ABSTRACT
This study is performed to evaluate the role of long noncoding RNA (lncRNA) LINC00619 in osteosarcoma through the PI3K-Akt signalling pathway by binding to HGF. Osteosarcoma and osteochondroma tissues from patients were collected. The relationship between lncRNA LINC00619 and HGF was proved by the dual-luciferase reporter gene assay. The expression patterns of lncRNA LINC00619 as well as the levels of proliferating cell nuclear antigen (PCNA), hepatocyte growth factor (HGF), phosphoinositide 3-kinase (PI3K), protein kinase B (Akt), Bax, Bcl-2, alkaline phosphatase (ALP), and osteopontin (OPN) were detected by RT-qPCR and Western blot analysis. In addition, MTT assay, flow cytometry, scratch test, and Transwell assay were performed to assess the cell proliferation, cell cycle distribution, apoptosis, cell migration, and invasion in each group, respectively. Osteosarcoma tissues presented with elevated positive expression rate of HGF, up-regulated expression levels of PCNA, HGF, PI3K, Akt, Bcl-2, ALP and OPN, and down-regulated expressions of Bax and LINC00619. HGF was verified as a target gene of lncRNA LINC00619. LINC00619 was found to down-regulate the expressions of PCNA, HGF, PI3K, Akt, Bcl-2, ALP, and OPN in osteosarcoma cells. Up-regulation of lncRNA LINC00619 decreased cell growth, migration intensity, and invasion ability, but enhanced the apoptosis rate of osteosarcoma cells. Our findings suggest that lncRNA LINC00619 inhibits proliferation, migration and invasion and improves apoptosis of osteosarcoma cells through the inhibition of the activation of the HGF-dependent PI3K-Akt signalling pathway.
Introduction
Osteosarcoma is a highly aggressive sarcoma of the bone and is accompanied by a very complicated karyotype and high level of chromosomal instability [Citation1]. Osteosarcoma is also the most prevalent malignancy in children and adults, accounting for 2.4% of all malignant tumours in paediatric patients and 20% of primary cancers [Citation2]. A large proportion of localized osteosarcoma patients is treated with surgery and routine chemotherapy, whereas only 33% of patients with detectable metastases are effectively cured [Citation3]. Although the prognosis remains poor if only treated by surgery, the relapse-free 5-year survival rate reported in the last three decades has increased to 60–80% with the advent of combinational neoadjuvant chemotherapy [Citation4]. Thus, finding novel methods to treat osteosarcoma to alleviate the burden bought on by the disease on patients and medical infrastructure is of great clinical significance.
Long noncoding RNAs (lncRNAs) are a heterogeneous group of non-coding transcripts exceeding lengths of 200 nt [Citation5]. Aberrant expressions of lncRNAs have long been regarded as original factor for the development of multiple tumours, such as colorectal cancer and pancreatic cancer [Citation5,Citation6]. Numerous lncRNAs, including HOTAIR, ANCR, AK055347, and UCA1, have also been reported to affect osteosarcoma by various mechanisms [Citation7–10]. Another such lncRNA, the long intergenic non protein coding RNA 619 (LINC00619), which is also termed C10orf136 or bA168P8.1, is located at Chromosome 10:43. Moreover, the hepatocyte growth factor (HGF) can initiate the epithelial–mesenchymal transition (EMT) under the regulation of lncRNA HOTAIR [Citation11]. Signal transduction and reduction of C-MET in HGF were also previously implicated in the induction of human osteosarcoma cells with low and high metastatic nature [Citation12]. Moreover, HGF also possesses the ability to elevate the expression of heparanase during the process of gastric cancer metastasis via the PI3K/Akt/NF-kappaB pathway [Citation13]. Upon receptor activation, MAPK, Akt, and Stat3 are targeted by C-MET, which regulates multiple biological processes [Citation14]. Furthermore, the PI3K/Akt pathway is regarded as a major cell survival pathway in osteosarcoma [Citation15]. By activating this pathway, lncRNA MALAT1 could accelerate the proliferation and metastasis of osteosarcoma cells [Citation16].
In the current study, initial analyses of the GEO dataset on the expression profiles of osteosarcoma (GSE37552) identified the HGF gene as the most significantly down-regulated gene in osteosarcoma. In addition, the Multi Experiment Matrix (MEM) website demonstrated that lncRNA LINC00619 and HGF are co-expressed in datasets. Furthermore, KEGG pathway analyses indicated that HGF regulates the PI3K-Akt pathway. Consequently, we speculated that lncRNA LINC00619 participates in osteosarcoma in relation to HGF and the PI3K/AKT pathway and performed a series of experiments to investigate the same, hoping to shed light on novel therapeutic targets for osteosarcoma.
Materials and methods
Ethical statements
All experimental protocols were approved by the Ethics Committee of the Linyi People’s Hospital, which was in accordance with the Declaration of Helsinki. Written informed consents were obtained from all participants prior to the study.
Study subjects
Seventy-two samples of osteosarcoma tissues were collected from osteosarcoma patients (43 males and 29 females; aged: 9–53 years, calculated mean age of 27.21 ± 9.34 years) that underwent operation at the Linyi People’s Hospital between September 2011 and September 2016. Additionally, 35 tissue samples were obtained from amputated patients with osteochondroma (benign bone lesions) as the control group, comprising 20 males and 15 females, aged 8–58 years, and a calculated mean age of 35.2 ± 10.88 years. Two groups of subjects were selected based on the following criteria: conventional osteosarcoma confirmed by clinic, imaging and diagnosis [Citation17]. The tumour was located in the extremities, without metastasis during diagnosis [Citation17]. All the patients received no chemotherapy or radiotherapy prior to the operation. Patients with secondary lesions, history of other malignancies, and complications were excluded. The collected tissue samples were frozen at −80 °C for later use.
