ABSTRACT
To evaluate the possible involvement of epigenetic modulation by HPV16-p16INK4a in oral potentially malignant disorder (OPMD). We generated DNA-methylation profiles, according to p16INK4a expression and HPV16 genotype (positive or negative), of OPMD samples and p16INK4a-HPV16 negative samples (used as control), using reduced-representation bisulphite sequencing (RRBS-Seq- Illumina) technology. Twelve samples, four for each group, as follows: 1) p16INK4a+ HPV16+; 2) p16INK4a+ HPV16-; 3) p16INK4a- HPV16-, were analysed in triplicate for DNA-methylation profiles. Fifty-four per cent of DMRs were hypermethylated and 46% were hypomethylated. An increase in methylation of loci in OPMD was independent of the presence of HPV. The hypermethylated genes in HPV+ samples were associated with signalling pathways such as NICD traffics to nucleus, signalling by NOTCH1 (p = 0.008), Interferon-gamma (p = 0.008) and Interleukin-6 signalling (p = 0.027). The hypomethylated genes in HPV infection were associated with TRAF3-dependent IRF activation pathway (p = 0.002), RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways (p = 0.005), TRAF6 mediated IRF7 activation (p = 0.009), TRIF-mediated TLR3/TLR4 signalling (p = 0.011) and MyD88-independent cascade release of apoptotic factors (p = 0.011). Protein association analysis of DMRs in OPMD revealed 19 genes involved in the cell cycle regulation, immune system, and focal adhesion. Aberrantly methylated loci in OPMD were observed in p16INK4a positive samples which suggests that a shift in global methylation status may be important for cancer progression. The results suggest that HPV infection in OPMD induces modulation of genes related to the immune system and regulation of the cellular cycle.
Introduction
Oral potentially malignant disorder (OPMD) includes lesions with altered epithelium that have a higher probability of progression to squamous cell carcinoma and these changes involve epithelial dysplasia [Citation1]. For OPMD, the prevalence of HPV has been found to be between 26.2% and 28% [Citation2,Citation3]. The risk of malignant transformation of OPMD to oral squamous cell carcinoma (OSCC) has been reported to be between 4.3% and 17.9% [Citation4].
OSCC is one of the most frequent cancers worldwide and represents more than 300.000 new cases annually [Citation5]. Likewise, OSCC has a significant burden of disease in terms of treatment and prognosis and most tumours are detected in advanced stages where the 5-year survival rate is almost 50–60% [Citation6]. Interestingly, these tumours develop in patients without a history of smoking and alcohol use [Citation7,Citation8], which could indicate the presence of an additional etiologic agent for human papillomavirus (HPV) [Citation9].
HPV-positive head and neck squamous cell carcinoma (HNSCC) differs distinctly from HPV-negative tobacco-related HNSCC, especially in the oropharyngeal region [Citation10]. However, OSCC does not show a clear difference between HPV-associated cancer and tobacco-associated cancer and its impact is still under study [Citation11]. Specific DNA-methylation patterns are showing increased promise as biomarkers, with some investigators claiming superiority to other markers with regards to establishing differences in clinical behaviour and prognosis [Citation12].
The transition from normal epithelium to premalignant tissue and carcinoma is partly caused by an accumulation of genetic, epigenetic, and cellular alterations [Citation13]. Oral carcinogenesis is a complex process involving genetic changes, changes in excitatory and inhibitory cell signals that generate rapid cell division, increase of blood vessels for greater growth and changes in signalling causing invasion of tissues [Citation14].
Somatic mutations and chromosomal copy number alterations with loss of heterozygosity (LOH) have been identified in OPMD progression, with LOH being the most effective factor in predicting progression to oral cancer [Citation15]. Several studies show the convergence of several genes and signalling pathways known for their oncogenic capabilities, in OPMD progression. Altered genes are involved in induction of signalling pathways related to proliferation, apoptosis, angiogenic, and immunomodulatory processes [Citation16–18].
Epigenetic factors such as HPV infection produce alterations in DNA methylation and play a key role in the malignant transformation of precancerous lesions; these changes lead to the silencing of tumour suppressor genes and the overexpression of oncogenes [Citation19]. Differential patterns of methylation in promoter regions in the tumour suppressor genes and greater levels of CpG methylation in HPV-positive tumours compared with HPV-negative oropharyngeal and oral cell carcinomas have been reported [Citation20,Citation21].
High-risk HPV16 is the most prevalent genotype found in OSCC [Citation22] and it is a strong risk factor for malignant transformation [Citation23]. Molecular profiling of HPV16 positive tumours of the head and neck and those that are HPV-associated is commonly associated with p16INK4a overexpression especially in oropharyngeal squamous cell carcinoma [Citation24]. Overexpression of p16 INK4a is caused by a viral E7 protein and it indicates that the virus is transcriptionally active, and it is often used as a surrogate marker for HPV infection and viral oncoprotein activity [Citation25]. Unlike normal cells, cancer cells exhibit alterations in methylation patterns that induce changes in cycle regulation and cell growth [Citation26]. p16/Rb/cyclin-D1 pathway is a key regulator of the cell cycle, which controls the passage of cells from G1 to S phase by binding cyclin D-dependent kinases (CDK4 and CDK6) and causes inactivation/phosphorylation of Rb (Retinoblastoma gene) [Citation27]. p16 gene is often mutated or epigenetically silenced in head and neck cancer and the hypermethylation of its promoter is a common finding in OSCC [Citation28,Citation29]. However, the role of HPV in the development of oral dysplasia and its subsequent malignancy is still unclear although p16INK4a expression may be an early predictor of malignant transformation [Citation30].
The association of HPV and epigenetic changes of specific genes in OPMD have been little studied. The aim of this study was to determine if p16INK4a positive OPMD associated with HPV-16 high-risk genotypes can modulate DNA methylation.
