accelerated aging. Aging Dis. [Internet] 2020 ; 11:93. [cited 2020 Mar 4]. Available from: http://www.aginganddisease.org/EN/https://doi.org/10.14336/AD.2019.0323
- Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647.
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. [Internet] 2009 ; 324 930–935. [cited 2020 Apr 3]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19372391
- He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307.
- Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303.
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453.
- Hahn MA, Szabó PE, Pfeifer GP. 5-hydroxymethylcytosine: A stable or transient DNA modification? Genomics. 2014;104:314–323.
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869.
- Liu L, Van Groen T, Kadish I, et al. Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clin Epigenetics. 2011;2:349–360.
- Mutze K, Langer R, Schumacher F, et al. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer. [Internet] 2011 ; 47:1817–1825. [cited 2020 Apr 16]. Available from. ;:. . : http://www.ncbi.nlm.nih.gov/pubmed/21458988
- Pfeifer GP, Tang MS, Denissenko MF. Mutation hotspots and DNA methylation. Curr Top Microbiol Immunol. 2000;249:1–19.
- Wang SC, Oeize B, Schumacher A. Age-specific epigenetic drift in late-ons,et Alzheimer’s disease. PLoS One. [Internet] 2008 ; 3:e2698. [cited 2020 Apr 16]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18628954
- Saini SK, Mangalhara KC, Prakasam G, et al. DNA Methyltransferase1 (DNMT1) Isoform3 methylates mitochondrial genome and modulates its biology. Sci Rep. [Internet] 2017 ; 7. [cited 2020 Apr 16]. Available from: www.nature.com/scientificreports
- Samikkannu T, Atluri VSR, Nair MPN. HIV and cocaine impact glial metabolism: energy sensor AMP-activated protein kinase role in mitochondrial biogenesis and epigenetic remodeling. Sci Rep. 2016;6:31784.
- Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880–891.
- Sung HY, Choi EN, Jo S A, et al. Amyloid protein-mediated differential DNA methylation status regulates gene expression in Alzheimer’s disease model cell line. Biochem Biophys Res Commun. [Internet] 2011 ; 414 700–705. [cited 2020 Apr 16]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22001921
- Pion M, Jaramillo-Ruiz D, Martínez A, et al. HIV infection of human regulatory T cells downregulates Foxp3 expression by increasing DNMT3b levels and DNA methylation in the FOXP3 gene. AIDS. [Internet] 2013 ; 27: 2019–2029. [cited 2020 Mar 4]. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00002030-201308240–00002
- Gao D, Zhu B, Sun H, et al. Mitochondrial DNA methylation and related disease. In: Advances in experimental medicine and biology. Springer New York LLC; 2017;1038:117–132.
- Lee C, Zeng J, Drew BG, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454.
- Lakhani CM. Neuronal brain region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Physiol Behav. 2019;176:139–148.
- Chomyn A, Mariottini P, Cleeter MWJ, et al. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature. 1985;314:592–597.
- Blanch M, Mosquera JL, Ansoleaga B, et al. Altered mitochondrial DNA methylation pattern in Alzheimer disease-related pathology and in Parkinson disease. Am J Pathol. 2016;186:385–397.
- McKenzie M, Duchen MR. Impaired cellular bioenergetics causes mitochondrial calcium handling defects in MT-ND5 mutant cybrids. PLoS One. 2016;11:1–12.