Immunohistochemical staining
Osteosarcoma and osteochondroma tissues were collected, paraffin-embedded, and sectioned into 3–4 μm-thick slices. The prepared slices were placed in 3% H2O2, dewaxed and hydrated. Following hydration, the slices were placed in a microwave at 80% power at 90 °C for 5 min for antigen repair. Subsequently, the slices were incubated with the monoclonal antibody rabbit-anti-human HGF (dilution ratio of 1:100, ab83760, Abcam Inc., Cambridge, MA, USA) at 4 °C overnight, and biotinylated secondary antibody mouse-anti-human IgG (dilution ratio of 1:1000, ab109489, Abcam Inc.) at 37 °C for 30 min. Haematoxylin was applied to stain the nuclei for 30 s, and diaminobenzidine was used for colour development. Afterwards, the slices were dehydrated with hydrochloric acid ethanol until no motley was observed, sealed by balata, observed and photographed. At last, five fields were randomly selected from each section to observe the positive expression of proteins.
RT-qPCR
TrizolTM kits (No. 16,096,020, Thermo Fisher Scientific, Waltham, MA) were applied to extract the total RNA content from the osteochondroma and osteosarcoma tissues. Primers were designed using Primer Premier 5 and Oligo 6 and synthesized by TaKaRa (Tokyo, Japan) (). U6 served as the internal reference of lncRNA LINC00619, and GAPDH for other genes. RNA samples of 10 μL were diluted 20 times with pure water containing no RNA enzyme. The concentration and purity of the diluted RNA were measured. Total RNA amounts V = 1.25/OD260 (μL). Subsequently, 5 μL of the Mix kit (No. 4,368,702, Tideradar Science and Technology Company, Beijing, China) was added to an EP tube. Then, 5 μL total RNA and 10 μL RNase Free H2O were added for centrifugation and mixing. RNA was placed into the qPCR instrument for the reaction. The generated complementary DNA (cDNA) was stored in a refrigerator at −20 °C. An ABI 7500 quantitative PCR instrument (No ABI 7500, Thermo Fisher Scientific) was applied to perform the RT-qPCR experiments. The relative quantitative method was used to calculate mRNA relative transcription levels: ΔΔCt = ΔCt model – ΔCt normal, and ΔCt = Ct target gene- Ct internal reference A. The relative transcription levels of target gene = 2−ΔΔCt.
Table 1. Prime sequence for RT-qPCR
Western blot analysis
Osteochondroma and osteosarcoma tissues were first lysed to obtain the total protein content. Next, the proteins were separated using 10% SDS-PAGE and transferred onto a nitrocellulose membrane. After blocking, the membrane was incubated with the following diluted primary antibodies from Abcam: Hepatocyte growth factor (HGF) (dilution ratio of 1:1000, ab83760), protein kinase B (AKT) (dilution ratio of 1:500, ab179463), proliferating cell nuclear antigen (PCNA) (dilution ratio of 1:2000, ab18197), p-phosphoinositide 3-kinase (PI3K) (dilution ratio of 1:1000, ab182651), alkaline phosphatase (ALP) (dilution ratio of 1:5000, ab83259), Bax (dilution ratio of 1:1000, ab32503), osteopontin (OPN) (dilution ratio of 1:1000, ab75285), Bcl-2 (dilution ratio of 1:1000, ab32124), p-AKT (dilution ratio of 1:1000, ab131443), PI3K (dilution ratio of 1:1000, ab86714), and GAPDH (dilution ratio of 1:2000, ab8245). The membrane was then incubated with the HRP-conjugated goat anti-rabbit IgG secondary antibody (dilution ratio of 1:1000, Boster Biological Technology, Ltd, Wuhan, China) for 1 h. Finally, the membrane was visualized with the help of an ECL reagent. The grey value ratio of target bands to interference bands was calculated using GAPDH as the control.
Primary cell isolation and culture
The frozen osteosarcoma tissues were stored in a freezing medium (CryoSure-DEX40, Hystemcell Company, Chongqing, China), and cancer tissues with no ulcers (approximately 30 mg) were added to DMEM (No. 190,040, GIBCO, Grand Island, NY, USA) containing 1% double antibodies. The tissues were then sectioned into 1–2 mm3 pieces, which were detached with 3 mL trypsin 37 °C for 40 min. Subsequently, after removing the digestive solution, the tissues were added with serum-free DMEM and triturated into single primary cells using a pipette gun. These isolated primary cells were cryopreserved for subsequent experiments. At last, the cells were passed through 40 meshes and stained with 1% trypan blue. Finally, the number of live cells in cell freezing medium was counted and recorded.
Cell treatment
The osteosarcoma cells were cultured in DMEM (No. 190,040, GIBCO), inoculated in a 6-well plate (density of 1 × 105 cells/well) and cultured at 37 °C with 5% CO2. Following culture, the osteosarcoma cells were assigned into the blank group (osteosarcoma cells), negative control (NC) group (sequence that was irrelevant to osteosarcoma), lncRNA LINC00619 vector group (lncRNA LINC00619 over-expression sequence), si-lncRNA LINC00619 group (lncRNA LINC00619 silencing sequence), si-HGF group (HGF silencing sequence), si-lncRNA LINC00619 + si-HGF group (lncRNA LINC00619 and HGF silencing sequence), HGF vector group (HGF over-expression sequence 5’-AGGATCCATGTGGGTGACCAAACTCC-3) [Citation18,Citation19], and lncRNA LINC00619 vector + HGF vector (lncRNA LINC00619 vector and HGF vector). All the aforementioned sequences were synthesized by Sangon Biotech (Shanghai, China) [Citation20]. Cell passage began 1 day before transfection, and the cells were transfected with lipofectamine 2000 transfection reagent (11,668,019, Invitrogen, Carlsbad, USA).
