ABSTRACT
Recently, an increasing incidence of HPV-induced oropharyngeal squamous cell carcinoma (OPSCC) has been observed. Moreover, locoregionally advanced stages require a combined modal approach, and the prognosis is poor. Therefore, it is essential to find early diagnostic and prognostic biomarkers. DNA methylation changes play a crucial role in the process of carcinogenesis and are often investigated as promising biomarkers in many types of cancer. For analysis of DNA methylation levels of selected tumour suppressor genes in HPV-positive and HPV-negative samples (including primary tumours and corresponding metastases of metastasizing OPSCCs, primary tumours of non-metastasizing OPSCCs, and control samples), methylation-specific MLPA and methylation-specific high-resolution melting analyses were used. A significant difference in methylation between OPSCCs and the control group was observed in WT1, PAX6 (P < 0.01) and CADM1, RARβ (P < 0.05) genes. CADM1 and WT1 hypermethylation was detected mostly in HPV-positive samples; all but one HPV-negative samples were unmethylated. Moreover, hypermethylation of PAX5 gene was observed in metastases compared with control samples and was also associated with shorter overall survival of all patients (P < 0.05). Associations described herein between promoter methylation of selected genes and clinicopathological data could benefit OPSCC patients in the future by improvement in screening, early detection, and prognosis of the disease.
Introduction
Worldwide, approximately 747,000 new cases of head and neck cancer were diagnosed in 2020, of which 98,000 were oropharyngeal carcinomas (OPCs) [Citation1]. More than 90% of cases are histologically designated as squamous cell carcinomas. OPC occurs more frequently in men than in women, with better survival rates among women [Citation2,Citation3]. The cause of male OPC patient predominance could be partly explained by men engaging more often in high-risk behaviours such as smoking, alcohol abuse, oral sex, and promiscuity [Citation4]. In addition, Chaturvedi et al. [Citation5] have shown that HPV vaccination has a lower effect on the male population due to the low HPV vaccine coverage compared to women. High-income regions such as North America and Northern Europe are characterized by higher incidence of HPV-related oropharyngeal cancer, which represents about 70–80% of all cases [Citation6]. Globally, 84.9% of HPV-related head and neck squamous cell carcinomas (HNSCCs) are attributed to HPV16/18 [Citation7].
Understanding the pathogenesis of HPV-positive OPC that is preceded by long-lasting high-risk HPV oropharyngeal infection is still limited, but the process seems to be different from the cervical cancer model. Pai and Westra [Citation8] reported that reticular epithelium of tonsillar crypts provides effective immune protection of mucosal membranes, allowing direct passage of immune cells like lymphocytes and antigen-presenting cells. However, this structure is advantageous for HPV because the virus can easily migrate through the epithelium and enter the basal cells where it replicates. It can even play a key role in cancer progression by promoting early invasion and metastasis.
DNA methylation is a covalent modification of DNA, which leads to the attachment of a methyl group to the fifth carbon of cytosine to form 5-methylcytosine. Cytosine methylation can occur in CpG-rich regions, known as CpG islands, where it plays an indispensable role in gene regulation and in maintaining cell function and cellular integrity [Citation9]. Aberrant methylation leads to significant changes in gene expression profiles, which can result in emergence of malignancies. Repression of tumour suppressor genes is associated with hypermethylation of their promotor sequences in tumour cells. On the other hand, global hypomethylation may lead to genomic instability and cellular transformation. Both methylation changes occur frequently in oropharyngeal cancer, accompanied by oncogene activation, loss of imprinting, and genomic instability and contribute subsequently to tumorigenesis [Citation10].
The purpose of this study was to examine methylation levels of selected tumour suppressor genes in OPSCC and control tissue and to find correlation between methylation levels of selected genes and clinicopathological characteristics.
