ABSTRACT
Adverse experiences in the perinatal period have been associated with the methylation of the human glucocorticoid receptor gene (NR3C1) and long-term diseases. We conducted a systematic review on the association between adversities in the perinatal period and DNA methylation in the 1 F region of the NR3C1 gene in newborns. We explored the MEDLINE, Web of Science, Scopus, Scielo, and Lilacs databases without time or language limitations. Two independent reviewers performed the selection of articles and data extraction. A third participated in the methodological quality assessment and consensus meetings at all stages. Finally, ten studies were selected. Methodological quality was considered moderate in six and low in four. Methylation changes were reported in 41 of the 47 CpG sites of exon 1 F. Six studies addressed maternal conditions during pregnancy: two reported methylation changes at the same sites (CpG 10, 13, 20, 21 and 47), and four at one or more sites from CpG 35 to 39. Four studies addressed neonatal parameters and morbidities: methylation changes at the same sites 4, 8, 10, 16, 25, and 35 were reported in two. Hypermethylation associated with stressful conditions prevailed. Hypomethylation was more often associated with protective conditions (maternal-foetal attachment during pregnancy, breast milk intake, higher birth weight or Apgar). In conclusion, methylation changes in several sites of the 1 F region of the NR3C1 gene in newborns and very young infants were associated with perinatal stress, but more robust and comparable results are needed to corroborate site-specific associations.
Introduction
Several studies show that environmental factors have a direct influence on the phenotypic manifestation of genes. This interaction between the environment and genes, causing gene expression changes without modifications in the DNA sequence and transmitted to other generations, is called epigenetics. DNA methylation is one of the most studied epigenetic patterns. A methyl group is added to the nucleotide cytosine linked to guanine on the same DNA strand in so-called CpG sites, reducing accessibility to transcription factors and silencing genes [Citation1,Citation2].
Human studies have reported methylation of the glucocorticoid receptor gene (NR3C1) associated with childhood adversities [Citation3,Citation4]. The result is an imbalance in the hypothalamic-pituitary-adrenal (HPA) axis and dysregulation of the hormonal response to stress [Citation5,Citation6]. Preclinical and clinical studies have demonstrated significant associations between stressful conditions and epigenetics alterations [Citation7–9]. The perinatal period has been considered a crucial period for the development of child’s brain, showing great susceptibility to epigenetic modifications that can influence the development of the HPA axis [Citation3,Citation10]. Exposure to psychological and environmental stressors stimulates the hypothalamus to secrete corticotrophin-releasing hormone (CRH), which in turn activates the anterior lobe of the pituitary gland to produce and secrete adrenocorticotropic hormone (ACTH), which promotes the release of glucocorticoids (GC) by the adrenal glands () [Citation11]. Glucocorticoids are involved in a wide range of immune and endocrine functions, including auto-regulation of the HPA axis [Citation12].
Figure 1. Overview of the HPA axis activity induced by perinatal stress and its GR epigenetic regulation. Perinatal stress triggers the over activity of the HPA axis by stimulating hypothalamus to release corticotropin releasing hormone (CRH), which acts on the anterior pituitary to stimulate the synthesis and secretion of adrenocorticotropic hormone (ACTH). ACTH then stimulates the production of glucocorticoids (GCs) by the adrenal cortex. In a classic negative feedback loop, GCs binding with glucocorticoid receptors (GRs) – which are encoded by the NR3C1 gene – across the body, to inhibit the release of CRH and ACTH, and thereby limit the magnitude of the GC increase. However, in response to perinatal stressors, epigenetic mechanisms reduce the ability of GRs to regulate the negative feedback loop, generating persistent HPA axis dysregulation and increased GC levels. These effects are mediated by transcriptionally silencing the NR3C1 via the epigenetic process of DNA methylation. At the cellular level, the inactive GR is located in the cytoplasm complexed with chaperones (Hsp70, Hsp90, and p23) until it becomes activated upon binding to GCs. At this point, GR dissociates from the multimeric protein complex and translocate to the nucleus. The GC-GR complex can stimulate or inhibit transcriptional responses by binding as a homodimer to glucocorticoid response elements (GREs) or possibly as a monomer to another transcription factor (TF) to enhance or down-regulate the transcription of many genes. When methylated, NR3C1 or other genes containing the GRE sequence inhibit TF binding and gene transcription, attenuating NR3C1 expression as well as an effective response to perinatal stress.
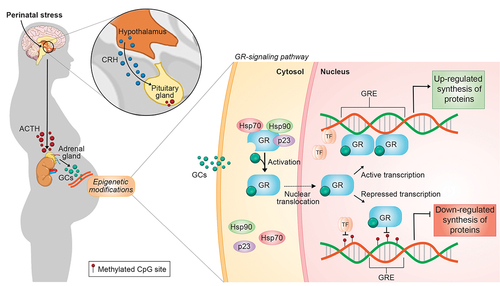
Conversely, disturbances in the HPA axis regulation during stress can result in the development of psychopathologies and have been associated with increased cortisol secretion and HPA-axis hyperactivity [Citation13–15]. Predominantly, cortisol is transported to the blood and released to the tissues through the action of the corticosteroid-binding globulin (CBG) [Citation16]. At the cellular level, cortisol, a lipophilic molecule, passively diffuses through the plasma membrane and acts by binding the glucocorticoid receptor (GR), a transcription factor encoded by the NR3C1 gene (). Once attached to cortisol, the inactive GR, connected to the chaperones proteins Hsp90, Hsp70, and p23, undergoes a conformational rearrangement that leads to GR dissociation from its activating chaperone machinery [Citation17–19]. Consequently, the exposure of nuclear localization signals (NLS) in the active GR protein quickly triggers GR translocation to the nucleus [Citation20,Citation21]. GR acts both as a transcription factor and as a repressor once in the nucleus, influencing several stress-related responses. In favour of eliminating the stress response signalling, GR can also mediate the negative-feedback regulation in the HPA axis. Thus, methylation at CpG-sequence sites in the NR3C1 gene, by decreasing its expression, is a critical epigenetic mechanism to impair the negative feedback loop’s sensitivity [Citation15]. This mechanism can create a bidirectional cycle between epigenetic regulation of NR3C1 and the functioning of the HPA axis, which further enhances cortisol levels and, subsequently, predisposes individuals to disease. Upon methylation, NR3C1 CpG sites cannot bind various transcription factors leading to reduced synthesis of proteins involved in stress response [Citation22].