Dual-luciferase reporter gene assay
The online prediction website RNA22 (https://cm.jefferson.edu/) was retrieved to analyse and predict the potential binding between LINC00619 and HFG. Then, for further verification, the HGF gene vector (pmiRRB-HGF-3 ‘UTR) was subsequently constructed and transfected into the osteosarcoma cells with the lncRNA LINC00619 vector, siRNA-lncRNA LINC00619, or NC, respectively. After 48 h, the cells were collected and lysed, and luciferase activity was detected using a Dual-Luciferase® Reporter Assay System (E1910, Promega, Madison, WI, USA).
MTT assay
Osteosarcoma cells were inoculated in a 96-well plate (density of 1 × 105 cells/well), and 8 parallel wells were set up. In addition, control wells only added with culture medium were also set. Cells were assessed at the 24th, 48th and 72th h for proliferation. Upon reaching 70% confluence, 10 μL of 5 mg/mL MTT solution (ST316, Beyotime Biotechnology Research Institute, Shanghai, China) was added to each well and incubated at 37 °C for 4 h. Next, each well was added with 100 μL DMSO (D5879, Sigma, St. Louis, MO, USA). Following that, the optical density (OD) was measured at a wavelength of 490 nm. Cell activity was calculated according to the following formula: (OD value at the experimental well – OD value at the blank well/OD value at the blank well).
Scratch test
The transfected cells were first inoculated in a 6-well plate (density of 1 × 105 cells/well). Upon reaching 90% confluence, a sterile tip was used to gently draw a cross on the central axis of the well. Serum-free medium was added for 0.5–1 h culture to restore cell proliferation. At the beginning of cell recovery or 24 h later, the cells were photographed, and the distance of cell migration was measured using the Image-Pro Plus Analysis software (Media Cybernetics, USA).
Transwell assay
Matrigel matrix (No. 356,234, BD, San Jose, CA, USA) was dissolved overnight at 4 °C and diluted with serum-free culture medium at a ratio of 1:3. Subsequently, 50 μL of the diluent was added to the apical chamber. Then, 1 × 105 cells/mL cell suspension was inoculated in the apical chamber containing serum-free medium, and medium containing 10% FBS was added to the basolateral chamber. Cells were migrated for 24 h. The number of cells passing through the Matrigel matrix was used as an index to evaluate their invasive ability.
Flow cytometry
Cells were gently triturated to prepare the cell suspension. The suspension was then centrifuged at 1000 r/min for 5 min, fixed with precooled 70% ethanol for 30 min, centrifuged and stained with 1% propidium iodide (PI) containing RNA enzyme for 30 min. Cells were subsequently analysed using a BD-Aria FACS Calibur flow cytometer (FACSCalibur, Beckman Kurt, USA) to assess the cell cycle.
After 48 h of transfection, the cells were treated with trypsin without EDTA. In accordance with the instructions of the Annexin-V- FITC apoptosis detection kit (K201-100, Biovision, Milpitas, CA, USA), Annexin-V-FITC, PI and HEPES buffer were mixed at a ratio of 1:2:50. Then, the cells were added with 1 mL HEPES buffer solution. The FITC and PI fluorescence were detected using a 525-nm and 620-nm band filter excited to a wavelength of 488 nm.
Statistical analysis
Measurement data were presented as mean ± standard deviation. Comparisons between two groups were examined by unpaired t-test, and comparisons among multiple groups were analysed using one-way analysis of variance (ANOVA). The data among different groups at different time points were analysed using two-way ANOVA. A value of p < 0.05 was considered to be statistically significant.
Results
LncRNA LINC00619 might play a role in osteosarcoma by regulating HGF-dependent PI3K-Akt pathway
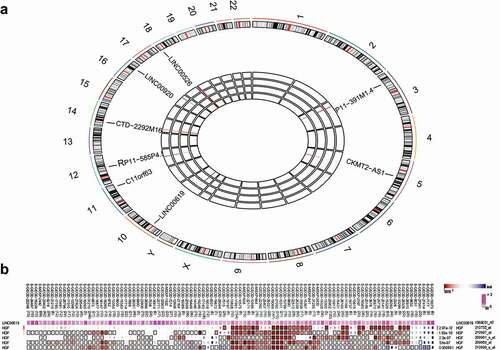
Initial analysis of the osteosarcoma-related gene expression dataset GSE37552 indicated that lncRNA LINC00619 was poorly expressed in osteosarcoma samples ()). Meanwhile, the MEM website suggested that lncRNA LINC00619 and HGF were co-expressed in multiple datasets ()), while KEGG pathway analyses insinuated that HGF gene could regulate the PI3K-Akt pathway (). These findings indicated that lncRNA LINC00619 could potentially play a role in osteosarcoma by regulating the HGF-dependent PI3K-Akt pathway.
Table 2. KEGG analysis of target genes of long noncoding RNA LINC00619
Figure 1. LncRNA LINC00619 is poorly expressed in osteosarcoma and coexpressed with HGF. A: A heat map of the aberrantly expressed lncRNAs in osteosarcoma. The characters (1–22, X, Y) outside the big circle represent chromosomes, with the location of aberrantly expressed lncRNAs noted in corresponding position. In the central part, each rectangle represents the corresponding lncRNA expression in each sample. Differential expression analysis is achieved through comparison with the expression of host cells prior to treatment. Red indicates high expression, whereas blue represents poor expression. B: lncRNA LINC00619 and HGF coexpression trend detected using the MEM website. The colour scale is from dark red to dark blue, and red indicates smaller ranks (smaller is better).