Materials and methods
A total number of 212 formalin-fixed, paraffin-embedded (FFPE) samples (74 primary tumours and 74 corresponding metastases of metastasizing OPSCCs, 20 primary tumours of non-metastasizing OPSCCs, and 44 control samples) were analysed in this study. The control samples of normal tissue were obtained from patients treated for a non-malignant diagnosis such as tonsillitis. The FFPE blocks were retrieved from the archives of the Fingerland Department of Pathology, University Hospital Hradec Králové, and Faculty of Medicine in Hradec Králové, Czech Republic. All malignant tumours of the oropharynx were diagnosed between the years 1999 and 2018. All slides were reviewed by an experienced head and neck pathologist (J. L.). Ethical approval was obtained from the Ethics Committee of the University Hospital in Hradec Králové. Information about gender, age of the patients at the time of diagnosis, tumour size, tumour localization (including oropharynx, the root of the tongue, palatine tonsils, and soft palate), and pathological stage according to 2017 version of TNM classification [Citation11] was recorded for each patient. During the follow-up period (until December 2020), data on recurrence, death, and tumour-related death were recorded. Treatment modalities for all patients were radical surgery with radiotherapy, and in 22 cases, chemotherapy was added. The HPV status was analysed at the Fingerland Department of Pathology as described by Laco et al. [Citation12]. Squamous cell carcinomas were classified according to the recently proposed criteria as keratinizing, non-keratinizing, and non-keratinizing with maturation. From these three categories, only keratinizing carcinomas were further graded as well, moderately, or poorly differentiated. Vascular invasion, perineural spread, and status of resection margins were also noted, with the latter being classified as positive (R1) or negative (R0). Due to the presence of lymph node metastases, vascular invasion was present in all patients with metastazing tumours, not regarding the difference between blood and lymphatic vessel invasion.
DNA extraction
DNA isolation was performed with a commercially available extraction kit (QIAamp DNA FFPE Tissue Kit, Qiagen, Hilden, Germany) according to the manufacturer’s protocol with xylene used for deparaffinization. Concentration and quality of DNA in all obtained samples were measured using the spectrophotometric measurement of absorbance on NanoDrop 1000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and the fluorometric method based on binding of dsDNA-selective fluorescent dyes on a Qubit Fluorometer (Thermo Fisher Scientific). The isolated DNA was stored at −20°C until further use.
MS-MLPA
For methylation analysis, a discovery subset of the study cohort comprised 101 samples (31 primary tumours, 31 corresponding metastases of OPSCCs, 20 non-metastasizing OPSCCs, and 19 control samples). Methylation-specific MLPA (MS-MLPA) (MRC Holland, Amsterdam, the Netherlands) was performed according to the manufacturer’s instructions for a set of 25 tumour suppressor genes (Probe mix ME002-C1). The choice for probe mix ME002-C1 was based on a thorough literature search, indicating that genes involved in this particular mix showed promising methylation patterns in head and neck cancer. Moreover, hypermethylation of some promoter regions was associated with HPV in cervical cancer [Citation13]. A short overview of the included genes is presented in https://www.mrcholland.com/products/25162/Probe%20sequences%20ME002%20C1-V01.xlsx
All runs were performed on a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA), and the thermal profile of PCR is shown in . Universal methylated DNA standard was used as positive (100% methylated) control (Zymo Research, Freiburg, Germany), and DNA from a healthy volunteer as a negative (unmethylated) control. Aliquots of PCR products (0.6 μl) were mixed with 0.2 μl of inner standard GeneScan™ – 500 LIZ® (Applied Biosystems) and 9.0 μl of formamide, subsequently denatured, and separated by electrophoresis on an ABI 3500 capillary sequencer (Applied Biosystems). Methylation levels of selected tumour suppressor genes were determined using Genemapper v4.1 software (Applied Biosystems) and CoffalyserNET analysis (MRC Holland) software.
Table 1. Thermal profile of MS-MLPA PCR.
The individual baseline methylation level was determined for each methylation-specific probe. The cut-off value to determine methylation status of each sample was calculated by taking the mean methylation of control samples per probe and adding two times the standard deviation as suggested in the manufacturer’s product description of the kit (https://www.mrcholland.com/products/25160/Product%20description%20ME002-C1%20Tumour%20suppressor%20mix%202-v01.pdf). A sample was then considered methylated if its methylation value was higher than the cut-off; conversely, it was considered unmethylated.