Materials and methods
Study design and samples
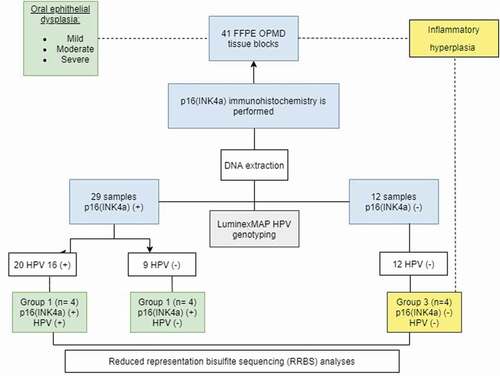
A prospective cross-sectional study of 41 patients was conducted from 2015 to 2017. Thirty-six individuals were diagnosed with OPMD using pre-established inclusion criteria: diagnosis of OPMD including histological dysplasia in oral lichen planus, leukoplakia, or erythroplakia in pre-establishing anatomical locations (tongue, gingiva, palate, cheek mucosa and oral cavity not specified, except for lesions of the oropharynx and soft palate or lesions of the floor of the mouth including floor of the tongue), with no previous cancer or smoking habit. Five samples were negative for dysplasia and showed a diagnosis of hyperplasia (, ). This study was approved by the Ethics Committee of the School of Dentistry of the Universidad El Bosque (Act. No. 012–2013). All samples evaluated were obtained with informed consent.
Table 1. Localization and clinical diagnosis by p16 VPH groups
Pathology and laboratory procedures
The laboratory procedures were carried out according to a well-validated methodology. Four to eight paraffin sections (10um) were obtained for each block and were processed under strict conditions to avoid DNA contamination. The tissue sections were stained with haematoxylin and eosin (H&E) for pathological revaluation, which included confirmation of OPMD or hyperplasia. Dysplasia was established when the epithelium exhibited cytologic and/or architectural alterations. Mild dysplasia is confined to the lower one-third of the epithelium, moderate dysplasia extends from the basal layer to the midportion of the spinous layer and severe dysplasia from the basal cells to the entire thickness of the epithelium. Hyperplasia without architectural or cytologic atypia was not considered an OPMD [Citation31].
Immunohistochemistry
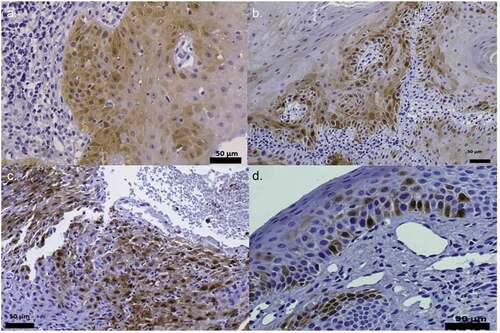
p16INK4a expression was evaluated on all cases using the CDKN2A antibody (Bond III, Leica Microsystems, INC, Buffalo Grove, IL, EEUU) following the manufacturer’s protocol using tumour tissue as a positive control. In this study, we chose, as before, the methodology developed by Kostopoulou et al., 2011 for p16INK4a expression [Citation32] and the Darragh et al. 2012 criteria where p16INK4a positivity is indicated by a cytoplasmic staining in a diffuse, faint, or continuous pattern, which we have observed in individual cells () [Citation33].
Figure 2. p16INK4a staining. Positive IHC pattern for p16INK4a expression obtained in the samples diagnosed with epithelial dysplasia HPV (+). (400x). Figures (a), (b) and (c) can be described, using Kospokolou 2011 patterns, as patchy positivity patterns and figure (d) as a focal positivity pattern
HPV-DNA detection and genotyping
For HPV-DNA detection and genotyping, FFPE samples were first treated with xylene and ethanol to remove paraffin, subsequently with 250 μl of freshly prepared proteinase K solution (Bioline) to extract DNA with QIAamp DNA FFPE Tissue Kit (QIAGEN). Real-time PCR using SybrGreen and consensus primers HPV GP5+/6+ and ß-globin gene fragment as a quality control was performed [Citation34,Citation35]. HPV genotypes were determined by Luminex technology (Optiplex Diamex). DNA extracted from FFPE samples was subjected to PCR amplification using sets of biotinylated primers for HPV L1 gene GP5+/GP6+, and a pair of primers for the amplification of a ß-globin gene fragment as a quality control for genomic DNA in the PCR. PCR products were added to the bead mix containing 26 distinct bead populations coupled to 24 HPV, one ß-globin and one hybridization control specific oligonucleotide probe. The fluorescence intensities of each sample were calculated as the median fluorescence intensity (MFI). The samples were categorized according to the presence or absence of HPV16.
HPV16-p16INK4a – HPV 16 Groups
Three groups were established based on the presence or absence of p16INK4a and HPV 16, as follows: Group 1) p16INK4a (+) HVP16 (+); 2) p16INK4a (+) HVP16 (-); Group 3) p16INK4a (-) HPV16 (-) (). Twelve samples, 4 per group, were analysed in triplicate for RRBS methylation.
Reduced-representation bisulphite sequencing (RRBS) methylation analysis
RRBS was performed on 12 samples with EpiNext™ High-Sensitivity Bisulphite-Seq Kit Illumina (EpiGentek) using 200 ng of DNA. The experimental protocol steps were as follows: (i) DNA digestion using the MspI restriction enzyme (NEB) for 16 hours at 37°C, which cuts DNA at its recognition site (C↓CGG) independent of the CpG methylation status; (ii) end repair and ligation of TruSeq adapters for Illumina sequencing (NEB); (iii) gel-based selection of DNA fragments with insert sizes ranging from 40 bp to 120 bp and 120–220 bp; (iv) bisulphite treatment; (v) 18 cycles of PCR amplification of the bisulphite-converted library; and (vi) 76 cycles of single-read sequencing using an Illumina Hiseq 2000 using the established single-end sequencing protocol. The sequences were aligned to the human genome assembly GRCh37 (hg19) using Bismark [Citation36]. Bisulphite conversion rates are calculated as the number of genomic cytosines outside a CpG context that are unconverted, divided by the total number of cytosines outside a CpG context.
Identification of the important genes based on differentially methylated regions (DMRs)
CLC Genomics Workbench 20 (Qiagen) was used to quantify the methylation level of each sample and for DMR identification. All the DMR-related genes were clustered based on the Gene Ontology (GO) annotation database and were analysed in DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources 6.83. Significant GO terms and pathways were selected after filtering with P < 0.01. After multiple testing corrections, we chose to deem pathways with a Q value ≤0.05 as significantly enriched with DMR-related genes. Considering the significant enrichment of pathways, we found the main biochemical pathways and signal transduction pathways in which the DMR-related genes were located.