HGF is positively expressed in osteochondroma tissues
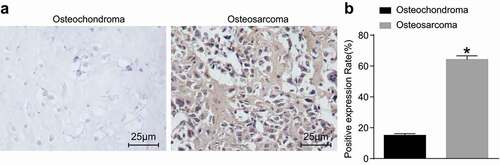
To explore whether HGF was specifically over-expressed in osteosarcoma tissues, osteochondroma tissues were considered as control. The results of immunohistochemistry, which was performed to measure the positive expression rate of HGF (), revealed that HGF protein was primarily located in tumour cells between the interstitial fibroblast cytoplasm and cell membrane. The protein stained as brown, and the staining intensity of HGF in osteosarcoma tissues was found to be elevated compared with the osteochondroma tissues. These findings demonstrated that the positive expression rate of HGF protein in osteosarcoma tissues was significantly higher relative to osteochondroma tissues.
Figure 2. Osteosarcoma tissues exhibit increased positive rates of HGF protein. A: Immunohistochemistry staining of osteochondroma tissues (n = 35) and osteosarcoma tissues (n = 72); B: HGF positive rate in osteochondroma and osteosarcoma tissues; *, p < 0.05 compared with osteochondroma tissues. Data were presented as mean ± standard deviation and the comparison between two groups was analysed by t-test.
Up-regulated levels of PCNA, HGF, PI3K, Akt, Bcl-2, ALP and OPN expression yet down-regulated levels of Bax and lncRNA LINC00619 in osteochondroma tissues
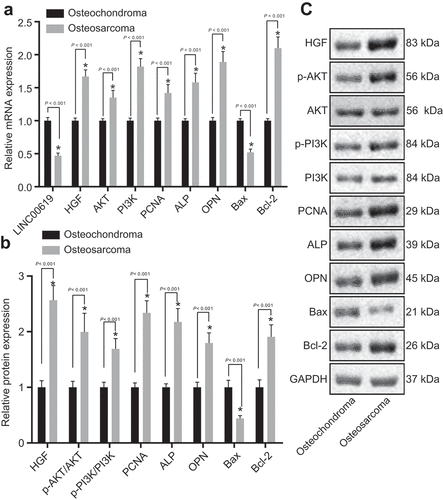
Compared with osteochondroma tissues, PCNA, HGF, AKT, PI3K, Bcl-2, ALP and OPN expression levels as well as AKT and PI3K phosphorylation were found to be elevated, while those of Bax and lncRNA LINC00619 were decreased in osteosarcoma tissues ().
Figure 3. PCNA, HGF, PI3K, Akt, Bcl-2, ALP and OPN expression is increased, whereas Bax and lncRNA LINC00619 expression is reduced. A: mRNA expressions of genes related to osteochondroma (n = 35) and osteosarcoma (n = 72) tissues; B: protein expressions of genes related to osteochondroma and osteosarcoma tissues; C: protein bands of related proteins in osteochondroma and osteosarcoma tissues; n = 3. *, p < 0.05 compared with osteochondroma tissues. Data were presented as mean ± standard deviation and the comparison between two groups was analysed by unpaired t-test.
HGF is a target gene of lncRNA LINC00619
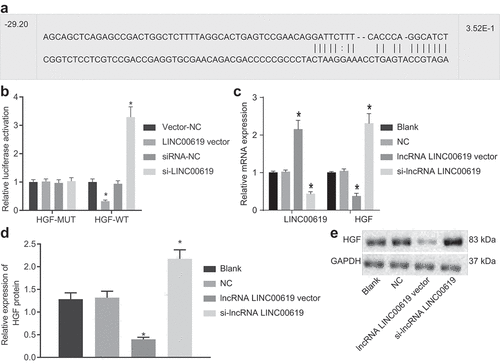
The bioinformatics website RNA22 predicted that HGF was a target gene of lncRNA LINC00619, and further indicated the presence of a specific binding site between lncRNA LINC00619 and HGF ()), which was then verified using a dual-luciferase reporter gene assay. As shown in ), compared with the vector NC group, the luciferase activity was obviously decreased in the lncRNA LINC00619 vector group; while compared with the siRNA-NC group, the luciferase activity was significantly increased in the siRNA-lncRNA LINC00619 group. Relative to the NC group, HGF expression levels were found to be decreased in the lncRNA LINC00619 vector group, while being elevated in the si-lncRNA LINC00619 group (). These findings verified that HGF was a target gene of lncRNA LINC00619.
Figure 4. HGF is a target gene of lncRNA LINC00619. A: Bioinformatics website predicted binding sites between HGF and lncRNA LINC00619; B: detection of luciferase activity by dual-luciferase reporter gene assay; HGF gene vector (pmiRRB-HGF-3ʹUTR) was synthetically constructed, which was cotransfected with lncRNA LINC00619 vector, siRNA-lncRNA LINC00619, vector NC, or siRNA-NC into osteosarcoma cells. Cells were collected for detection of luciferase activity. C: RT-qPCR of lncRNA LINC00619 expression and HGF mRNA expression in osteosarcoma cells; D: western blot analysis of HGF protein expression in osteosarcoma cells. E: protein bands of HGF in osteosarcoma cells. n = 3. *, p < 0.05 compared with the NC group. Data were presented as mean ± standard deviation and the comparison among multiple groups was analysed ANOVA.
