ABSTRACT
Sodium bisulphite conversion of DNA to separate methylated from unmethylated cytosines is a standard for methylation analysis. This study evaluated a direct cell conversion protocol on cervical samples as alternative to isolated genomic DNA as input.
Clinician-collected cervical samples (n = 120) were subjected to a direct conversion protocol, or genomic DNA was isolated with a fixed amount used for subsequent bisulphite conversion. Converted samples were compared for ACTB control gene and methylation of FAM19A4 and miR124-2 genes using quantitative methylation-specific PCR (QIAsure Methylation Test).
Direct conversion resulted in a high success rate, i.e., 119/120 (99.2%) samples reported a valid test result. ΔΔCq values of FAM19A4 and miR124-2 were significantly correlated between both protocols (Spearman Rho 0.708 and 0.763, respectively, all p-values = 0.000). Agreement between both the bisulphite protocols was demonstrated by Bland–Altman plots.
A direct cell conversion protocol shows good technical and analytical performance and offers a streamlined workflow for methylation analysis.
INTRODUCTION
Cervical cancer is the fourth most commonly occurring cancer in women [Citation1]. The usually slow progression from cervical high-risk human papillomavirus (HPV) infection to cancer via precancerous changes called cervical intraepithelial neoplasia (CIN) provides opportunities for prevention and early detection [Citation1]. Cytology-based screening has been a cornerstone of cervical cancer prevention strategies for decades. In recent years, HPV-based screening has been adopted in several countries given a better protection against cervical cancer and precancer than cytology [Citation2]. With the implementation of HPV-based screening, triage testing has become important to increase specificity and positive predictive value, while retaining accurate identification of women with high-grade CIN, who require follow-up management. Epigenetic biomarkers have a strong potential to be implemented as molecular triage tool. At present, the most widely studied epigenetic alteration is the methylation of DNA at CpG dinucleotides (5-methylcytosine), which are usually highly concentrated in CpG islands within the promoter regions of human genes [Citation3]. Studies have reported a gradual increase in DNA methylation of specific host–cell genes with higher grade of CIN, reaching highest levels in cervical cancer [Citation4–6]. A well-studied methylation marker panel with host–cell genes FAM19A4 (currently known as TAFA-4) and miR124-2 showed good triage performance in HPV-positive women [Citation7–9], as recently validated in a large cross-sectional, multicentre European cohort study across four different countries [Citation10].
A commonly used approach for DNA methylation analysis is genomic DNA isolation from a clinical specimen followed by sodium bisulphite treatment prior to PCR amplification. Treating DNA with sodium bisulphite converts unmethylated cytosines to uracil, while methylated cytosines remain unchanged. This conversion thus generates specific changes in the DNA sequence that depend on the methylation status of the individual cytosines and can be measured by subsequent quantitative methylation-specific PCR (qMSP) [Citation11,Citation12]. This protocol could be made more efficient by bisulphite conversion directly on cells, without the need for prior genomic DNA isolation and normalization of DNA input. Here, we evaluated a direct cell conversion protocol on a series of clinician-collected cervical samples. We compared results of ACTB control gene and methylation of FAM19A4 and miR124-2 genes on bisulphite-treated DNA derived from direct cell conversion to those from a protocol involving prior DNA isolation and normalization (reference protocol). Direct cell conversion protocols could further improve efficiency and considerably enhance the practicability and operations of methylation analysis in diagnostic and screening settings.
METHODS
Cervical samples
Clinician-collected cervical samples in PreservCyt solution (Hologic Inc., San Diego, CA, USA) were obtained from the Scottish HPV Archive of the University of Edinburgh, Scotland, according to the Research Tissue Bank status as approved by the Lothian NRS BioResource (15/ES/0094). Aliquots of 2–4 mL were available for this study. In light of the HPV triage application, the series consisted of cervical samples of 120 HPV-positive women, comprising of 14 women with histologically confirmed CIN3 (median age 38; range 32–45; IQR 32–45), 10 women with histologically confirmed CIN2 (median age 33.5; range 25–54; IQR 27.8–42.3) and 96 women defined as controls (median age 35; range 22–63; IQR 28–44), who either had histologically confirmed CIN0 (n = 28) or CIN1 (n = 10) or were considered to have no evidence of CIN2 or worse (CIN2+) as they displayed HPV clearance combined with normal cytology in follow-up (n = 58).
DNA isolation
Extraction of DNA from 1 mL PreservCyt sample was performed using an automated extraction system (NucleoMag 96 tissue kit, Macherey-Nagel GmbH&Co. KG, Düren, Germany) and a Microlab Star robotic system (Hamilton, Gräfelfing, Germany) according to manufacturer’s protocol. The concentration of extracted DNA was measured using a Qubit fluorometer (Qubit, ThermoFisher Scientific, MA, USA).