Motif discoveries of differential methylated sites/regions
We performed motif discovery analysis using MEME suite online software for both DMSs and DMRs. The MEME suite calculated the statistical significance of motif occurrences for observed regions in a large set of known TF (transcription factors) motif usages. Only statistically significant ones matching a known TF motif were reported. Peak and gene associations were performed using GREAT by applying the basal plus extension settings [Citation37]. GREAT assigns biological meaning to genomic regions by analysing the annotations of nearby genes and regulatory elements wherein genomic distances are divided into bins relative to transcription start site (TSS) as [0, 5 kb], [5 kb, 50 kb], [50 kb, 500 kb], [500 kb, infinity]. Values on the Y-axis represent percent of peaks found in each genomic region. Values displayed on bar graphs represent the number of genes associated with peaks in each genomic window. Genic annotations were performed with annotatr package (that include 1–5Kb upstream of the TSS, the promoter (<1Kb upstream of the TSS), UTRs, exons, and CDS).
Statistical analysis
Descriptive statistics were carried out for categorical variables and clinical data and p16INK4a expression by HPV were compared using chi squarer/fisher tests. RRBS DNA-methylation analysis was performed with the bisulphite sequencing plugin in CLC Genomics Workbench 20 (Qiagen)-. The assessment of differential methylation in cases-sample vs control group was carried out using the ANOVA test with a significance of 5% using the STATA® Software version 12.
Results
Characteristics of the study population
describes the design and the samples collected and tested for p16INK4a staining and HPV16 genotype. A total of 41 cases were selected, of which twenty-nine samples (70.7%) were p16INK4a positives and twelve (29.3%) were positive for HPV16. Only one sample with p16INK4a negative was positive for HPV. The samples of the control group corresponded to hyperplasia that were negative for the p16 immunohistochemistry tests and were negative for the identification of HPV.
Sixty-three per cent of the patients were women with an age average of 55.2 (range was 30 to 82 years). Twenty-two lesions (53.76%) had mild dysplasia, 13 (31,71%) showed moderate dysplasia, and only 2 (4.88%) had severe dysplasia. Four samples (9,76%) that did not have dysplasia and showed histologic hyperplasia and were negative for p16 and for HPV were included as control (). The clinical diagnosis was variable; leukoplakia was the most frequent lesion followed by lichen planus and erythroplakia; no statistical differences were observed between the groups (p = 0.104). Four unspecific samples with no infection overgrowth were p16INK4a-HVP-16 negatives and were included as control samples. Localization was variable although samples from the tongue were more frequently p16INK4a positive (p = 0.009) (). In the p16INK4a positive groups, the samples selected showed moderate or severe dysplasia, whereas in the p16INK4a-HPV16 negative group the samples had histological hyperplasia.
p16INK4A expression in oral lesions
matches p16INK4A expression in oral lesions according to HPV infection by immunohistochemical analysis. It was demonstrated that 3/29 samples (10.3%) of OPMD p16INK4a (+) were stained in small patches and 26/29 of cases (86.7%) demonstrated focal positivity. However, the control samples were p16INK4a negative and these results showed a strong association between overexpression of p16 INK4a protein and oral lesions with malignancy potential (p < 0.0001). Interestingly, OPMD infected with HPV16 gave a diffuse and focal pattern (), but p16INK4a over-expression in OPMD did not correlate with clinical variables such as age, sex, and lesion site (data not shown). p16INK4a+ with HPV-negative had sporadic p16INK4a stain.
Table 2. p16INK4a expression by HPV groups
Fourteen out of the 29 p16 INK4a positive samples showed DNA from some HPV genotype and only nine had high-risk HPV16 genotype. Only one out of 12 p16INK4a negative samples had HPV and it was HPV16. Four random samples were selected for DNA methylation according to the presence or absence of HPV16 and p16INK4a ().
Genome-wide DNA-methylation changes in OPMD
Twelve samples were randomly selected according to p16INK4a and HPV16 results and were processed for DNA-methylation profiles analysis in order to investigate the epigenomic landscape of OPMD by RRBS analysis (supplementary Table 1). The clinical characteristics of the samples are included in supplementary material (see supplementary Table 1). To assess variability, three biological replicates were generated for each p16INK4a+HPV+ sample, p16INK4a+HPV- sample or p16INK4a-HPV- sample as control. To test the reproducibility of our results, we calculated Pearson correlation coefficients between these replicates with high reproducibility within our libraries (r = 0.87–0.89). Bisulphite conversion efficiencies of 12 samples ranged from 98% to 99% with 20X read coverage. The per-sample average percentages of cytosine methylation rate in CHG and CHH sites were ranged between 45%–63% and 50%–57%, respectively.
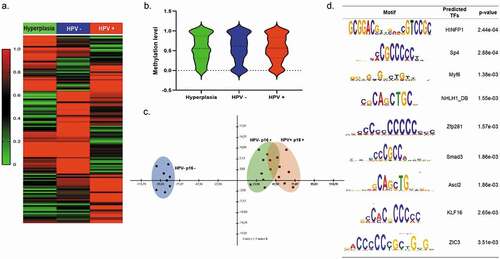
Methylation profiles of DMRs present in OPMD samples were normalized to normal-appearing oral mucosa HPV- from control groups with hyperplasia. The heat-map showed distinct methylation profiles of DMRs identified in OPMD p16INK4a +HPV+ and p16INK4a + HPV- tissues ()). A total of 270 regions were differentially methylated, 142 (54%) of these DMRs were hypermethylated, while 128 (46%) were hypomethylated indicating a tendency towards increased methylation at aberrantly methylated loci in OPMD and suggesting that a shift in global methylation status may be important for cancer progression. As shown in ), methylation level data were clearly separated by principal component analysis (PCA) indicating distinct methylation profiles between hyperplasia and dysplasia samples. However, no statistically significant differences in global methylation level were found between hyperplasia, OPMD HPV+ and HPV- tissues, as shown in violin plot ()). The novo motif search revealed several sequences that were highly enriched in DMRs that corresponded to consensus binding sites for known TFs, including HINFP1, Sp4, Myf6, NHLH1_DB, Zfp281, Smad3, Ascl2, KLF16, and ZIC3. These motifs were enriched with high significance and showed a clear enrichment in unmethylated or methylated CpG island regions, respectively ()).