LncRNA LINC00619 inhibits PCNA, HGF, PI3K, Akt, bcl-2, ALP and CPN expression in osteosarcoma cells
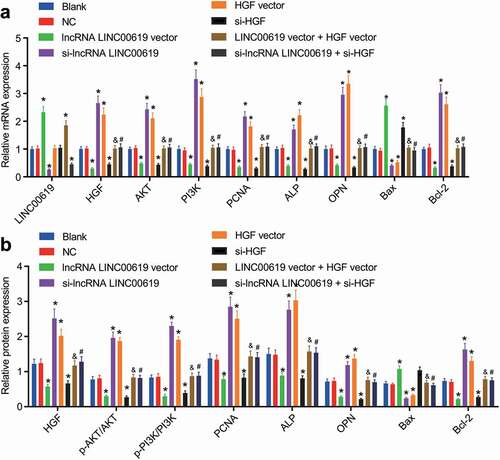
ALP is widely used in clinical practice nowadays, and regarded as a standard tumour biomarker for osteosarcoma [Citation21]. Moreover, the AKT pathway is also known to play a significant role in osteosarcoma [Citation22]. The results of RT-qPCR showed that lncRNA LINC00619 expression was not significantly different between the blank and NC groups. Compared with the blank and NC groups, lncRNA LINC00619 expression levels were found to be increased in the lncRNA LINC00619 vector and lncRNA LINC00619 vector + HGF groups, while being reduced in the si-lncRNA LINC00619 and si-lncRNA LINC00619 + si-HGF groups. Meanwhile, up-regulated expression levels of Bax, and down-regulated PCNA, HGF, AKT, PI3K, bcl-2, ALP and CPN and AKT and PI3K phosphorylation were noted in the lncRNA LINC00619 vector and si-HGF groups. Yet in the si-lncRNA LINC00619 vector and HGF vector groups, Bax mRNA and protein expression levels were decreased, whereas those of PCNA, HGF, AKT, PI3K, bcl-2, ALP and CPN and AKT and PI3K phosphorylation were augmented. Compared with the lncRNA LINC00619 vector group, expression levels of PCNA, HGF, AKT, PI3K, bcl-2, ALP and CPN, and AKT and PI3K phosphorylation were increased, and those of Bax mRNA and protein were diminished in the lncRNA LINC00619 vector + HGF vector group. In contrast to the si-lncRNA LINC00619 group, expression levels of PCNA, HGF, AKT, PI3K, bcl-2, ALP and CPN, and AKT and PI3K phosphorylation were down-regulated, but those of Bax were elevated in the si-lncRNA LINC00619 + si-HGF group (). These findings suggested that lncRNA LINC00619 inhibited PCNA, HGF, PI3K, Akt, bcl-2, ALP and CPN expression in osteosarcoma cells.
Figure 5. PCNA, HGF, PI3K, Akt, bcl-2, ALP and CPN expression in osteosarcoma tissues is downregulated by lncRNA LINC00619. Osteosarcoma cells were treated with lncRNA LINC00619, HGF, si-lncRNA LINC00619 and si-HGF vectors alone or in combination. A: RT-qPCR analysis of PCNA, HGF, PI3K, Akt, bcl-2, ALP and CPN expression in osteosarcoma cells; B, western blot analysis of protein expression of PCNA, HGF, PI3K, Akt, bcl-2, ALP and CPN and the extent of PI3K and Akt phosphorylation in osteosarcoma cells. n = 3. *, p < 0.05 indicated significant difference of gene or protein expression when compared with the blank group or the empty vector group; &, p < 0.05 indicated significant difference of gene or protein expression when compared with the lncRNA LINC00619 vector group, #, p < 0.05 indicated significant difference of gene or protein expression when compared with the siRNA-lncRNA LINC00619 group. Data were presented as mean ± standard deviation and the comparison among multiple groups was analysed by ANOVA.
LncRNA LINC00619 suppresses proliferation of osteosarcoma cells
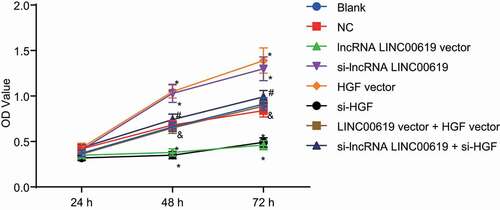
Cell proliferation was assessed in the treated cells with the help of MTT assay (). Compared with the blank and NC groups, cell growth was found to be significantly elevated at 48 h and 72 in the si-lncRNA LINC00619 vector and HGF vector groups, while being retarded in the lncRNA LINC00619 vector and si-HGF groups. Relative to the si-lncRNA LINC00619 group, cell growth in the si-lncRNA LINC00619 + si-HGF group was notably reduced at 48 h and 72 h. On the other hand, compared with the lncRNA LINC00619 vector group, cell growth in the lncRNA LINC00619 vector + HGF vector was significantly accelerated at 48 h and 72 h. These findings suggested that lncRNA LINC00619 suppressed the proliferation of osteosarcoma cells.
Figure 6. LncRNA LINC00619 represses the proliferation of osteosarcoma cells. Osteosarcoma cells were treated with lncRNA LINC00619, HGF, si-lncRNA LINC00619 and si-HGF vectors alone or in combination. *, p < 0.05 compared with the blank group; &, p < 0.05 compared with the lncRNA LINC00619 vector group, #, p < 0.05 compared with the siRNA-lncRNA LINC00619 group. Data were presented as mean ± standard deviation. The data obtained by multiple measurements at different time points were analysed using two-way ANOVA.