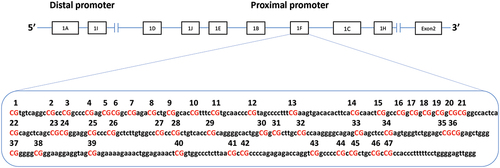
The region known as exon 1 F on the NR3C1 gene promoter contains 47 CpG sites (). It is homologous to region 17 of rodents, described as hypermethylated in puppies subjected to maternal neglect [Citation23] and has been drawing special attention for that. The results in rodents support the idea that epigenetic alterations in CpG sites of exon 1 F can alter transcription. At least four transcription factors have been identified to bind CpG sites at the exon 1 F region in NR3C1, including activator protein (AP-1), specificity protein (Sp-1), glucocorticoid receptor DNA binding factor 1 (GRF-1), and nerve growth factor (NGFI-A) [Citation24,Citation25]. Regarding NGFI-A, in vitro studies have demonstrated increased site-specific methylation of the exon 1 F of the NR3C1 in suicide victims with a history of childhood abuse [Citation25]. Interestingly, the differential methylation of the human NR3C1 diminished NGFI-A binding and NGFI-A-induced gene transcription, indicating a relationship between gene expression, differences in methylation status, and transcription factor binding [Citation25]. Turecki and Meaney [Citation8] carried out a systematic review including 40 case-control or cohort studies with a control group since 2004, 13 in animals and 27 in humans. They sought to correlate childhood adversities and the methylation of the NR3C1. Seven of the human studies considered perinatal stress, but only three of them followed the inclusion criteria of the present systematic review [Citation26–28]. In 89% of human studies and 70% of animal studies, increased methylation of the NR3C1 gene has been reported in individuals who have experienced traumatic childhood experiences. Exon 1 F was the most studied in humans.
The foetal period and the first 2 years of life are critical for developing and modulating brain architecture. The so-called first 1000 days represent a phase of significant vulnerability and susceptibility to external stimuli, whether positive or negative. Imbalance in the functioning of genes at this stage can cause diseases in adulthood, such as alcoholism, depression, diabetes, and cardiovascular problems [Citation29,Citation30]. Psychological or environmental stress during pregnancy or neonatal period, such as maternal depression, anxiety, malnutrition or violence [Citation31–33], as well as premature birth and admission to the Neonatal Intensive Care Unit (NICU) [Citation34–36], can generate intense and toxic stress. With technological advances in the perinatal area, more and more premature newborns survive and need complex and invasive therapies, with long periods of hospitalization in NICU [Citation37]. It is a hostile environment for the newborn since they remain away from the mother and undergo various procedures, often painful [Citation38,Citation39]. Studies on the association of methylation of the NR3C1 gene in newborns with perinatal stress and early neurological and behavioural development are still scarce. Small samples and wide variation in the studied genetic sites’ amplitude make it difficult to interpret the results.
This systematic review aims to synthesize the existing data on the association between stress during the perinatal period and methylation of the exon 1 F region of the NR3C1 gene promoter in newborns.
Materials and methods
We conducted this systematic review according to the recommendations of PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) [Citation40].
Bibliographic research
Extensive bibliographic research was carried out up to 31 January 2020, in the databases of MEDLINE, SCOPUS, Scielo, LILACS, and Web of Science, without date or language restriction. According to the PICO strategy, we define the following criteria were to answer the review question: (P) participants – pregnant women and/or newborns; (I) intervention (exposure) – perinatal stress (during pregnancy or in the neonatal period); (C) comparison – not applied and (O) outcome – methylation of the exon 1 F region of the NR3C1 gene. We used the following terms and their synonyms in the search strategy: preterm OR newborn OR infant OR child OR pregnant women OR prenatal AND epigenetic OR methylation (complete strategy is available in Supplementary material).
Selection of studies and data extraction
Two independent reviewers (GC and MBGO) selected the articles according to the inclusion and exclusion criteria in three stages: first by the title, then reading the abstract, and finally reading the full text. Then the same reviewers performed the data extraction in their forms. At the end of each stage, the reviewers discussed the divergences concerning selecting and extracting data to obtain consensus. A third reviewer (MCMB) solved disagreements between the two reviewers.
Inclusion and exclusion criteria
Articles should meet all the following inclusion criteria: 1) original articles with any design; 2) addressing stress in the perinatal period, during pregnancy, or in the neonatal period; 3) assessing methylation of exon 1 F of the NR3C1 gene promoter in biological samples of newborns or very young infants admitted to the NICU. Exclusion criteria: 1) systematic or other reviews; 2) assessing only biological material from a pregnant woman or placenta; 3) not including the NR3C1 gene among the evaluated genes.
Studies analysing methylation only in placenta were not included in this review. As a maternal-foetal organ, different patterns of methylation were reported and not comparable to those in cord blood [Citation41–43].
Quality assessment of included studies
Three independent reviewers (GC, MCMB, and MBGO) assessed the studies’ methodological quality and elaborated a final consensus. The studies were evaluated for the risk of bias using the instruments available on the US National Institute of Health (https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools), selected according to the design of the included studies: observational, cohort, or cross-sectional studies. Each risk of bias related to selection, measurement, confounding, and specification was independently classified as low, moderate, or high. Control for confounders and adjustment for multiple tests were taken into account. Each study’s final methodological quality was assigned as low, moderate, or high, considering the amount and magnitude of all types of risk of bias. This review’s protocol was submitted to the International Prospective Registry for systematic reviews in November 2019 under the number 158,285, and the title ‘Perinatal stress and methylation of the NR3C1 gene in newborns: Systematic Review.’
Data synthesis
The data were synthesized and presented as figures and tables.