Figure 3. Genome-wide DNA-methylation changes in hyperplasia, OPMD HPV+ and HPV- samples (a) Hierarchical clustering and heatmap of DMRs defined by difference ≥15%, false discovery rate (FDR)-adjusted q-value ≤ 0.05 and read coverage ≥10 in OPMD tissues. The colour indicates the percentage of methylation: hyper (in red) and hypo (in green). Heat maps for tissues were generated by complete linkage clustering using a Pearson correlation. (b) Violin plot showing the median methylation level (horizontal line) and distribution patterns (density and IQR) of the identified DMRs in hyperplasia, OPMD HPV+ and HPV- samples (c) Principal component analysis (PCA) of DNA-methylation changes demonstrates a clear separation of HPV-p16m- samples (hyperplasias) (blue) and dysplasia samples (green and red) (d) DNA sequences motifs identified to be enriched in DMRs by MEME suite software. The right column contains the transcription factor similar to these motifs using JASPAR database and p value
In order to identify the biological pathways that are significantly affected by HPV infection, we conducted gene ontology (GO) analysis (). A total of 360 GO terms were significantly represented in DMRs (p = 0.001), and these were associated with viral reproductive process, cell cycle regulation, chromatin, modification, metabolic process, MAP kinase activity, and cytoskeleton, as shown in . The hypermethylated genes in HPV+ samples are associated with the signalling pathways related to: NICD traffics to nucleus p = 0.0003, NOTCH1 signalling p = 0.008, interferon-gamma signalling p = 0.008 and Interleukin-6 signalling p = 0.027 ()). Hypomethylated genes were significantly associated with TRAF3-dependent IRF activation pathway p = 0.00247, RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways p = 0.00544, TRAF6 mediated IRF7 activation p = 0.00978, TRIF-mediated TLR3/TLR4 signalling p = 0.0112, MyD88-independent cascade p = 0.0115 and Release of apoptotic factors from the mitochondria p = 0.0108 ()) (supplementary ).
Figure 4. DNA-methylation patterns were identified by RRBS analysis in OPMD samples (a) Interactome integrated network analysis of DMRs involved in OPMD HPV+ and HPV – (b) Gene ontology (GO) analysis of DMRs in OPMD samples. The GO terms were clustered into biological processes (BP), molecular function (MF) and cellular component (CC). (c). Enriched reactome pathways for DMRs in OPMD tissues involved in immune system regulation
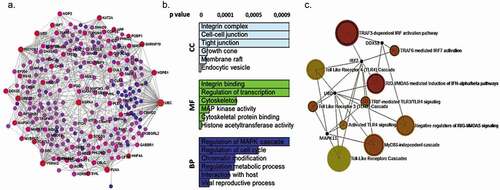
The protein association analysis of DMRs in OPMD revealed 19 genes that are central to the network, as highlighted in blue and purple in ). These genes include polyubiquitin precursors and proteins which contain two ubiquitin-like domains (UBC, UBD, UBE2D3), heat shock proteins (HSPB1, HSPA4), adapter proteins (YWHAG, -GRB2, YWHAQ), histone acetyltransferase, and histone deacetylase (KAT2A, EP300, HDAC2, CHD4), tyrosine-protein kinase (SRC, IKBKG, p53), cytoskeleton (FLNA), transcriptional regulator of type I interferon (IRF7), receptor proteins (NOTCH1) and spliceosome (SNRNP70). This suggests that HPV infection modulated genes are involved in the cell cycle, immune system, and focal adhesion. Protein association network of hypermethylated genes in HPV+ OPMD samples revealed eight genes that are central to the network: PLEC, KAT2A, PQBP1, MAP3K11, SOCS3, TCF7L2, NOTCH1 and ARID1A (Supplementary ).
Overall, these last results suggest that HPV infection in OPMD induces modulation of genes related to the immune system ()). However, while GO analysis revealed biological processes that are significantly affected by HPV infection, it does not provide information on interactions among genes. Therefore, we further examined the interaction among individual proteins using the online tools STRING and InnateDB [Citation36]. DNA methylation surrounding the transcription start site (TSS) is much more tightly linked to transcriptional silencing. As shown in , the detected DMRs were mostly located at the nearest distance to TSS in gene bodies. We then examined the genomic distribution of DMRs and found that ∼18% of the DMRs were located within TSS 50 kb (1,501–50,000 bp upstream from the TSS) and ∼25%–30% of all DMRs were located >50 kb from the nearest transcript and considered intergenic ()). Furthermore, 43% of all DMRs were located in promoters, 35.84% in exons, 12.90% in CDS, and 8.24% were in UTRs ()).