LncRNA LINC00619 reduces migration of osteosarcoma cells
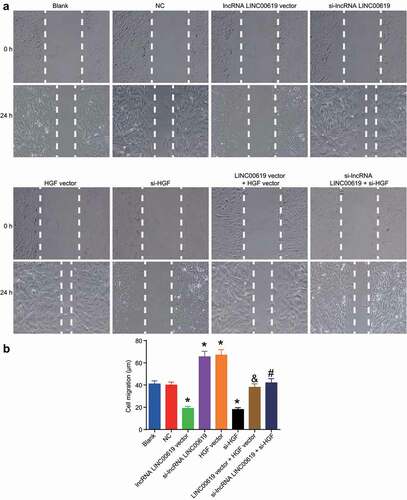
Migration was assessed in the treated cells using scratch test at the 0 h and 24 h (). Compared with the blank and NC groups, migration intensity was found to increase in the si-lncRNA LINC00619 and HGF vector groups, while exhibiting a notable decrease in the lncRNA LINC00619 vector and si-HGF groups. Compared with the si-lncRNA LINC00619 group, the migration intensity of the si-lncRNA LINC00619 + si-HGF group was diminished. Relative to the lncRNA LINC00619 vector group, the migration intensity in the lncRNA LINC00619 vector + HGF vector group was increased. These findings revealed that lncRNA LINC00619 reduced the migration of osteosarcoma cells.
Figure 7. LncRNA LINC00619 downregulates cell migration in osteosarcoma cells. Osteosarcoma cells were treated with lncRNA LINC00619, HGF, si-lncRNA LINC00619 and si-HGF vectors alone or in combination. A: representative images of cell migration in each group (× 100); B: migration distance for each group, *, p < 0.05 compared with the blank group; &, p < 0.05 compared with the lncRNA LINC00619 vector group, #, p < 0.05 compared with the siRNA-lncRNA LINC00619 group. Data were presented as mean ± standard deviation and the comparison among multiple groups was analysed by ANOVA.
LncRNA LINC00619 inhibits invasion of osteosarcoma cells
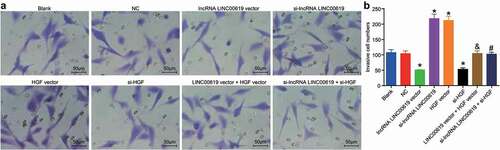
Cell invasive ability was assessed in the treated groups using Transwell assay (). No significant differences in terms of invasive ability were found between the blank and NC groups. Compared with the blank and NC groups, the number of cells transferred from the Transwell apical chamber to the basolateral chamber was markedly reduced in the lncRNA LINC00619 vector and si-HGF groups, while being increased in the si-lncRNA LINC00619 and HGF vector groups. Compared with the si-lncRNA group, the invasive ability in the si-lncRNA LINC00619 + si-HGF group was diminished. However, compared with the lncRNA LINC00619 vector group, the invasive ability in the lncRNA LINC00619 vector + HGF vector group was increased. These findings demonstrated that lncRNA LINC00619 inhibited the invasion of osteosarcoma cells.
Figure 8. LncRNA LINC00619 inhibits the invasive ability of osteosarcoma cells. Osteosarcoma cells were treated with lncRNA LINC00619, HGF, si-lncRNA LINC00619 and si-HGF vectors alone or in combination. A: representative images of cell invasion (× 200); B: number of invasive cells, *, p < 0.05 compared with the blank group; &, p < 0.05 compared with the lncRNA LINC00619 vector group, #, p < 0.05 compared with the siRNA-lncRNA LINC00619 group. Data were presented as mean ± standard deviation and the comparison among groups was analysed by ANOVA.
LncRNA LINC00619 promotes apoptosis of osteosarcoma cells
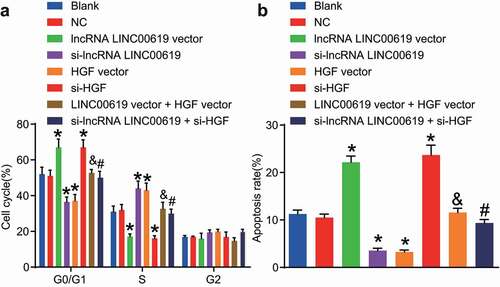
Apoptosis was assessed in the treated cells using flow cytometry (). In comparison with the blank and NC groups, the proportion of cells at the G1 phase was increased, whereas the proportion of cells at the S phase was notably decreased in the lncRNA LINC00619 vector and si-HGF groups, indicating apoptosis was enhanced. In the siRNA-LINC00619 and HGF vector groups, the proportion of cells at the G1 phase was reduced, while that at the S phase cells was increased indicating weakened apoptosis rate. Compared with the lncRNA LINC00619 vector group, the percentage of cells at the G1 phase in the lncRNA LINC00619 vector + HGF vector group was decreased, whereas the percentage at the S phase cells was elevated, demonstrating that the apoptosis rate was lowered. Compared to the siRNA-lncRNA LINC00619 group, the percentage of cells at the G1 phase in the si-lncRNA LINC00619 + si-HGF group was increased, but the percentage of S phase cells was reduced. These findings indicated that lncRNA LINC00619 promoted the apoptosis of osteosarcoma cells.
Figure 9. LncRNA LINC00619 enhances osteosarcoma cell apoptosis. Osteosarcoma cells were treated with lncRNA LINC00619, HGF, si-lncRNA LINC00619 and si-HGF vectors alone or in combination. A: cell cycle distribution in each group; B: cell apoptosis rate in each group; *, p < 0.05 compared with the blank group; &, p < 0.05 compared with the lncRNA LINC00619 vector group, #, p < 0.05 compared with the siRNA-lncRNA LINC00619 group. Data were presented as mean ± standard deviation and the comparison among multiple groups was analysed by ANOVA.