Bisulphite treatment
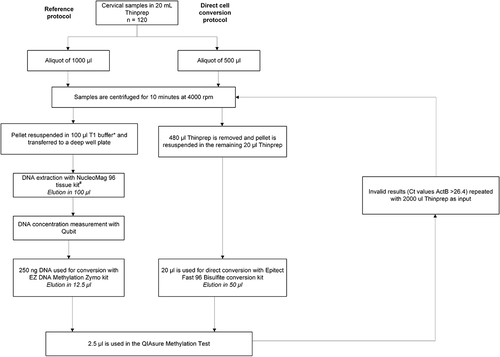
For bisulphite conversion of DNA, the reference protocol (EZ DNA Methylation Zymo kit; Zymo Research, CA, USA) was performed according manufacturer’s recommendation using 250 ng of isolated genomic DNA as input material [Citation13]. This bisulphite-conversion kit is verified for use with the QIAsure Methylation Test [Citation14], but not compatible with direct cell input. A direct conversion protocol on the cervical cells used the Epitect Fast 96 Bisulphite conversion kit (QIAgen, Hilden, Germany) [Citation15]. Cervical cells from 500 µl (1/40th of the original sample) PreservCyt sample was used as input. For invalid samples, an input of 2 mL (1/10th of the original sample) PreservCyt sample was used. Samples were centrifuged and the cell pellet was resuspended in 20 µl PreservCyt prior to bisulphite conversion, performed according to manufacturer’s instructions, except for elution in 50 μl Elution Buffer and omission of carrier RNA. Details on both protocols are outlined in .
DNA methylation analysis
FAM19A4/miR124-2 methylation analysis was performed using the QIAsure Methylation Test (QIAgen, Hilden, Germany) according to the manufacturer’s instructions [Citation14]. Sample input in the assay, for both the reference and direct conversion protocol, is 2.5 μl bisulphite-converted DNA. The assay was performed on a Rotor-Gene Q Mdx 5plex HRM instrument (QIAgen, Hilden, Germany). AssayManager software (QIAgen, Hilden, Germany) controls amplification as well as data analysis and reporting using a fixed assay profile. The housekeeping gene β-Actin (ACTB) was used to verify DNA quality and successful bisulphite conversion. A sample was considered to have a valid test result when Cq value of ACTB was below 26.4. ΔCq values were calculated for each target separately (i.e., FAM19A4 and miR124-2) as the difference between the Cq value of the target and the Cq value of the reference (ACTB). This ΔCq is a relative quantitative value for the promoter methylation level of the FAM19A4 or miR124-2 gene. For normalization, the ΔCq value of a calibrator sample that is included in the QIAsure Methylation Test was subtracted from the ΔCq of the target resulting in a ΔΔCq value. The calibrator is a standardized low-copy plasmid DNA sample with known copy number of the three targets (i.e., FAM19A4, miR124-2 and ACTB). A lower ΔΔCq value corresponds to a higher methylation level of the respective target.
Data and statistical analysis
The direct cell bisulphite conversion protocol was compared with the reference protocol for results of ACTB control gene (Cq values) and methylation of FAM19A4 and miR124-2 genes (ΔΔCq values). Paired evaluation of ΔΔCq values of FAM19A4 and miR124-2 for both bisulphite protocols was done by Spearman rank analysis. Agreement between ΔΔCq values derived from the two protocols was visualized using Bland–Altman plots. In these plots, the difference of two methods is plotted against the average of both methods. We computed the mean of the differences (i.e., bias), the standard deviation (SD), and the 95% limits of agreement (=bias ±1.96 × SD) [Citation16].
All statistical analyses were performed in IBM SPSS Statistics version 26.0 (International Business Machines Corp., Armonk, New York, USA), and p-values are two-sided with 0.01 as significance threshold.
RESULTS
To assess whether bisulphite conversion directly on cervical cells performs equally well as the reference protocol, 120 cervical samples were subjected to both protocols. For the reference protocol, genomic DNA was isolated and DNA concentrations measured. DNA yield of the samples varied between 0.18 μg and 9.75 μg. Accordingly, 108/120 (90.0%) samples reached the normalized input of 250 ng for bisulphite conversion. All 120 samples had a valid test result, with ACTB Cq values ranging from 22.53–26.11.
With 1/40th sample input volume in the direct conversion protocol, 114/120 (95.0%) had a valid test result, with ACTB Cq values ranging from 20.67 to 26.26. Of the six initially invalid samples (ACTB Cq values 26.41–31.77), five samples had sufficient material available for repeat testing with 1/10th sample input volume, which all generated valid test results, with ACTB Cq values ranging from 24.48 to 25.68.