Figure 5. DNA-methylation patterns were identified by RRBS analysis in OPMD samples (a) Heat maps of DMRs in OPMD HPV+ and HPV – tissues. (b). Enriched reactome genes for DMRs in OPMD tissues involved in immune system regulation
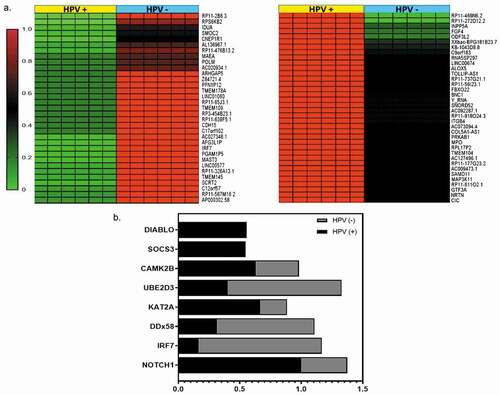
Figure 6. Distribution of Methylated Regions (MRs) annotated to specific genomic regions. (a). Peak and gene associations obtained by analysing the annotations of nearby genes and regulatory elements wherein genomic distances are divided into bins relative to transcription start site (TSS) as [0, 5 kb], [5 kb, 50 kb], [50 kb, 500 kb], [500 kb, infinity]. Values on the Y-axis represent per cent of peaks found in a given genomic region (b). Genic annotations of DMRs include 1–5Kb upstream of the TSS, the promoter (< 1Kb upstream of the TSS), UTRs, exons and CDS regions. The schematic above illustrates the positions of genic regions
![Figure 6. Distribution of Methylated Regions (MRs) annotated to specific genomic regions. (a). Peak and gene associations obtained by analysing the annotations of nearby genes and regulatory elements wherein genomic distances are divided into bins relative to transcription start site (TSS) as [0, 5 kb], [5 kb, 50 kb], [50 kb, 500 kb], [500 kb, infinity]. Values on the Y-axis represent per cent of peaks found in a given genomic region (b). Genic annotations of DMRs include 1–5Kb upstream of the TSS, the promoter (< 1Kb upstream of the TSS), UTRs, exons and CDS regions. The schematic above illustrates the positions of genic regions](/cms/asset/ff06929c-8cce-459d-a2ae-abdaa6811f92/kepi_a_1834923_f0006_oc.jpg)
Discussion
The modulation of DNA-methylation patterns has been shown to play an important role in the pathogenesis of HPV-related oral cancer [Citation38]. However, the epigenetic landscape at the earliest stages of oral neoplasia has not yet been reported. In this study, we present the first analysis of RRBS DNA methylation in OPMD samples.
We found that HPV16 was the most prevalent genotype in OPMD samples and p16INK4a positive patterns were observed in the different degrees of dysplasia, mainly associated with focal and diffuse characteristics, as reported by Fregonesi et al., 2003 [Citation39]. Consequently, p16INK4a protein overexpression has been proposed as a surrogate marker of HPV infection even if not all studies confirm its prognostic significance in OPMD and the role of HPV16 in OPMD is still under debate [Citation25]. The control samples in this study were inflammatory lesions with a diagnosis of hyperplasia and these did not show HPV16 or p16INK4a positive patterns; these samples were chosen as control group taking into account that they are a common group of reactive lesions that may be encountered during routine dental examinations since, because of ethical reasons, it was not possible to obtain healthy tissue samples [Citation40]. However, only nine out of twenty-seven OPMD with p16INK4a expression were HPV-positive, indicating that non-HPV factors also lead to p16INK4a overexpression in oral neoplasia. The differences in expression of p16INK4A may be caused by different genetic and epigenetic alterations [Citation41]. Usually, these alterations occur before lesions are clinically and morphologically detectable and increase with the advancement of the disease from carcinogenesis processes [Citation42]. The environmental conditions of the oral cavity, with its special microbiome and chemical irritations, are a risk factor that can lead to the development of chronic inflammatory stress. The interplay of an inflammatory microenvironment and genetic and epigenetic alterations can lead to cancer development [Citation43].
Disruption of genetic and epigenetic signatures in tumours has been associated with transcriptional silencing of genes that lead to malignant alterations in cells [Citation44], which involves enhanced function of several protooncogenes, oncogenes and/or the deactivation of tumour suppressor genes. Also, DNA methylation has been frequently found in oral cancer [Citation45]. Several studies reported that HPV infection may be related to hyper-or hypomethylation of genes [Citation46]. Globally, our results show that HPV has a specific and genome-wide effect in the DNA methylome of OPMD samples, such as reports of aberrant hypermethylation in gene promoters of HPV (+) oropharyngeal cancer (OPC) [Citation17,Citation47]. van Kempen et al. reported distinct patterns of DNA methylation between HPV-positive OPC compared to HPV-negative samples [Citation21].
Tumour-specific DNA methylation observed in HPV may be associated with distinct clinical-pathological characteristics [Citation48]. We found epigenetic deregulation by hypermethylation of NOTCH1 signalling in OPMD. Tripathi et al. observed nuclear Notch-1 down-expression in HPV16 positive precancerous lesions. This supports the hypothesis that down-modulation of Notch-1 in OPMD exerts specific protective effects against HPV-induced transformation [Citation49] and HPV E6 protein decreases NOTCH1 expression [Citation50]. The NOTCH family of genes regulate cell–cell interactions and is considered to have a bimodal role as a tumour suppressor and an oncogene [Citation51]. Besides, exome-sequencing studies on HNSCC have consistently revealed inactivating mutations in NOTCH1 [Citation52,Citation53]. Izumchenko et al. identified a higher incidence of mutated NOTCH1 in patients with oral leukoplakia diagnosed with mild dysplasia and also demonstrated a substantial frequency of NOTCH1 alterations, thus confirming it is crucial role in early premalignant transition [Citation54].
Host response to viral infection is known to be regulated by multiple transcription factors, whose downstream targets include genes associated with apoptotic, immune, and pro-inflammatory responses. Several of these transcription factors play critical roles in the activation of anti-viral gene programs by acetylation [Citation55]. Depletion of lysine acetyltransferase 2A (KAT2A), a histone acetyltransferase (HAT), in proliferative keratinocytes triggered changes in cell morphology, colony clustering and led to the premature expression of early and late differentiation markers in the absence of initiating signals [Citation56]. Several studies have started to uncover the means by which HDACs are recruited to viral protein-associated complexes and viral genomes. HDAC1 and HDAC2 act as repressors of host gene transcription within the nucleus. Upon HSV-1 infection HDACs promote expression of immediate-early viral genes to facilitate HCMV replication [Citation57,Citation58]. The oncoprotein HPV/E6 mediates TP53 degradation by binding to HAT EP300, inhibiting EP300-mediated TP53 acetylation and promoting MDM2-mediated degradation of TP53 [Citation59,Citation60]. We found that many host pathways already targeted by HPV oncoproteins were also modulated by epigenetic alterations. This included the polyubiquitin precursors and proteins which contain two ubiquitin-like domains-mediated targeting to the proteasome (UBC, UBD, UBE2D3). The Ub‐proteasome system is targeted by viruses for the sake of their replication and to escape host immune response. E6 viral oncogene functions as an adaptor protein to redirect the E6AP/UBE3A HECT Ub ligase to target some cellular proteins such as P53 for proteasome‐dependent degradation [Citation61]. On the other hand, HPV16 E7 has been detected in a compound with RB1 and a Cullin 2‐based E3 Ub ligase [Citation62] and has been shown to interact with the substrate recognition factor (SRF) Zer1 protein for RB1 degradation [Citation63].