Discussion
Initial findings in our study demonstrated that lncRNA LINC00619 was poorly expressed and HGF was highly expressed in osteosarcoma. LncRNAs are known to exhibit developmental and tissue-specific expression patterns, and also play important regulatory roles in various diseases, including cancer [Citation23]. More specifically, lncRNAs have been proposed to act as potential therapeutic markers for the diagnosis and prognosis of osteosarcoma [Citation8]. For instance, owing to its interaction with EZH2 and effects on p21 expression, lncRNA-ANCR was previously highlighted to regulate the growth of osteosarcoma cells [Citation10]. Meanwhile, both lncRNA MEG3 and lncRNA TUSC7 have been documented to be poorly expressed in osteosarcoma, while their high expressions were cited as independent predictors of satisfactory survival for patients affected by osteosarcoma [Citation24]. The other prime focus of the current study, as a multipotent endogenous repair factor, HGF is primarily secreted by mesenchymal cells and affects cells expressing MET, its receptor [Citation25]. More importantly, the MET/HGF receptor is known to be overexpressed in human osteosarcoma [Citation26]. Not just osteosarcoma, the MET/HGF receptor has also been found to be over-expressed in other malignancies, such as thyroid carcinomas that originate from follicular cells [Citation27]. Further expanding on the role of lncRNA LINC00619 and HGF in osteosarcoma, our findings demonstrate that the expression of lncRNA LINC00619 was negatively correlated with the HGF expression and the PI3K-Akt pathway. The PI3K/Akt/mTOR pathway is a key component in the functioning of lncRNA HULC in tumorigenesis by activating TGF-β1 [Citation28]. Besides, HGF proliferation was abrogated in cells pretreated with LY294002 prior to HGF stimulation, which indicates that the PI3K/AKT pathway plays a regulatory role in the biological effect of HGF [Citation29]. Together, these evidences and findings stress that lncRNA LINC00619 is poorly expressed and HGF is highly expressed in osteosarcoma, and lncRNA LINC00619 regulates the HGF-dependent PI3K/AKT pathway.
Additionally, our findings revealed that over-expression of lncRNA LINC00619 brought about diminished expression levels of PCNA, AKT, PI3K, bcl-2, ALP and OPN, and increased those of Bax, which suggests that augmenting lncRNA LINC00619 blocked the PI3K/Akt pathway, inhibited proliferation and enhanced apoptosis of osteosarcoma cells. The PI3K/Akt pathway is one of the most important signal transduction pathways, and plays a major role not only in tumour development but also in the tumour’s potential response to cancer treatment [Citation16,Citation30]. Furthermore, the pathway is regarded as a pivotal regulator of osteosarcoma as it is activated by many factors, including SLC3A2, Grifolin and VEGF [Citation31–33]. Amongst the factors investigated in the current study, OPN is a non-collagenous, sialic acid-rich protein that plays critical roles in controlling cancer progression, including osteosarcoma, functioning by mediating cell-matrix interactions and cellular signalling through binding with integrins and CD44 receptors [Citation34]. Similarly, PCNA and ALP are also regarded as important factors in osteosarcoma [Citation35,Citation36]. Meanwhile, the Bax and bcl-2 genes are also known to play predominant roles in cell apoptosis, and their expression exerts an effect on primary osteosarcoma where increased Bax/bcl-2 protein expression ratio is associated with a decreased 4-year survival and disease free survival [Citation37]. Overall, these results indicate that the over-expression of lncRNA LINC00619 represses the PI3K/Akt pathway, impairs proliferation and accelerates apoptosis in osteosarcoma.
Furthermore, we uncovered that the up-regulation of lncRNA LINC00619 inhibited the HGF expression and regulated the PI3K-Akt pathway to inhibit osteosarcoma cell proliferation, invasion and migration and promote its apoptosis. LncRNAs are functional RNAs longer than 200 nucleotides, and recent evidence indicates that they play a role in the regulation of diverse cellular processes [Citation10]. More importantly, literature has also illustrated that lncRNA-p21 contributes to retarded osteosarcoma cell proliferation by repressing activation of the AKT pathway [Citation38]. In addition, lncRNA MALAT1 was previously indicated to affect the proliferation and metastasis of osteosarcoma as an oncogenic lncRNA by stimulating the PI3K/Akt pathway, which is very much in accordance with our findings [Citation16]. LncRNA HOTAIR is known to serve as an EMT regulator, and EMT can be initiated by transcription factors, including HGF, which highlights the role of lncRNA HATAIR in the regulation of HGF [Citation11]. Furthermore, HGF plays a pivotal role in the regulation of cell proliferation [Citation29]. When HGF binds to the receptor, a variety of signalling events induce several cellular processes, including proliferation, migration, morphogenesis, and cell survival [Citation25]. HGF regulates the expression of heparanase, an endoglucuronidase that promotes the shedding of associated cytokines in different types of tumours by activating PI3K/Akt, thus promoting tumour metastasis in gastric cancer [Citation39].
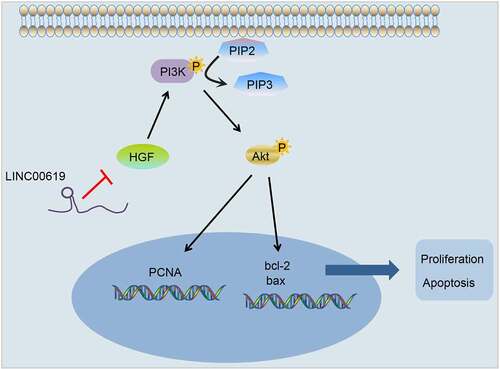
Altogether, the current study evidenced that lncRNA LINC00619-mediated down-regulation of HGF inhibits osteosarcoma progression via the PI3K-Akt pathway (), providing a novel avenue for the treatment of osteosarcoma. Nevertheless, a larger cohort and in vivo experiments should be conducted in the future to further elucidate the underlying mechanism of lncRNA LINC00619 in osteosarcoma.
Supplemental Material
Download Zip (128.5 KB)Acknowledgments
We would like to acknowledge the helpful comments on this paper received from our reviewers.
Disclosure statement
The authors declare no competing financial interests.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- Maire G, Martin JW, Yoshimoto M, et al. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 2011;204(3):138–146.