Results
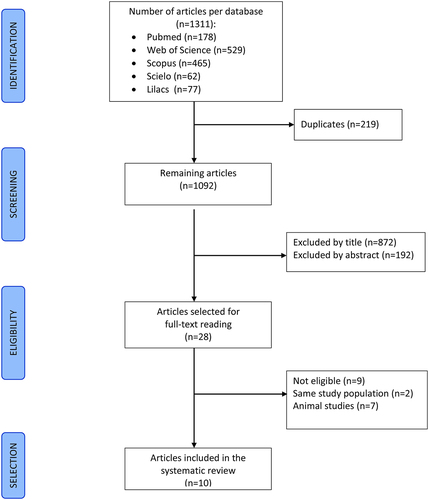
The search strategy generated 1311 articles. After excluding duplicates and reading the title and abstract, 28 articles remained for full-text reading. Eighteen studies were further excluded: seven performed on animals, nine not eligible, two covering the same populations of other studies. Ten articles to synthesize results ().
The reviewers assessed the methodological quality as low in four studies [Citation27,Citation36,Citation44,Citation45] and moderate in six [Citation26,Citation28,Citation34,Citation46–48] (). Due to the great heterogeneity in the way of identifying the studied CpG sites in the studies included in this review, we performed a mapping of the exon 1 F sites reported by each author and created a standard numbering from 1 to 47, indicating the correspondence with the sites referred in the articles (). Next, we will always refer to these sites according to this standard numbering.
Table 1. Methodological quality assessment of articles included in the systematic review.
Six studies evaluated the association of methylation in exon 1 F of the NR3C1 gene at birth with adverse exposures during pregnancy (maternal anxiety and depression) [Citation26–28,Citation44–46]; three with adverse neonatal exposures (morbidities in premature newborns admitted to the NICU) [Citation34,Citation36,Citation47] and one with foetal growth and birth weight in term newborns [Citation48] ().
Table 2. Characteristics of the studies on the association of NR3C1 methylation in newborn infants and perinatal condition.
Methylation analysis in newborns was performed predominantly in umbilical cord blood, but buccal swab and peripheral blood were also used. The most used technique for analysing the methylation of the NR3C1 gene was pyrosequencing using bisulphite-treated genomic DNA. All studies examined the region of exon 1 F of the NR3C1 gene promoter, but there was great variation in the number and sites analysed. Hompes et al. (2013) also evaluated exons 1B and 1D [Citation28] and Non et al. (2014) conducted a genome-wide study [Citation44], but these other genes were not the focus of the presente review ().
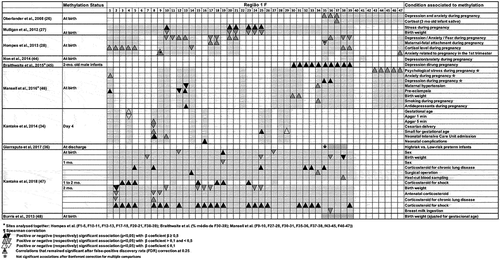
shows the association between methylation of CpG sites of exon 1 F and maternal or infant condition in the perinatal period. Most studies analysed the association between variables through linear regressions. For the purpose of comparison between the studies, we arbitrarily stratified the strength of the association in three categories: as β ≤ 0.1, β > 0.1 and < 0.5 and β ≥ 0.5. Hypermethylation of various CpG sites was the most frequent finding associated with stressful conditions in the perinatal period; hypomethylation was reported in high-risk preterm newborn with more medical morbidities [Citation36], in association with maternal-foetal attachment during pregnancy [Citation28], breast milk ingestion [Citation47] and birth weight [Citation27,Citation34,Citation47].
Maternal mental condition during pregnancy and NR3C1 DNA methylation in infants
Sites CpG 1 to 29
Three studies on maternal mental condition during pregnancy included sites located in the region from CpG 1 to 29 of exon 1 F in their methylation analysis in the cord blood.
Methylation rate at sites 10, 20, 21, 23, 24 and 25 were positively associated with intense stress in the prenatal period and negatively associated with birth weight [Citation27]; In bivariate analysis, mood in pregnancy (anxiety or depression or fears related to childbirth/foetal well-being) was positively associated with methylation rate at sites 1–5, 9, 12–13, and 20–21 and negatively associated with methylation rate at sites 10–11, and 17–18, but after false-positive discovery rate (FDR) correction at 0.25 to deal with multiple tests, only the correlation between hypermethylation at site 9 and anxiety related to pregnancy remained significant [Citation28]. Hypermethylation at site 1 and 13 were respectively associated with anxiety and antidepressants use during pregnancy, but none of these associations remained significant after Bonferroni correction for multiple comparisons; hypermethylation at both sites (Citation1 and Citation13) were also associated with pre-eclampsia, while hypomethylation at site 12 and 13 were respectively associated with pre-eclampsia and maternal hypertension [Citation46] ().
Briefly, methylation rate at sites 10, 13, 20 and 21 in the cord blood were associated with stressful conditions during pregnancy in at least two studies [Citation27,Citation28]. Hypermethylation prevailed, but hypomethylation was mainly associated to birth weight [Citation27].
Sites 30 to 39
Six studies analysed methylation rate at sites included in the region from CpG 30 to 39 of exon 1 F, five in the cord blood [Citation26–28,Citation44,Citation46] and one in the newborn buccal swabs [Citation45].
Hypermethylation at sites 30 to 39, analysed together, in male infants at two months of age was significantly associated with depression during pregnancy [Citation45]. One or more of the sites 35, 36 and 37 were hypermethylated in newborns in association with one or more conditions during pregnancy, such as depression and/or anxiety and/or fears related to childbirth/foetal well-being [Citation26,Citation28,Citation45,Citation46] while hypermethylation at 34 was associated with maternal clinical conditions, such as smoking during pregnancy and hypertension [Citation46]. Hypermethylation at site 37 was associated with increase in cortisol level in the infant in response to stress at 3 months of age [Citation26], while hypermethylation at sites 38 and 39 was associated with increase in cortisol level during pregnancy [Citation28]. Hypomethylation at site 35 was associated with a good maternal-foetal bond in the first trimester of pregnancy and hypomethylation at sites 38 and 39 was associated with fear of changes during pregnancy [Citation28], although this last association could not stand FDR correction at 0.25 for multiple tests.
Briefly, methylation rate at sites 35 to 39 in the cord blood or infant buccal swab was more often positively associated with negative maternal psychological or clinical conditions during pregnancy in at least four studies [Citation26,Citation28,Citation45,Citation46], although not always in the same direction. Hypomethylation at site 35 was associated with a positive maternal attitude [Citation28].