MS-HRM
Based on the results from MS-MLPA and a thorough literature search focused on possible clinical use of the assay, methylation status of CADM1 was selected for validation by a more specific method. Two hundred and seven initially enrolled samples of 133 patients (74 primary tumours and 74 corresponding metastases of OPSCCs, 20 non-metastatic OPSCCs, and 39 control samples) were intended to be analysed using methylation-specific high-resolution melting (MS-HRM) analysis. Due to the insufficient amount of obtained tissue or poor quality of isolated DNA 79 samples were excluded from the analysis. First, bisulphite conversion was performed using an EZ DNA Methylation-Gold™ Kit (Zymo Research) according to the manufacturer’s instruction. Next, the PCR amplification and HRM analysis were performed using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) according to the protocol in . The selection of primers was based on Fisser et al. [Citation14]. In each run, bisulphite-converted universal methylated DNA, universal unmethylated DNA, and 10% standard were used, with the latter serving as a cut-off reference for methylation status.
Table 2. Protocol for MS-HRM analysis of the CADM1 amplicon.
Data were analysed using the CFX maestro™ for qPCR interpretation and Precision Melt Analysis™ Software for HRM parameter determination (Bio-Rad Laboratories).
Statistical analysis
The statistical analysis was carried out using STATISTICA (data analysis software system) version 12 (StatSoft, Inc., Tulsa, OK, USA) and GraphPad Prism software version 8.0.0 (GraphPad Software, San Diego, CA, USA). All statistical tests were two-sided, and statistical significance was claimed for P < 0.01 or <0.05. Association between clinicopathological factors and DNA methylation status was analysed using Fisher’s exact test and the chi-square test. For analysis of the overall survival rate, the Kaplan-Meier method and the log-rank test were used.
Results
MS-MLPA
In general, a low level of promoter methylation was detected in the discovery set ()). Using a different methylation cut-off value for each gene, TP73, GSTP1, CHFR, BRCA2, VHL, CDKN2A, ATM, RB1, PYCARD, BRCA1, and STK11 genes exhibited low methylation levels in all studied samples. In contrast, a high methylation rate (~ 50%) was detected in MSH6, CDH13, WT1, and GATA5 genes regardless of the disease status (control vs. tumour). More importantly, we observed significantly higher frequency of methylation of WT1, PAX6 (P < 0.01) and CADM1, RARβ (P < 0.05) genes in tumour samples compared to control samples ()). For CADM, WT1, and PAX6 genes, no methylation in control samples was present. Besides that, our results also showed hypermethylation of PAX5 (P < 0.05) in metastases samples compared to the control group.
Figure 1. MS-MLPA results of DNA methylation: A) heat map representing the methylation profile of selected tumour suppressor genes in the analysed samples (% of methylation) and B) promoter hypermethylation of 25 studied tumour suppressor genes (% of methylated samples).
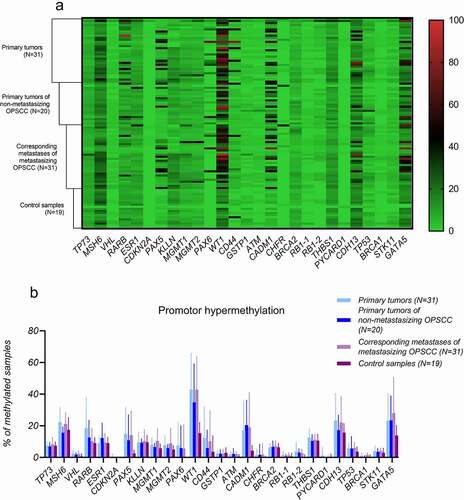
Tumour samples were also stratified according to HPV status. Whilst tumour hypermethylation of RARβ, PAX5, and PAX6 promoters was not associated with HPV status, CADM1 and WT1 genes were significantly hypermethylated in HPV-positive tumours compared to HPV-negative OPSCCs, which were almost all unmethylated (P < 0.01) () Finally, methylation status of examined genes in primary tumours and their corresponding metastases of metastasizing OPSCCs was the same in most cases.
Table 3. Clinicopathological data versus count of methylated samples based on MS-MLPA results.