- Huang J, Ni J, Liu K, et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72(1):230–238.
- Gill J, Ahluwalia MK, Geller D, et al. New targets and approaches in osteosarcoma. Pharmacol Ther. 2013;137(1):89–99.
- Yamaguchi U, Honda K, Satow R, et al. Functional genome screen for therapeutic targets of osteosarcoma. Cancer Sci. 2009;100(12):2268–2274.
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874.
- Zhang YH, Fu J, Zhang ZJ, et al. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–5297.
- Zhou Q, Chen F, Fei Z, et al. Genetic variants of lncRNA HOTAIR contribute to the risk of osteosarcoma. Oncotarget. 2016;7(15):19928–19934.
- Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5(2):80–85.
- Fan H, Liu G, Zhao C, et al. Transcription factor Oct4 promotes osteosarcoma by regulating lncRNA AK055347. Oncol Lett. 2017;13(1):396–402.
- Zhang F, Peng H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J Orthop Surg Res. 2017;12(1):103.
- Liu YW, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802.
- Husmann K, Ducommun P, Sabile AA, et al. Signal transduction and downregulation of C-MET in HGF stimulated low and highly metastatic human osteosarcoma cells. Biochem Biophys Res Commun. 2015;464(4):1222–1227.
- Yue B, Sun B, Liu C, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106(10):1323–1332.
- Matsumura A, Kubota T, Taiyoh H, et al. HGF regulates VEGF expression via the c-Met receptor downstream pathways, PI3K/Akt, MAPK and STAT3, in CT26 murine cells. Int J Oncol. 2013;42(2):535–542.
- Zhao G, Cai C, Yang T, et al. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8(1):e53906.
- Dong Y, Liang G, Yuan B, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36(3):1477–1486.
- Group ESESNW. Bone sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(3):113–123.
- Makarevich PI, Dergilev KV, Tsokolaeva ZI, et al. Angiogenic and pleiotropic effects of VEGF165 and HGF combined gene therapy in a rat model of myocardial infarction. PLoS One. 2018;13(5):e0197566.
- Tu YJ, Ye AF, Pan ZM, et al. Regulation of expression of HGF in BM-MSCs by baculovirus-mediated transduction. Cell Biol Int. 2013;37(7):659–668.
- Wang B, Zhao RM, Zhang J, et al. Rigid aromatic linking moiety in cationic lipids for enhanced gene transfection efficiency. Eur J Med Chem. 2017;136:585–595.
- Kim SH, Shin KH, Moon SH, et al. Reassessment of alkaline phosphatase as serum tumor marker with high specificity in osteosarcoma. Cancer Med. 2017;6(6):1311–1322.
- Zhou L, Yu Y, Sun S, et al. Cry 1 regulates the clock gene network and promotes proliferation and migration via the Akt/P53/P21 pathway in human osteosarcoma cells. J Cancer. 2018;9(14):2480–2491.
- Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. Febs J. 2012;279(17):3159–3165.
- Wang Y, Huang Y, Xiang P, et al. LncRNA expression and implication in osteosarcoma: a systematic review and meta-analysis. Oncol Targets Ther. 2017;10:5355–5361.
- Mungunsukh O, Day RM. Transforming growth factor-beta1 selectively inhibits hepatocyte growth factor expression via a micro-RNA-199-dependent posttranscriptional mechanism. Mol Biol Cell. 2013;24(13):2088–2097.
- Ferracini R, Di Renzo MF, Scotlandi K, et al. The Met/HGF receptor is over-expressed in human osteosarcomas and is activated by either a paracrine or an autocrine circuit. Oncogene. 1995;10(4):739–749.
- He JX, Gendron JM, Yang Y, et al. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in arabidopsis. Proc Natl Acad Sci U S A. 2002;99(15):10185–10190.
- Zhu Y, Zhang X, Qi L, et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440.
- Okano J, Shiota G, Matsumoto K, et al. Hepatocyte growth factor exerts a proliferative effect on oval cells through the PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2003;309(2):298–304.
- Fresno Vara JA, Casado E, De Castro J, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204.
- Wang C, Yang C, Ji J, et al. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int J Oncol. 2017;50(4):1136-1146.
- Jin S, Pang RP, Shen JN, et al. Grifolin induces apoptosis via inhibition of PI3K/AKT signalling pathway in human osteosarcoma cells. Apoptosis. 2007;12(7):1317–1326.
- Peng N, Gao S, Guo X, et al. Silencing of VEGF inhibits human osteosarcoma angiogenesis and promotes cell apoptosis via VEGF/PI3K/AKT signaling pathway. Am J Transl Res. 2016;8(2):1005–1015.
- Liu SJ, Zhang DQ, Sui XM, et al. The inhibition of in vivo tumorigenesis of osteosarcoma (OS)-732 cells by antisense human osteopontin RNA. Cell Mol Biol Lett. 2008;13(1):11–19.
- Berner K, Bjerkehagen B, Bruland OS, et al. Extraskeletal osteosarcoma in Norway, between 1975 and 2009, and a brief review of the literature. Anticancer Res. 2015;35(4):2129–2140.
- Nakashima H, Nishida Y, Sugiura H, et al. Telomerase, p53 and PCNA activity in osteosarcoma. Eur J Surg Oncol. 2003;29(7):564–567.
- Kaseta MK, Khaldi L, Gomatos IP, et al. Prognostic value of bax, bcl-2, and p53 staining in primary osteosarcoma. J Surg Oncol. 2008;97(3):259–266.
- Han W, Liu J. LncRNA-p21 inhibited the proliferation of osteosarcoma cells via the miR-130b/PTEN/AKT signaling pathway. Biomed Pharmacother. 2018;97:911–918.
- Hao NB, Tang B, Wang GZ, et al. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015;361(1):57–66.