Sites CpG 40 to 47
Methylation at one or more sites in this region were evaluated in four studies [Citation26,Citation28,Citation44,Citation46] and altered in two [Citation28,Citation46]. Hypermethylation at sites 43 to 45 and 46–47 analysed together were observed in mothers under psychological stress during pregnancy [Citation46], and hypermethylation at site 47 in mothers with fear of changes in the third trimester of pregnancy [Citation28], though this last association could not stand FDR correction at 0.25 ().
Briefly, hypermethylation at site 47 associated with negative maternal psychological condition during pregnancy was reported in at least two studies [Citation28,Citation46].
Neonatal parameters or morbidities and NR3C1 methylation in infants
Sites CpG 1 to 39
Four studies analysed associations between neonatal parameters or morbidities and methylation rate at sites located in the region from CpG 1 to 39 [Citation34,Citation36,Citation47,Citation48]. At birth, methylation rate in preterm newborns admitted to the NICU was lower at sites 1, 5, 8 and higher at site 4, compared to healthy term newborns, but no significant correlations were observed between those sites and prenatal parameters, such as gestational age, birth weight, SGA, antenatal steroid, and mode of delivery [Citation34]. In preterm newborns with chronic lung disease (CLD), methylation rate at sites 12,19 and 27 at birth were associated with sex in variable directions and methylation rate at sites 25, 36 and 38, with birth weight, in negative direction [Citation47]. In term newborns, no associations between methylation rate at sites 35 to 39 and birth weight were observed [Citation47]. At day 4, methylation rate was significantly higher in preterm newborns admitted to the NICU compared to healthy term newborns. Increase at 11 CpG sites (sites 1, 2, 8, 9, 10, 14, 16, 25, 26, 28, 29) and decrease at site 4 were observed between birth and the 4th day of life, while term newborns kept methylation rates stable in the period [Citation34]. Some of these changes (day 4 to day 1 methylation ratio) were associated with several perinatal parameters: CpG4 positively associated with gestational age and negatively associated with Apgar at 1 min; CpG 8 positively associated with Apgar at 5 min, small for gestational age (SGA), and admission to the NICU and negatively associated with caesarian delivery; CpG 10 positively associated with admission to the NICU, and CpG 25 and 29 positively associated with SGA. Hypermethylation at CpG 16 on day 4 was positively associated with neonatal complications [Citation34]. In premature newborns with CLD, between birth and 1 month, methylation rates were stable at sites 1 to 39. At the age of 1 month, hypomethylation at site 19 was associated with sex and hypermethylation at sites 5, 8, 25, 32, 33 and 35, with corticosteroid treatment for chronic lung disease (CLD); between 1 and 2 months of age, methylation rate increased at nine sites (CpG 3, 4, 5, 11, 15, 18, 19, 23 and 27) and hypermethylation at sites 3, 4, 5, 15, 18, 19 and 23 were associated with corticosteroid for shock; at 2 months of age, hypermethylation at 22 of the 39 sites studied (Citation2–5, Citation8–10, Citation12, Citation15–19, Citation23–26, Citation31–34, and Citation39) were associated with the use of corticosteroids for shock [Citation47]. In this same population of preterm newborns with chronic lung disease, hypomethylation at several sites were associated with the following parameters: sites 2, 10, 15, 16, 17, 24 and 25 with birth weight; sites 2, 8 and 9 with the use of antenatal corticosteroids and site 36 with breastfeeding [Citation47] (). Hypomethylation at site 35 was also observed in high-risk compared to low-risk preterm infants classified by the Neonatal Therapeutic Intervention Score System (NTISS) score at discharge from the NICU [Citation36].
Briefly, in at least two studies hypermethylation at sites 4, 8, 10, 16 and 25 [Citation34,Citation47], and changes in methylation rate in both directions at site 35 [Citation36,Citation47] were associated with parameters or morbidities during the first months of life in preterm newborns admitted to the NICU. Hypermethylation associated to negative neonatal conditions prevailed, while hypomethylation at various sites was associated with good or protective conditions.
Discussion
In this review, we addressed epigenetic changes in the NR3C1 gene in newborns and very young infants, specifically the methylation at CpG sites in the 1 F region, related to perinatal stress, whether in the foetal period or throughout the first months of life. Methylation changes were reported in 41 of the 47 CpG sites of exon 1 F. In general, hypermethylation was the predominant change associated with stressful situations, while hypomethylation was more often associated with protective conditions or better outcomes, such as good maternal-foetal bonding, breast milk ingestion and higher birth weight. However, we could not define very consistent patterns of site-specific changes associated with maternal or infant perinatal conditions. Several sources of heterogeneity among studies, such as the type and methods of measuring exposure and outcome, number and region of CpG sites analysed, type of tissue sample used, methods of methylation analysis and presentation of findings hindered the comparison and synthesis of results.
The adversities experienced by the mother during pregnancy and the possible epigenetic repercussions on the foetus and childhood are a topic widely studied due to its importance for child neurodevelopment [Citation49]. Since the pioneering study by Weaver et al. (2004) in rodents, associating the intensity of maternal care with changes in methylation at sites 17 and adjacent, homologous to CpG sites 35, 36, and 37 in humans [Citation23], several studies have focused on exon 1 F of the NR3C1 gene, particularly in this region. Methylation of exon 1 F NR3C1 promotor can affect NGFI-A-induced gene transcription, since canonical NGFI-A binding sites are between −3215 and −3204 (concerning the ATG translation initiation site), containing sites 37 and 38 and also −3361 to 3345, which includes 16 to 21. The noncanonical binding-sites located between −3405 and −3390, contains CpG-units 12 and 13 and the second noncanonical binding site, located between −3276 and −3250, contains CpG-units 30 to 32 [Citation50]. In the present review, hypermethylation at sites 35 to 39 in newborns or very young infants were associated with depression and/or anxiety in four studies [Citation26,Citation28,Citation45,Citation46], but hypomethylation at site 38 was also reported in one study [Citation28]. Methylation changes in cord blood in the region containing sites 12, 13 and 16 to 21 were associated with negative maternal psychological or clinical conditions during pregnancy in three studies [Citation27,Citation28,Citation46], but the direction varied. These contradictory directions may be partially explained by the great heterogeneity in the assessment of maternal well-being during pregnancy, not only regarding the number of times (one or more times) and the moment of the assessment (first, second, third trimester, or at delivery), but also the assessed condition (depression/anxiety or extreme stress, such as war situations or very unfavourable socioeconomic conditions) and the different measurement scales used.