Correlation of MS-MLPA results with clinicopathological features
The analysed carcinoma group consisted of 37 males and 14 females. The median age of oropharyngeal carcinoma patients at the time of diagnosis was 58 years (range 45–80 years). The follow-up period (until December 2020) ranged from 3 to 180 months (median 82 months). Tumour recurrence occurred in 3/31 (9.7%) patients with available follow-up data. During the follow-up period, 14/31 (45.2%) patients died, of whom 3/31 (9.7%) died due to the tumour. HPV positivity was found in 41/51 (80.4%) tumour samples. Clinicopathological characteristics were compared with methylation status of OPSCC tumour samples ().
The RARβ gene showed significantly higher frequency of methylation in patients who consumed alcohol (P < 0.05), and PAX6 hypermethylation was associated with tumour localization in oropharynx (P < 0.05).
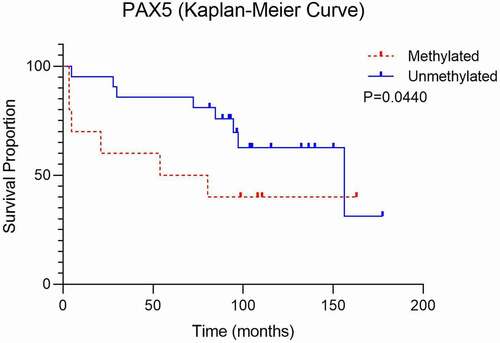
The Kaplan-Meier survival curve shows that patients with higher methylation levels of PAX5 gene had impaired survival compared to those with the unmethylated PAX5 gene (P < 0.05) (). Also, methylation frequency of PAX5 was significantly higher in T1 size of the tumour (tumour was 2 cm or smaller, P < 0.05). In samples with non-keratinizing tumour typing, higher WT1 methylation (P < 0.01) was observed. From the selected tumour suppressor genes, correlation between HPV status and hypermethylation was found in CADM1 and WT1 genes (P < 0.01).
MS-HRM of CADM1 gene
Based on literature search, CADM1 promoter was selected for further validation by an independent technique. CADM1 methylation status in bisulphite-converted DNA was detected using MS-HRM in 128 samples: 35 primary tumours and 35 corresponding metastases of OPSCCs, 20 non-metastatic OPSCCs, and 38 control samples. The cut-off value to assign sample’s methylation status was set to 10%, based on the value detected previously for this gene in the MS-MLPA analysis (11%) ().
Figure 3. Representative plots from MS-HRM of CADM1 gene. Plots show curves of standards (100% red, 10% orange, unmethylated template green), methylated sample (purple), and unmethylated sample (blue).
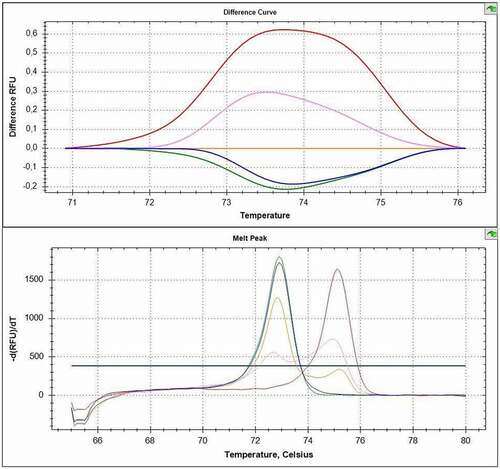
Methylation was detected in 36.4% (20/55) of all primary tumour samples and none of the controls (0/38). Methylation status of tumours and their associated metastases corresponded in all cases. CADM1 hypermethylation was detected only in the samples affected by HPV infection; 45.5% (20/44) of HPV-positive samples were hypermethylated (.).
Table 4. CADM1 hypermethylation and prevalence of HPV in studied groups using MS-HRM.