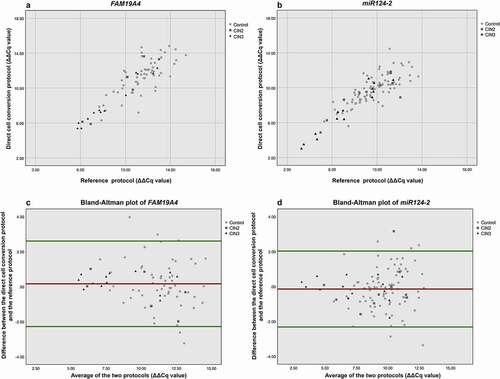
ΔΔCq values of FAM19A4 and miR124-2 were significantly correlated between both protocols ( and ; Spearman Rho 0.708 p = 0.000 and 0.763, p = 0.000, respectively), with particularly strong correlation in CIN2+ (Spearman’s Rho 0.965, p = 0.000 for FAM19A4, and 0.889, p = 0.000 for miR124-2). In further investigation of the agreement between methylation results from each protocol, Bland–Altman plots were prepared ( and ). Overall, there was a good agreement between ΔΔCq values obtained using the direct cell conversion protocol and the reference protocol for both FAM19A4 () and miR124-2 (), with a few outliers observed mainly at higher pairwise average ΔΔCq values.
Figure 2. Correlation of ΔΔCq values of FAM19A4 (a) and miR124-2 (b) between direct cell conversion and reference protocol. For both x- and y-axis: a lower ΔΔCq value represents a higher methylation value. Bland–Altman plot for FAM19A4 (c) and miR124-2 (d). The horizontal red line shows the mean of the difference in ΔΔCq value (=bias) between the two protocols, being 0.167 for FAM19A4 and −0.142 for miR124-2. The horizontal green lines show the upper and lower 95% limits of agreement (= bias ±1.96 × SD), being −2.274 and +2.608, respectively, for FAM19A4 and −2.316 and +2.031, respectively, for miR124-2.
DISCUSSION
In this study, we report on a direct cell bisulphite conversion protocol that can be performed on cervical samples, without the need of prior genomic DNA isolation and input normalization. The direct cell conversion protocol had a high valid rate and performed analytically equally well on cervical samples as the reference protocol using normalized DNA as input.
Bisulphite conversion is an essential step in many DNA methylation assays. A direct cell conversion protocol may overcome some laborious steps in the workflow of DNA methylation analysis, i.e., genomic DNA isolation, DNA concentration measurement, and standardization of DNA input, and thereby is of great interest for implementation of DNA methylation assays in service laboratories that have a cervical screening and/or diagnostic remit. Clinician-collected cervical samples are taken under relatively standardized conditions and in that way likely well suited for protocols with standardized input volume, as is also the case in many HPV test platforms. With the introduction of more sample-to-result HPV assays, a direct cell conversion protocol is highly beneficial as a physical DNA isolate may not be generated as a result of the standard HPV testing workflow in cervical screening. Automated protocols could further improve efficiency and considerably enhance the practicability of methylation analysis in diagnostic and screening settings associated with high-throughput. Of note, an automated solution for preparation of purified bisulphite-converted DNA from direct conversion of plasma and urine has been reported [Citation17]. Our findings provide the basis for development of streamlined and automated workflows for methylation analysis.
The direct conversion protocol with 1/40th of the original sample volume as input used basically similar input amount in qMSP, i.e., 1/800th of original sample volume, as the protocol with prior DNA isolation, when considering a typical DNA concentration of about 20 ng/μl as seen in this study. Three of the six initially invalid samples with the direct protocol had ACTB Cq values close to the threshold for valid test results (i.e., <0.5 Cq difference). Three corresponded with a low DNA concentration (<5 ng/μl) in the reference protocol suggesting a low amount of cells in the original PreservCyt sample. Using four times more input of these samples in the direct cell conversion protocol generated a valid result.
Overall, our data support congruence between the two different bisulphite protocols. We found a particularly strong correlation of ΔΔCq values of FAM19A4 and miR124-2 between both protocols in samples associated with CIN2 +. The divergence between results was slightly larger at lower levels of methylation, corresponding to higher Cq values in PCR. A higher variation seen with higher Cq values is inherent to the PCR process, which becomes more stochastic in the presence of less template. Of interest, when we exploratively applied the assay’s cut-off to score a sample hypermethylation-positive or -negative [Citation14], the direct cell conversion protocol also showed a high agreement (108/119, 90.8%, Cohen’s kappa 0.711) with the reference protocol. The percent agreement and kappa value are well in line with an earlier study reporting on the intra- and inter-laboratory agreement of the FAM19A4/miR124-2 methylation test [Citation18]. In that study, an inter-laboratory workflow (i.e., bisulphite conversion and assay combined) agreement of 90.0% and kappa score of 0.76 were found. The data presented herein support further studies to validate the clinical performance of the new protocol [Citation19].