Many elements of the tumour microenvironment (TME) impact the progression of HNSCC. Genetic and epigenetic alterations induced by risk factors related to tobacco and alcohol or the HPV virus infection initiate the sequence of events that trigger the transformation of stromal cells, immune suppression, and chronic inflammation [Citation64]. The oral inflammation caused by HPV infection is associated with OPMD development, indicating that a deregulated microenvironment can affect the occurrence of premalignant lesions [Citation65]. Genes associated with modulation of the immune system in OPDM were hypo and hypermethylated. Suppression of cytokine signalling 3 (SOCS3) was silenced by promoter hypermethylation which gives a growth advantage to cancer cells, enhances tumour cell survival and radioresistance degradation [Citation66]. Moreover, activation of the host anti-viral IFN signalling pathway is countered by HPV E6, E7, and E5 proteins to promote infection persistence [Citation67]; these viral proteins down-regulate expression of the dsDNA pattern recognition receptor TLR9 [Citation68] and repress transcription mediated by IFN regulatory factors 1, 3, and 9 (IRF1, IRF3, and IRF9) [Citation69].
Interestingly, Interferon regulatory factor 7 (IRF7) was hypomethylated in HPV OPMD samples contrary to negative samples for virus. Activation of IRF7 is a prerequisite for its function as a transcription factor. Pathogenic infection triggers IRF7 phosphorylation and translocation into the nucleus, where, along with other co-activators, it forms a transcriptional complex that binds to the promoter regions of target genes to activate transcription and play key roles in the activation of the innate immune response to viral infections [Citation70]. Oldak et al. showed that IRF-3 and IRF-7 are differentially expressed in human epidermis and regulate the transcriptional activity of HPV by binding to the HPV-DNA non-coding region inducing HPV8 late promoter [Citation71]. Thus, IRF7 is not only part of an antiviral response but may also have positive effects on HPV gene expression. TLR9 and TLR3 convey their signals via the adapter molecule MyD88, the KCs recognize HPV activating NF-kB, resulting in the production of proinflammatory cytokines and the activation of signalling pathways, particularly type I interferons and transcription factors IRF7/3 [Citation72].
Persistent infection and tumorigenesis by HPVs require viral manipulation of innate immune responses. HPV16/E6 antagonizes the activation of the cytoplasmic innate immune sensor RIG-I signalling cascade which is important for an antiviral response, although there is evidence that RIG-I, whose role in sensing RNA virus infections has been well characterized, also plays a crucial role in the antiviral host response to small DNA viruses of the Papillomaviridae family [Citation73]. The hypomethylation of genes associated with RIG-1 pathways (IRF7, DDX58, UBE2D3) shows that HPVs have evolved multiple mechanisms to evade innate immune responses.
Furthermore, HPV infection plays an important role in host cell interactions, such as the cell cycle process and apoptosis (MAP3K11, DIABLO, YWHAG), especially the M phase (CYP26B1, PBRM1, SMC1A), which is important for viral infection [Citation74]. Also, the hypomethylation of the ubiquitin-conjugating enzyme UBE2D3 has been associated with a phenotype linked to cell cycle progression, reduced apoptosis, and telomere stability [Citation75].
On the other hand, methylated genes might be associated with genomic region-specific DNA-methylation patterns [Citation76]. In this study, CpG islands localized in promoter and exon regions suggested that methylation levels in OPMD were higher in both regions (). It is well known that DNA methylation in a promoter is correlated with the transcription of a target gene [Citation77]. These results suggested that CpG islands located in different genic features displayed effects on the methylation patterns of the associated genes [Citation78].
These results support the idea that the molecular profiles that distinguish HPV-positive from HPV-negative OSCCs were reflected in different mutational signatures in cancer genomes [Citation79]. Thus, HPV oncoproteins and host genomic alterations cooperatively disrupt genomic stability, epithelial differentiation, apoptosis, transcriptional regulation and the anti-viral immune response, and induce cellular proliferation [Citation60,Citation80,Citation81]. These observations represent the mechanisms of HPV-related carcinogenesis in OPMD. The significant progress in the understanding of HPV-related carcinogenesis in oral early lesions reviewed here will serve as a foundation towards the development of novel diagnostic and prognostic markers and individualized therapies for this emerging viral malignancy.
Supplemental Material
Download MS Word (23.8 KB)Supplemental Material
Download MS Word (29.4 KB)Supplemental Material
Download MS Excel (9.3 KB)Acknowledgments
The authors express their gratitude to the Vice-Rectory of Research at Universidad El Bosque.
Disclosure statement
The authors have no conflict of interest in this publication.
Supplemental material
Supplemental data for this article can be accessed here.
References
- Sarode SC, Sarode GS, Karmarkar S, et al. A new classification for potentially malignant disorders of the oral cavity. Oral Oncol. 2011;47(9):920–921.
- Perdomo-Lara SJ, Buenahora MR, Álvarez E, et al. Human papilloma virus genotypes in dysplasia and epithelial hyperplasia of oral cavity using the luminex xmap technology. A multicenter study. Med Oral Patol Oral Cir Bucal. 2020 1;25:e61–e70. Published 2020 Jan 1. .
- Ishibashi M, Kishino M, Sato S, et al. The prevalence of human papillomavirus in oral premalignant lesions and squamous cell carcinoma in comparison to cervical lesions used as a positive control. Int J Clin Oncol. 2011;16(6):646–653.
- Liu W, Wang YF, Zhou HW, et al. Malignant transformation of oral leukoplakia: a retrospective cohort study of 218 Chinese patients. BMC Cancer. 2010;10:685. Published 2010 Dec 16. .
- Du M, Nair R, Jamieson L, et al. Incidence Trends of Lip, Oral Cavity, and Pharyngeal Cancers: Global Burden of Disease 1990-2017. J Dent Res. 2020;99(2):143–151.