The NICU represents a highly hostile environment for the newborn. They remain away from their mother and family, receive invasive therapies, undergo numerous painful procedures under excessive handling and continuous light and sound stimuli. Many have transient adrenal insufficiency in the first week of life, and around the 14th day, there is adaptation and adequate response of the HPA axis. Neonates who suffer prolonged intrauterine hypoxaemia or hypoxic-ischaemic encephalopathy may remain with elevated cortisol levels for extended periods [Citation51]. Premature birth and NICU admission are associated with methylation of genes related to neurological and behavioural development, compared to full-term newborns [Citation52,Citation53], especially in those exposed to more painful procedures [Citation54] and those with greater severity [Citation34,Citation47], in addition to reducing the volume of brain regions on MRI [Citation55–57]. Imaging studies have shown that premature newborns admitted to the NICU [Citation58] and children victims of abuse, neglect, and extreme poverty [Citation59] may present a reduction in the parietal and frontal regions, areas rich in glucocorticoid receptors, which can cause emotional problems [Citation6]. On the other hand, animal studies suggest that quality maternal care can reverse methylation associated with stress [Citation60].
The results of the present review corroborate these literature findings. Hypermethylation associated with neonatal comorbidities predominated at several sites in the region involving sites 1 to 39. Remarkably, hypomethylation at some sites were associated with positive conditions, suggesting protective effects: sites 35 and 36 respectively associated with good maternal-foetal bonding during pregnancy [Citation28], and breast milk ingestion [Citation47]; hypomethylation at various sites associated with better birth weight [Citation27,Citation47] and first minute Apgar [Citation34]. However, contradictory methylation changes at site 35 was also reported in association with infant conditions: hypomethylation in sicker preterm infants [Citation36] and hypermethylation in preterm infants receiving corticosteroids for CLD [Citation47]. We can point out several differences between these two studies: Kantake et al. (2018) performed a retrospective longitudinal analysis of methylation at 39 sites in premature newborns with CLD, at birth, one and 2 months of age associated with clinical conditions [Citation47]. Giarraputo et al. (2017) conducted a cross-sectional analysis on the association between methylation at four sites (CpG 35–39) and high versus low-risk preterm newborns, classified according to the Neonatal Therapeutic Intervention Score System (NTISS) [Citation61]; both conditions (methylation and severity) were assessed at the same time, before hospital discharge [Citation36]. However, in this study, both groups were of preterm newborns admitted to the NICU, different from studies that compared preterm versus term newborns. Besides, the moment of hospital discharge and, therefore, of methylation assessment was very different among premature infants classified as high and low risk (mean 99 days x 44 days of life, respectively). If epigenetic changes are transient and some factors can reverse them [Citation62], other analyzes at similar hospitalization times should have been performed to compare the groups. Also, there were differences in the type of tissue collected (cord blood, peripheral blood, and saliva). Peripheral tissues are more accessible than brain, and in general the observed genomes can be compared [Citation63]. However, the use of peripheral tissues and different cell types as brain methylation markers is controversial. Several studies have reported a correlation between methylation levels in peripheral blood [Citation64,Citation65], buccal cells [Citation66], saliva [Citation67], umbilical cord blood [Citation68] and the brain. Otherwise, some authors observed great variation in the methylation of different CpG sites and different genes comparing peripheral and cerebral tissues [Citation69,Citation70]. Besides, variations of DNA methylations in different cell types and tissues of the same individual and between different individuals [Citation69,Citation71,Citation72], in addition to factors such as changes in blood T cell count and macrophages [Citation73] and response to external stimuli, such as drug use, must be considered [Citation74].
The NR3C1 gene plays a central role in regulating the hormonal stress response. Understanding the relationship between epigenetic changes and the balance of the HPA axis and glucocorticoid metabolism is essential for understanding long-term psychological, emotional disorders and diseases [Citation75]. The most acceptable theory is that prenatal psychological conditions impact on foetal development via increased maternal cortisol levels. In conditions such as depression, anorexia nervosa, and panic syndrome, the hormones produced by the hypothalamus, pituitary and adrenal remain increased in the systemic circulation [Citation76,Citation77]. Increased exposure to cortisol in foetuses can cause an imbalance in the HPA axis [Citation78] and epigenetic changes in DNA involving several genes related to its functioning, such as NR3C1, FKBP5, SLC6A4, 11B-HSD2, and others [Citation49]. However, results on the association between maternal depression and cortisol levels in pregnancy are still controversial [Citation79]. Three studies included in this review measured cortisol in mothers [Citation28,Citation45] or babies [Citation26]. Association between its increase and hypermethylation, especially in the region of the sites 37 [Citation26], 38 and 39 [Citation28] was found only in two studies. Moreover, maternal psychological conditions were not associated with cortisol levels during pregnancy [Citation28,Citation45], suggesting that cortisol did not mediate the effects of maternal mood on epigenetic outcomes in the newborn. Cortisol levels were not measured in newborns admitted to the NICU in the studies included in this review. It would be necessary for better understanding the relationship between disease severity in the neonatal period, hormonal imbalance, and epigenetic changes [Citation34,Citation36,Citation47]. Kantake et al. (2018) demonstrated a strong positive association between the postnatal use of glucocorticoid for CLD or circulatory shock in preterm infants and increased methylation at various CpG sites of exon 1 F of the NR3C1 gene. On the other hand, antenatal glucocorticoid administration had a moderate negative effect on NR3C1 gene methylation. The authors suggest that glucocorticoid demand due to relative adrenal insufficiency may result in NR3C1 methylation, not glucocorticoid administration [Citation47].
Maternal risk factors such as age, multiple pregnancies, infections, chronic diseases such as high blood pressure and diabetes, nutritional status, smoking, alcohol, and drug use can contribute to premature birth and epigenetic changes in the offspring [Citation80–82]. In the present review, preeclampsia, maternal arterial hypertension and smoking during pregnancy were associated with methylation changes [Citation46]. Birth conditions and newborn characteristics such as gender, birth weight, gestational age, type of delivery, Apgar, admission to the NICU were also factors with repercussions on the epigenome [Citation27,Citation34,Citation45,Citation47,Citation83]. Animal and human studies suggest a greater male epigenetic vulnerability in response to prenatal and postnatal experiences, considered a critical factor of stress impact on long-term pregnancy [Citation84–86] which is corroborated by the findings of one study in this review, that was the association between depression in pregnancy and hypermethylation only in male infants [Citation45].