Discussion
Hypermethylation of CADM1 is one of the principal causes of gene silencing in many types of cancers [Citation15–17]. It has been demonstrated that CADM1 inhibits proliferation and invasion of squamous carcinoma cells, both the fundamental processes for disease progression [Citation18]. According to Steenbergen et al. [Citation19], hypermethylation of CADM1 caused by HPV16 and HPV18 was observed in cervical cancer. However, little is known about the role of CADM1 hypermethylation in oropharyngeal cancer. Using MS-MLPA, our results showed higher methylation in the CADM1 gene in patients with oropharyngeal carcinoma compared with the control group. Hypermethylation of CADM1 gene additionally correlated with HPV status, all but one HPV-negative samples were unmethylated. CADM1 methylation is associated with squamous carcinoma cells and HPV infection in cervical cancer [Citation19]. Since more than 90% of oropharyngeal cancer cases are histologically squamous cell carcinomas and, in some regions, the most common cause is HPV infection, we used MS-HRM to validate CADM1 hypermethylation in oropharyngeal cancer samples. Using primers based on the study by Fisser et al. [Citation14], we were able to detect methylation of the CADM1 promoter region in a significant number of HPV-positive oropharyngeal cancer samples included in our study. Methylation was present only in samples with recorded HPV positivity, and we were not able to detect any hypermethylation in HPV-negative tissue samples. These results suggest CADM1 as a possible biomarker of initial carcinogenic processes in the tissue harbouring HPV viral infection. Analysis by also MS-HRM confirmed that all control samples were unmethylated, implying that methylation of the CADM1 promoter may play an important role in carcinogenesis of oropharyngeal cancer. However, it does not seem to affect the metastatic process, as the corresponding metastases had the same CADM1 methylation status as their primary tumours. These data correlate with the research results in the study by van Kempen et al. [Citation20,Citation21], where HPV-positive OPSCCs showed a significantly higher cumulative methylation index compared to HPV-negative OPSCCs. Also, patients with HPV-positive OPSCC generally respond more favourably to chemotherapy and radiation than patients with negative HPV status. Data from the study by Woo et al. [Citation22] also indicate that HPV infection may play a crucial role in the loss of CADM1 gene expression via aberrant DNA methylation.
PAX5 is widely expressed in normal adult and embryonic tissues, as well as in various neoplasms including HNSCC [Citation23]. In our study, PAX5 gene was frequently found methylated amongst oropharyngeal metastases compared to control tissue samples. A higher level of methylation was detected in HPV-positive tumours than in HPV-negative tumours. Guerrero-Preston et al. [Citation23] also compared PAX5 methylation in HPV-positive and HPV-negative HNSCCs and additionally demonstrated that tumours from patients with a history of tobacco exposure had a similar frequency of PAX5 methylation as the patients with no smoking history. Our data confirmed their results; the frequency of PAX5 methylation was comparable in smokers and non-smokers. Our study also showed that PAX5 hypermethylation was associated with the advanced stage of tumour (P < 0.05). Finally, the Kaplan-Meier survival curve shows that patients with higher methylation levels of PAX5 promoter had a worse survival in comparison to those with the unmethylated PAX5, highlighting it as a potential prognostic marker. Results of another study showed that PAX5 gene methylation can be used as a molecular marker for surgical margin analysis and as a prognostic marker of HNSCCs [Citation24].
The WT1 gene plays a role in regulating cell proliferation, growth, differentiation, and apoptosis and can be both a tumour suppressor and a carcinogenic inducer [Citation25,Citation26]. We found significantly higher methylation of the WT1 gene in most tumour cases, compared to no hypermethylated controls. WT1 gene also showed significantly higher methylation in patients with HPV-positive tumours and with non-keratinizing squamous cell carcinoma. Our data showed a higher methylation level in stages T1 and T2 of the tumours, too. The latter agrees with the study by Ribeiro et al. [Citation27]; using MS-MLPA, WT1 methylation was associated with T1 and T2. Conversely, methylation levels of control samples were <20%, much lower than that in our study, which did not correlate with our results. This discrepancy may be due to using a different MS-MLPA kit, which means we analysed different CpG sites. Oji et al. [Citation28] suggested that WT1 could serve as a prognostic factor, as they found that high WT1 expression significantly correlated with a poor tumour differentiation and, consequently, an advanced tumour stage. They observed overexpression of the WT1 gene in 42 of 56 (75%) cases of examined HNSCC. Herein, there were only a few advanced cases, and thus, it was not possible to evaluate this association.