- Jemal A, Bray F, Center MM, et al. Global cancer statistics [published correction appears in CA. 2011 Mar-Apr;61(2):134]. CA Cancer J Clin. 2011;61(2):69–90.
- Mohideen K, Krithika C, Jeddy N, et al. Meta-analysis on risk factors of squamous cell carcinoma of the tongue in young adults. J Oral Maxillofac Pathol. 2019;23(3):450–457.
- Katada C, Yokoyama T, Yano T, et al. Alcohol consumption and multiple dysplastic lesions increase risk of squamous cell carcinoma in the esophagus, head, and neck. Gastroenterology. 2016;151(5):860–869.e7.
- Ajila V, Shetty H, Babu S, et al. Human papilloma virus associated squamous cell carcinoma of the head and neck. J Sex Transm Dis. 2015;2015:791024.
- Zhao YH, Bai YP, Mao ML, et al. Zhonghua Bing Li Xue Za Zhi. 2019;48(2):127–131.
- Husain N, Neyaz A. Human papillomavirus associated head and neck squamous cell carcinoma: Controversies and new concepts. J Oral Biol Craniofac Res. 2017;7(3):198–205.
- Lindsay C, Seikaly H, Biron VL. Epigenetics of oropharyngeal squamous cell carcinoma: opportunities for novel chemotherapeutic targets. J Otolaryngol Head Neck Surg. 2017;46(1):9. Published 2017 Jan 31. .
- Torres-Rendon A, Stewart R, Craig GT, et al. DNA ploidy analysis by image cytometry helps to identify oral epithelial dysplasias with a high risk of malignant progression. Oral Oncol. 2009;45(6):468–473.
- Wils LJ, Poell JB, Evren I, et al. Incorporation of differentiated dysplasia improves prediction of oral leukoplakia at increased risk of malignant progression. Mod Pathol. 2020;33(6):1033–1040.
- William WN Jr, Papadimitrakopoulou V, Lee JJ, et al. Erlotinib and the risk of oral cancer: the Erlotinib Prevention of Oral Cancer (EPOC) randomized clinical trial. JAMA Oncol. 2016;2(2):209–216.
- Banerjee AG, Bhattacharyya I, Vishwanatha JK. Identification of genes and molecular pathways involved in the progression of premalignant oral epithelia. Mol Cancer Ther. 2005;4(6):865–875.
- Németh CG, Röcken C, Siebert R, et al. Recurrent chromosomal and epigenetic alterations in oral squamous cell carcinoma and its putative premalignant condition oral lichen planus. PLoS One. 2019;14(4):e0215055. Published 2019 Apr 9
- Carenzo A, Serafini MS, Roca E, et al. Gene expression clustering and selected head and neck cancer gene signatures highlight risk probability differences in oral premalignant lesions. Cells. 2020;9(8):E1828. Published 2020 Aug 3. .
- Soto D, Song C, McLaughlin-Drubin ME. Epigenetic alterations in human papillomavirus-associated cancers. Viruses. 2017;9(9):248. Published 2017 Sep 1. .
- Sartor MA, Dolinoy DC, Jones TR, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6(6):777–787.
- van Kempen PM, van Bockel L, Braunius WW, et al. HPV-positive oropharyngeal squamous cell carcinoma is associated with TIMP3 and CADM1 promoter hypermethylation. Cancer Med. 2014;3(5):1185–1196.
- Spector ME, Sacco AG, Bellile E, et al. E6 and E7 antibody levels are potential biomarkers of recurrence in patients with advanced-stage human papillomavirus-positive oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2017;23(11):2723–2729.
- van der Waal I. Oral potentially malignant disorders: is malignant transformation predictable and preventable? Med Oral Patol Oral Cir Bucal. 2014;19(4):e386–e390. Published 2014 Jul 1. .
- Langendijk JA, Psyrri A. The prognostic significance of p16 overexpression in oropharyngeal squamous cell carcinoma: implications for treatment strategies and future clinical studies. Ann Oncol. 2010;21(10):1931–1934.
- Grønhøj Larsen C, Gyldenløve M, Jensen DH, et al. Correlation between human papillomavirus and p16 overexpression in oropharyngeal tumours: a systematic review. Br J Cancer. 2014;110(6):1587–1594.
- Lechner M, Fenton T, West J, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5(2):15. Published 2013 Feb 5. .
- Gaździcka J, Gołąbek K, Strzelczyk JK, et al. Epigenetic modifications in head and neck cancer. Biochem Genet. 2020;58(2):213–244.
- Dvojakovska S, Popovic-Monevska D, Grcev A, et al. Promotor hypermethylated genes: Prospective diagnostic biomarkers in oral cancerogenesis. J Craniomaxillofac Surg. 2018;46(10):1737–1740.
- Allameh A, Moazeni-Roodi A, Harirchi I, et al. Promoter DNA methylation and mRNA expression level of p16 gene in oral squamous cell carcinoma: correlation with clinicopathological characteristics. Pathol Oncol Res. 2019;25(4):1535–1543.
- Wu W, Wang Z, Zhou Z. Role of the human papillomavirus in malignant transformation of oral leukoplakia distinct from oropharyngeal squamous cell carcinoma: A study of 76 patients with internal-control specimens. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;128(3):273–279.
- Müller S. Oral epithelial dysplasia, atypical verrucous lesions and oral potentially malignant disorders: focus on histopathology. Oral Surg Oral Med Oral Pathol Oral Radiol. 2018;125(6):591–602.
- Kostopoulou E, Samara M, Kollia P, et al. Different patterns of p16 immunoreactivity in cervical biopsies: correlation to lesion grade and HPV detection, with a review of the literature. Eur J Gynaecol Oncol. 2011;32(1):54–61.
- Darragh TM, Colgan TJ, Thomas Cox J, et al. The lower anogenital squamous terminology standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology [published correction appears in Int J Gynecol Pathol. 2013 Jul;32(4):432][published correction appears in Int J Gynecol Pathol. 2013 Mar;32(2):241]. Int J Gynecol Pathol. 2013;32(1):76–115.
- Shikova E, Todorova I, Ganchev G, et al. Detection and Typing of Human Papillomaviruses by PCR. Biotechnol Biotechnol Equip. 2009;23(sup1):877–880.