This review has strengths and limitations. The lack of standardization in the designation of CpG sites in the 1 F region of the NR3C1 gene hindered the initial understanding and comparability of the results of the sites studied by the various authors. The careful mapping of the sites referred to in each article and standardization of these sites’ numbering represents a contribution to the understanding of the relationship between adverse perinatal conditions and changes in the methylation of the NR3C1 gene. The main limitation was the great heterogeneity among the studies, related to several aspects, such as design, exposures and outcomes, varied scales for their measurement, some not validated, as well as the moment and the number of times they were applied, location and number of the analysed CpG sites, type of tissue sample used, methods of methylation analysis and way of reporting findings. All these sources of variability made it difficult to carry out a meaningful synthesis of the results. The wide diversity of cell types, all with great transcriptional activity of the NR3C1 gene, and possible effects that can go in opposite directions of activation or silencing its expression according to its function in the organism’s different cells make the comparability between the findings a complex task. Besides, most of the existing studies are of limited methodological quality. No study reported sample size justification, power description nor precision estimates. Moreover, considering the observational nature of the studies and the possibility of intergenerational transmission of epigenetic changes, we cannot be sure about all confounders involved nor about the direction of the association (exposure vs. outcome), so that results do not yield strong enough evidence to support causal inferences. Finally, we must consider the possibility of positive publication bias, since only two of ten studies reported no associations.
Many questions still await evidence-based answers and may be the subject of future research. Few studies investigating the association between adversities, changes in the response of the HPA axis and methylation of the NR3C1 gene have measured cortisol levels so that the association between endocrine and neural imbalances, epigenetic alterations and diseases can be established. Are these epigenetic changes really mediated by circulating cortisol? Should other potential mediators be considered in future research? Differences in DNA methylation may be related to inter-individual and intra-individual tissue and cell type difference. How to interpret DNA methylation results observed in different peripheral tissues? Can adversity trigger specific methylation patterns in different cell types and tissues? Can the intensity of the stress experienced generate epigenetic changes in multiple tissues? Future research should aim to standardize cell types and peripheral tissues that better represent the central nervous system, as well as the identification of the sites, facilitating the comparability of the results. Besides, statistical adjustments of the cellular composition of the samples would increase the methodological quality of NR3C1 gene methylation studies. Very few studies evaluate the impact of admission to the NICU, one of the earliest, most intense, and prolonged negative experiences in an individual’s life. What is the impact of prematurity and admission to the NICU on the epigenome? Do the early observed epigenetic changes persist throughout childhood and into adulthood, or are they transitory? What factors could reverse the epigenetic marks caused by stressful conditions experienced in the foetal and neonatal period? Moreover, transmission of methylation profiles from parents to the offspring must also be considered. Few studies have an intergenerational approach. Research on stress in the perinatal period assess changes in DNA at birth or during pregnancy. Do the methylation sequences observed in pregnant women and newborns, represent maternal or paternal marks transmitted to the offspring or constitute new marks acquired by them? Epigenetic mechanisms and changes can be considered excellent candidate biomarkers and promising tools for diagnosis and prognosis. The scientific literature today is enriched by studies that associate DNA methylation with clinical parameters, but they are still unable to extrapolate most of biomarkers to clinical practice. Which site-specific methylation change can predict a particular outcome? Which protective intervention can mitigate it? Finally, research on epigenetic alterations in specific genes and sites provides detailed, but sometimes confusing and contradictory results. Will genome-wide research be more enlightening, by revealing the possible interactions between different genes?
In conclusion, we are still at the frontier of knowledge on the impact of adverse perinatal conditions on epigenetic changes, affecting the lives of individuals and their offspring in the short, medium, and long term. In this review, we were interested in answering how perinatal stress affects methylation of the NR3C1 gene in newborns. Understanding how stressors or protective conditions affect the methylation of this gene in a phase of intense brain development and knowledge acquisition may help reduce short and long-term morbidities. Other reviews in this topic included other genes or analysed only maternal tissues, like blood or placenta, which do not reflect methylation in the newborn and may confound results. Moreover, in these previous studies, each investigator identified the CpG sites in their own way, making it difficult to understand the role of methylation at specific sites. We thought that narrowing the focus on the methylation of specific CpG sites and standardizing the designation of the already studied sites could bring some new light. Although this review does not make it possible to conclude about specific CpG sites, we believe that it contributes to corroborating evidence that adversities experienced in the perinatal period are associated with epigenetic changes in exon 1 F of the NR3C1 gene. Still, many gaps remain to be elucidated in future research. We believe that the standardization presented in this review may help to systematize the description of future results.
Authors’ contribution
GC, MCMB and APB conceived and designed the study. GC, MBGO and MCMB conducted the selection of articles, data extraction and assessment of the methodological quality of the studies. GC, AAB, MMR and MCMB drafted the manuscript. MMR, MCMB, MSS, AJLAC and APB critically reviewed the manuscript. All authors approved the final version of the manuscript.
Disclosure of interest
The authors report no conflict of interest.
Financial support:
This study received no financial support.
Supplemental Material
Download MS Word (16.1 KB)Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
- Epigenetics: HR. A historical overview. Epigenetics. 2006;1(2):76–80.
- Van Der Knaap LJ, Riese H, Hudziak JJ, et al. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence the TRAILS study. Transl Psychiatry. 2014;4(4):e381.
- Romens SE, Mcdonald J, Svaren J, et al. Associations between early life stress and gene methylation in children. Child Dev. 2015;86(1):303–309.
- Hatfield LA, Hoffman RK, Polomano RC, et al. Epigenetic modifications following noxious stimuli in infants. Biol Res Nurs. 2018;20(2):137–144.
- Papadopoulou Z, Vlaikou A-M, Theodoridou D, et al. Stressful newborn memories: pre-conceptual, in utero, and postnatal events. Front Psychiatry. 2019;10:220. doi:10.3389/fpsyt.2019.00220. eCollection 2019.