Various bisulphite conversion kits, including those with direct cell input, have been evaluated before [Citation20], but this is the first study comparing a direct cell protocol with a protocol including DNA isolation and normalization, on cervical samples. Preliminary findings also support the direct conversion protocol for use with self-collected cervico-vaginal samples. Self-sampling is seen increasingly as a credible approach to improve cervical screening coverage and particularly gaining attention in view of the new challenges imposed by the COVID-19 pandemic [Citation21].
In conclusion, we showed that a direct cell conversion protocol demonstrates a high success rate and good analytical performance on cervical samples as compared with a protocol using normalized genomic DNA as input. Direct cell conversion provides a practical workflow, and the results shown here may form the basis for effective high-throughput DNA methylation analysis to support a fully molecular solution to cervical cancer screening.
Author contributions
Study design: DAMH, RDMS, AF, AH, CJLMM
Data collection: SD, RB, KC
Data management: LV, AF
Laboratory experiments: SD
Statistical analysis: LV
Data interpretation: LV, DAMH
Writing first draft of manuscript: LV, DAMH
All authors were involved in writing the manuscript and gave final approval of the submitted and published version of the manuscript.
Data availability
The data that support the findings of our study are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank Ms Elia Alcaniz Boada (Scottish HPV Archive) for preparation of samples for the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953
- Ronco G, Dillner J, Elfstrom KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383(9916):524–532.
- Wentzensen N, Sherman ME, Schiffman M, et al. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112(2):293–299.
- Kelly H, Benavente Y, Pavon MA, et al. Performance of DNA methylation assays for detection of high-grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta-analysis. Br J Cancer. 2019;121(11):954–965.
- Steenbergen RD, Snijders PJ, Heideman DA, et al. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14(6):395–405.
- Bierkens M, Sm W, WN VW, et al. Chromosomal profiles of high-grade cervical intraepithelial neoplasia relate to duration of preceding high-risk human papillomavirus infection. Int J Cancer. 2012;131(4):E579–585.
- LMA DS, Berkhof J, Steenbergen RDM, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124- methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow-up. Int J Cancer. 2018;143(6):1541–1548.
- Dick S, Kremer WW, LMA DS, et al. Long-term CIN3+ risk of HPV positive women after triage with FAM19A4/miR124-2 methylation analysis. Gynecol Oncol. 2019;154(2):368–373.
- Vink FJ, Lissenberg-Witte BI, Meijer C, et al. FAM19A4/miR124-2 methylation analysis as a triage test for HPV-positive women: cross-sectional and longitudinal data from a Dutch screening cohort. Clin Microbiol Infect. 2021 Jan;27(1):125.e1-125.e6.
- Bonde J, Floore A, Ejegod D, et al. Methylation markers FAM19A4 and miR124-2 as triage strategy for primary HPV screen positive women; A large European multi-center study. Int J Cancer. 2020.
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
- Feng L, Lou J. DNA Methylation Analysis. Methods Mol Biol. 2019;1894:181–227.
- EZ DNA Methylation(TM) Kit - Instruction manual Version 1.2.6; https://www.zymoresearch.com/collections/ez-DNA-methylation-kits/products/ez-DNA-methylation-kit, Accessed on June 8, 2021
- QIAsure Methylation Test Instructions for use (handbook) Version 1 (March 2017). https://www.qiagen.com/nl/resources/resourcedetail?id=d4851ddc-405d-4d93-be9a-a343c8576d64&lang=en; Accessed on June 8, 2021.
- Fast E 96 Bisulfite Conversion Handbook (July 2020) https://www.qiagen.com/us/resources/resourcedetail?id=96b87a18-4894-432a-ae20-6c18dfb9ba20&lang=en; Accessed on June 8, 2021
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310.
- Rausch S, Hasinger O, Konig T, et al. An automated high throughput solution for DNA extraction and bisulfite-conversion from high volume liquid biopsy specimens: sample preparation for epigenetic analysis. BMC Res Notes. 2019;12(1):551.
- Floore A, Hesselink A, Ostrbenk A, et al. Intra- and inter-laboratory agreement of the FAM19A4/mir124-2 methylation test: results from an international study. J Clin Lab Anal. 2019;33(4):e22854.
- Arbyn M, Ronco G, Cuzick J, et al. How to evaluate emerging technologies in cervical cancer screening? Int J Cancer. 2009;125(11):2489–2496.
- Holmes EE, Jung M, Meller S, et al. Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS One. 2014;9(4):e93933.
- Ajenifuja KO, Belinson J, Goldstein A, et al. Designing low-cost, accurate cervical screening strategies that take into account COVID-19: a role for self-sampled HPV typing2. Infect Agent Cancer. 2020;15(1):61.