- Cubie HA, Seagar AL, McGoogan E, et al. Rapid real time PCR to distinguish between high risk human papillomavirus types 16 and 18 [published correction appears in Mol Pathol 2001 Jun;54(3):200]. Mol Pathol. 2001;54(1):24–29.
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572.
- Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33(15):2381–2383.
- Degli Esposti D, Sklias A, Lima SC, et al. Unique DNA methylation signature in HPV-positive head and neck squamous cell carcinomas. Genome Med. 2017;9(1):33. Published 2017 Apr 5. .
- Fregonesi PA, Teresa DB, Duarte RA, et al. p16(INK4A) immunohistochemical overexpression in premalignant and malignant oral lesions infected with human papillomavirus. J Histochem Cytochem. 2003;51(10):1291–1297.
- Awange DO, Wakoli KA, Onyango JF, et al. Reactive localised inflammatory hyperplasia of the oral mucosa. East Afr Med J. 2009;86(2):79–82.
- Tang KD, Menezes L, Baeten K, et al. Oral HPV16 prevalence in oral potentially malignant disorders and oral cavity cancers. Biomolecules. 2020;10(2):223. Published 2020 Feb 3. .
- Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8(9):11884–11894.
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545.
- Jain A. Molecular pathogenesis of oral squamous cell carcinoma, squamous cell carcinoma - hallmark and treatment modalities. Hamid Elia Daaboul: IntechOpen; May 31 2019. DOI:https://doi.org/10.5772/intechopen.85650
- Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7(1):77–82.
- Di Domenico M, Giovane G, Kouidhi S, et al. HPV epigenetic mechanisms related to oropharyngeal and cervix cancers. Cancer Biol Ther. 2018;19(10):850–857.
- Lleras RA, Smith RV, Adrien LR, et al. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin Cancer Res. 2013;19(19):5444–5455.
- Slebos RJ, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(3 Pt 1):701–709.
- Tripathi R, Rath G, Jawanjal P, et al. Clinical impact of de-regulated Notch-1 and Notch-3 in the development and progression of HPV-associated different histological subtypes of precancerous and cancerous lesions of human uterine cervix. PLoS One. 2014;9(6):e98642. Published 2014 Jun 6. .
- Kranjec C, Holleywood C, Libert D, et al. Modulation of basal cell fate during productive and transforming HPV-16 infection is mediated by progressive E6-driven depletion of Notch. J Pathol. 2017;242(4):448–462.
- Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27(38):5115–5123.
- Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157.
- Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160.
- Izumchenko E, Sun K, Jones S, et al. Notch1 mutations are drivers of oral tumorigenesis. Cancer Prev Res (Phila). 2015;8(4):277–286.
- Chen H, Ning X, Jiang Z. Caspases control antiviral innate immunity. Cell Mol Immunol. 2017;14(9):736–747.
- Moris N, Edri S, Seyres D, et al. Histone acetyltransferase KAT2A stabilizes pluripotency with control of transcriptional heterogeneity. Stem Cells. 2018;36(12):1828–1838.
- Terhune SS, Moorman NJ, Cristea IM, et al. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog. 2010;6(6):e1000965. Published 2010 Jun 24. .
- Park JJ, Kim YE, Pham HT, et al. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J Gen Virol. 2007;88(Pt 12):3214–3223.
- Zimmermann H, Degenkolbe R, Bernard HU, et al. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73(8):6209–6219.
- Gillison ML, Akagi K, Xiao W, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019;29(1):1–17.
- Zanier K, Charbonnier S, Sidi AO, et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013;339(6120):694–698.
- Huh K, Zhou X, Hayakawa H, et al. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81(18):9737–9747.
- White EA, Sowa ME, Tan MJ, et al. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A. 2012;109(5):E260–E267.
- Curry JM, Sprandio J, Cognetti D, et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. 2014;41(2):217–234.
- Gupta S, Gupta S. Role of human papillomavirus in oral squamous cell carcinoma and oral potentially malignant disorders: A review of the literature. Indian J Dent. 2015;6(2):91–98.
- Zhou H, Miki R, Eeva M, et al. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res. 2007;13(8):2344–2353.
- Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Res. 2017;231:21–33.
- Hasan UA, Zannetti C, Parroche P, et al. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the Toll-like receptor 9 promoter. J Exp Med. 2013;210(7):1369–1387.
- Hong S, Laimins LA. Manipulation of the innate immune response by human papillomaviruses. Virus Res. 2017;231:34–40.
- Yang H, Lin CH, Ma G, et al. Interferon regulatory factor-7 synergizes with other transcription factors through multiple interactions with p300/CBP coactivators. J Biol Chem. 2003;278(18):15495–15504.
- Oldak M, Tolzmann L, Wnorowski A, et al. Differential regulation of human papillomavirus type 8 by interferon regulatory factors 3 and 7. J Virol. 2011;85(1):178–188.
- Nees M, Geoghegan JM, Hyman T, et al. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75(9):4283–4296.
- Chiang C, Pauli EK, Biryukov J, et al. The human papillomavirus E6 oncoprotein targets USP15 and TRIM25 to suppress RIG-I-mediated innate immune signaling. J Virol. 2018;92(6):e01737–17. Published 2018 Feb 26. .
- Pyeon D, Pearce SM, Lank SM, et al. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5(2):e1000318.
- Yang H, Wu L, Ke S, et al. Downregulation of Ubiquitin-conjugating enzyme UBE2D3 promotes telomere maintenance and radioresistance of Eca-109 human esophageal carcinoma cells. J Cancer. 2016;7(9):1152–1162. Published 2016 Jun 6. .
- Raza MA, Yu N, Wang D, et al. Differential DNA methylation and gene expression in reciprocal hybrids between Solanum lycopersicum and S. pimpinellifolium. DNA Res. 2017;24(6):597–607.
- Niesen MI, Osborne AR, Yang H, et al. Activation of a methylated promoter mediated by a sequence-specific DNA-binding protein, RFX. J Biol Chem. 2005;280(47):38914–38922.
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186.
- Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15(3):266–282.
- Hoppe-Seyler K, Bossler F, Braun JA, et al. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26(2):158–168.
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582.