- Johnstone SE, Baylin SB. Stress and the epigenetic landscape : a link to the pathobiology of human diseases? Nat Publ Gr. 2010;11(11):806–812.
- Turecki G, Meaney MJ. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: a Systematic Review. Biol Psychiatry. 2016;79(2):87–96.
- Peña CJ, Nestler EJ. Progress in Epigenetics of Depression. Prog Mol Biol Transl Sci. 2018;157:41–66.
- Murgatroyd C, Spengler D. Epigenetics of early child development. Front Psychiatry. 2011; 2:16.doi: 10.3389/fpsyt.2011.00016. eCollection 2011.
- Van Bodegom M, Homberg JR, Henckens MJAG. Modulation of the hypothalamic-pituitary-adrenal axis by early life stress exposure. Front Cell Neurosci. 2017 Apr 19;11:87.doi: 10.3389/fncel.2017.00087. eCollection 2017 .
- Palma-gudiel H, Leza JC. Glucocorticoid receptor gene (NR3C1) methylation processes asmediators of early adversity in stress-related disorders causality: a critical review. Neurosci Biobehav Rev. 2015;55:520-35.
- Kamin HS, Kertes DA. Cortisol and DHEA in Development and Psychopathology. Horm Behav. 2017;89:69-85. .
- Stetler C, Miller GE. Depression and Hypothalamic-Pituitary-Adrenal Activation : a Quantitative Summary of Four Decades of Research. Psychosom Med. 2011;73(2):114–126.
- Efstathopoulos P, Andersson F, Melas PA, et al. NR3C1 hypermethylation in depressed and bullied adolescents. Transl Psychiatry. 2018;8(1):121. doi:10.1038/s41398-018-0169-8.
- Seckl JR. 11 b -hydroxysteroid dehydrogenases : changing glucocorticoid action. Curr Opin Pharmacol. 2004;4(6):597–602.
- Grad I, Picard D. The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol. 2007;275(1–2):2–12.
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones *. Endocrinol Rev. 1997;18(3):306–360.
- Kirschke E, Goswami D, Southworth D, et al. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157(7):1685–1697.
- Smoak KA, Cidlowski JA. Mechanisms of glucocorticoid receptor signaling during inflammation. Mech Ageing Dev. 2004;125(10–11):697–706.
- Argentieri MA, Nagarajan S, Seddighzadeh B, et al. Epigenetic Pathways in Human Disease: the Impact of DNA Methylation on Stress-Related Pathogenesis and Current Challenges in Biomarker Development. EBioMedicine. 2017;18:327–350.
- Labonte B, Yerko V, Gross J, et al. Differential glucocorticoid receptor exon 1 B, 1 C, and 1 H expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72(1):41–48.
- Weaver ICG, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854.
- Turner JD, Alt SR, Cao L, et al. Transcriptional control of the glucocorticoid receptor: cpG islands, epigenetics and more. Biochem Pharmacol. 2010;80(12):1860–1868.
- McGowan PO, Sasaki A, D’Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348.
- Oberlander TF, Weinberg J, Papsdorf M, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106.
- Mulligan CJ, D’Errico NC, Stees J, et al. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7(8):853–857.
- Hompes T, Izzi B, Gellens E, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res. 2013;47(7):880–891.
- Center on the Developing Child at Harvard University. From Best Practices to Breakthrough Impacts: A Science-Based Approach to Building a More Promising Future for Young Children and Families. [Internet]. 2016. Available from: http://www.developingchild.harvard.edu. Accessed on September 6, 2021.
- National Scientific Council on the Developing Child. Early Experiences Can Alter Gene Expression and Affect Long-Term Development: Working Paper No. 10. [Internet]. 2010. Available from: http://www.developingchild.harvard.edu. Accessed on September 6, 2021. h ttp:// do i.wiley.co m/10.11 11/j.11 51-2 91 6.191 8.tb172 32.x
- Cardenas A, Faleschini S, Hidalgo AC, et al. Prenatal maternal antidepressants, anxiety, and depression and offspring DNA methylation: epigenome-wide associations at birth and persistence into early childhood. Clin Epigenetics [Internet]. 2019 Mar [cited 2019 Oct 24];11. Available from 10.1186/s13148-019-0653-x
- Conradt E, Adkins DE, Crowell SE, et al. An epigenetic pathway approach to investigating associations between prenatal exposure to maternal mood disorder and newborn neurobehavior. Dev Psychopathol. 2018;30(3):881–890.
- Vidal AC, Neelon SEB, Liu Y, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet Epigenet. 2014;6:37–44.
- Kantake M, Yoshitake H, Ishikawa H, et al. Postnatal epigenetic modification of glucocorticoid receptor gene in preterm infants: a prospective cohort study. BMJ Open. 2014;4(7):e005318.
- Provenzi L, Montirosso R. Epigenethics in the neonatal intensive care unit: conveying complexity in health care for preterm children. JAMA Pediatr. 2015;169(7):617–618.
- Giarraputo J, DeLoach J, Padbury J, et al. Medical morbidities and DNA methylation of NR3C1 in preterm infants. Pediatr Res. 2017;81(1):68–74.
- Blencowe H, Cousens S, Chou D, et al. Born Too Soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013;10(S1/S2):1–14.
- Casavant SG, Cong X, Moore J, et al. Associations between preterm infant stress, epigenetic alteration, telomere length and neurodevelopmental outcomes: a systematic review. Early Hum Dev. 2019;131:63–74.
- Casavant SG, Cong X, Fitch RH, et al. Biomarkers of Stress in the Preterm Infant: an Integrative Review. Biol Res Nurs. 2019;21(2):210–223.
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:7.
- De Carli MM, Baccarelli AA, Trevisi L, et al. Epigenome-wide cross-tissue predictive modeling and comparison of cord blood and placental methylation in a birth cohort. Epigenomics. 2016;9(3):231–240.
- Wang X-M, Tian F-Y, Fan L-J, et al. Comparison of DNA methylation profiles associated with spontaneous preterm birth in placenta and cord blood. BMC Med Genomics. 2019;12(1):1–14.
- Ma B, Allard C, Bouchard L, et al. Locus-specific DNA methylation prediction in cord blood and placenta. Epigenetics. 2019;14(4):405–420.
- Non AL, Binder AM, Kubzansky LD, et al. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics. 2014;9(7):964–972.
- Braithwaite EC, Kundakovic M, Ramchandani PG, et al. Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics. 2015;10(5):408–417.
- Mansell T, Vuillermin P, Ponsonby AL, et al. Maternal mental well-being during pregnancy and glucocorticoid receptor gene promoter methylation in the neonate. Dev Psychopathol. 2016;28(4):1421–1430.
- Kantake M, Ohkawa N, Iwasaki T, et al. Postnatal relative adrenal insufficiency results in methylation of the glucocorticoid receptor gene in preterm infants: a retrospective cohort study. Clin Epigenetics. 2018;10(1):66. eCollection 2018 doi:10.1186/s13148-018-0497-9.
- Burris HH, Braun JM, Byun H-M, et al. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics. 2013;5(3):271–281.
- Sosnowski DW, Booth C, York TP, et al. Maternal prenatal stress and infant DNA methylation: a systematic review. Dev Psychobiol. 2018;60(2):127–139.
- McGowan PO, Sasaki A, D’Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348.
- Ng PC. Effect of stress on the hypothalamic-pituitary-adrenal axis in the fetus and newborn. J Pediatr. 2011;158(2):e41–3.
- Arpón A, Milagro FI, Laja A, et al. Methylation changes and pathways affected in preterm birth: a role for SLC6A3 in neurodevelopment. Epigenomics. 2018;10(1):91–103.
- Sparrow S, Manning JR, Cartier J, et al. Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Transl Psychiatry. 2016;6(1):e716.
- Provenzi L, Fumagalli M, Sirgiovanni I, et al. Pain-related stress during the Neonatal Intensive Care Unit stay and SLC6A4 methylation in very preterm infants. Front Behav Neurosci. 2015;9:99. doi:10.3389/fnbeh.2015.00099.eCollection 2015.
- Montirosso R, Provenzi L, Fumagalli M, et al. Serotonin Transporter Gene (SLC6A4) Methylation Associates With Neonatal Intensive Care Unit Stay and 3-Month-Old Temperament in Preterm Infants. Child Dev. 2016;87(1):38–48.
- Fumagalli M, Dessimone F, Sirgiovanni I, et al. From early stress to 12-month development in very preterm infants: preliminary findings on epigenetic mechanisms and brain growth. PLoS One. 2018;13(1):1–15.
- Lester BM, Marsit CJ, Giarraputo J, et al. Neurobehavior related to epigenetic differences in preterm infants. Epigenomics. 2015;7(7):1123–1136.
- Ranger M, Chau CMY, Garg A, et al. Neonatal Pain-Related Stress Predicts Cortical Thickness at Age 7 Years in Children Born Very Preterm. PLoS One. 2013;8(10):1–12.
- Hanson JL, Nacewicz BM, Sutterer MJ, et al. Behavioral Problems After Early Life Stress: contributions of the Hippocampus and Amygdala. Biol Psychiatry. 2015;77(4):314–323.
- Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28(9):456–463.
- Gray JE, Richardson DK, McCormick MC, et al. Neonatal Therapeutic Intervention Scoring System: a therapy-based severity-of-illness index. Pediatrics. 1992;90(4):561–567.
- Montirosso R, Provenzi L. Implications of epigenetics and stress regulation on research and developmental care of preterm infants.J Obstet Gynecol Neonatal Nurs. 2015;44(2):174–182
- Byun H-M, Siegmund KD, Pan F, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18(24):4808–4817.
- Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet Part B Neuropsychiatr Genet. 2013;162(7):595–603.
- Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43.
- Lowe R, Gemma C, Beyan H, et al. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8(4):445–454.
- Smith AK, Kilaru V, Klengel T, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence for tissue specificity and relatedness to brain. Am J Med Genet Part B-NEUROPSYCHIATRIC Genet. 2015;168B(1):36–44.
- Nemoda Z, Massart R, Suderman M, et al. Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Transl Psychiatry. 2015;5:e545.
- Braun PR, Han S, Hing B, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019;9(1):1.
- Walton E, Hass J, Liu J, et al. Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophr Bull. 2016;42(2):406–414.
- Heijmans BT, Mill J. The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2011;6:74–78.
- Ziller MJ, Gu H, Müller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477-481.
- Uddin M, Aiello AE, Wildman DE, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107(20):9470–9475.
- Klengel T, Mehta D, Anacker C, et al. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41.
- TureckiG, Ota VK, Belangero SI, et al. Early life adversity, genomic plasticity, and psychopathology. The Lancet Psychiatry.2014;1(6): 461–6.
- Pervanidou P, Chrousos GP. Early-Life Stress: from Neuroendocrine Mechanisms to Stress-Related Disorders. Horm Res Paediatr. 2018;89(5):372–379.
- Zannas AS, Chrousos GP. Epigenetic programming by stress and glucocorticoids along the human lifespan. Mol Psychiatry. 2017;22(5):640–646.
- Maccari S, Krugers HJ, Morley-Fletcher S, et al. The consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. J Neuroendocrinol. 2014;26(10):707–723.
- Seth S, Lewis AJ, Galbally M. Perinatal maternal depression and cortisol function in pregnancy and the postpartum period: a systematic literature review. BMC Pregnancy Childbirth. 2016;16(5):1–19.
- Knight AK, Smith AK. Epigenetic biomarkers of preterm birth and its risk factors. Genes (Basel). 2016;7(4):1–26.
- Knopik VS, Marceau K, Bidwell LC, et al. Prenatal substance exposure and offspring development: does DNA methylation play a role? Neurotoxicol Teratol. 2019:50–63.
- Joubert BR, Håberg SE, Nilsen RM, et al. 450K Epigenome-Wide Scan Identifies Differential DNA Methyla tion in Newborns Related to Maternal Smoking during Pregnancy. Environ Health Perspect. 2012;120(10):1425–1431.
- Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012;7(7):735–746.
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28(36):1421–1430.
- Goel N, Bale TL. Examining the intersection of sex and stress in modelling neuropsychiatric disorders. J Neuroendocrinol. 2009;21(4):415–420.
- Bale TL. Sex differences in prenatal epigenetic programing of stress pathways. Stress Int J Biol Stress. 2011;14(4): 348–356.