The RARβ gene is a member of the thyroid-steroid hormone receptor superfamily of nuclear transcriptional regulators. It binds retinoic acid, a biologically active form of vitamin A that mediates cell signalling in embryonic morphogenesis, cell growth, and differentiation. It is assumed that this protein limits growth of many cell types by regulating gene expression. Lotan [Citation29] confirmed that retinoic acid suppresses carcinogenesis and inhibits the growth of human HNSCC. A loss of retinoids and their receptors has been associated with malignant progression in HNSCC [Citation30]. In our study, some oropharyngeal carcinoma samples were methylated at the studied CpG site, whilst almost all control non-malignant samples were unmethylated. RARβ gene was significantly methylated in patients who consume alcohol. Other studies have also pointed to the importance of RARβ methylation. Maruya et al. [Citation31] identified methylation of RARβ in 15/32 (46.9%) samples of primary oral malignant diseases. Histologically normal mucosa showed a 50% level of methylation. In the study by Youssef et al. [Citation32], promoter hypermethylation of RARβ gene in HNSCCs was a malignant marker.
The PAX6 encodes a transcription factor and may play an important role in tumorigenesis [Citation33]. To date, no data are available regarding the role of PAX6 aberrant methylation in oropharyngeal carcinoma. Our data showed significantly higher methylation in the PAX6 gene in samples of patients with oropharyngeal carcinoma compared to the control group. We also examined the correlation between methylation status and clinicopathological data. Significantly higher methylation of the PAX6 gene was associated with tumour localization in oropharynx. Since there was no methylation in the control samples, hypermethylation could serve as a potential tumour biomarker, further studies are needed to understand the role of the PAX6 gene in the pathogenesis of OPSCC.
To sum up, significant association between clinicopathological data and promoter methylation of selected genes could potentially positively impact screening, early detection and prognosis of the oropharyngeal cancer, and approach to tailored therapy and individualization of the treatment.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- INTERNATIONAL AGENCY FOR RESEARCH ON CANCER. Global Cancer Observatory: GLOBOCAN 2020. 2021 Jan 5th. December, 2020. http://gco.iarc.fr/
- Saba NF, Goodman M, Ward K, et al. Gender and ethnic disparities in incidence and survival of squamous cell carcinoma of the oral tongue, base of tongue, and tonsils: a surveillance, epidemiology and end results program-based analysis. Oncology. 2011;81(1):12–20.
- de Souza DL, Bernal MM, Jerez Roig J, et al. Oropharyngeal Cancer Survival: a Population-Based Study of Patients Diagnosed between 1978 and 2002. ISRN Oncol. 2012;39(6):207263.
- Osazuwa-Peters N, Adjei Boakye E, Rohde RL, et al. Understanding of risk factors for the human papillomavirus (HPV) infection based on gender and race. Sci Rep. 2019 Jan 22;9(1):297.
- Chaturvedi AK, Graubard BI, Broutian T, et al. Effect of Prophylactic Human Papillomavirus (HPV) Vaccination on Oral HPV Infections Among Young Adults in the United States. J Clin Oncol. 2018 01;36(3):262–267.
- Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011 Nov;29(32):4294–4301.
- Castellsagué X, Alemany L, Quer M, et al. HPV Involvement in Head and Neck Cancers: comprehensive Assessment of Biomarkers in 3680 Patients. J Natl Cancer Inst. 2016 Jun;108(6):djv403.
- Pai SI, Westra WH. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol. 2009;4(1):49–70.
- Chmelarova M, Palicka V. Epigenetics in cancer: a promising path to follow? Clin Chem Lab Med. 2019 Jun 26;57(7):927–931.
- Bhat S, Kabekkodu SP, Noronha A, et al. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie. 2016Feb;121:298–311.
- Westra WH, Lewis JS. Update from the 4th Edition of the World Health Organization Classification of Head and Neck Tumours: oropharynx. Head Neck Pathol. 2017 Mar;11(1):41–47.
- Laco J, Sieglová K, Vošmiková H, et al. The presence of high-risk human papillomavirus (HPV) E6/E7 mRNA transcripts in a subset of sinonasal carcinomas is evidence of involvement of HPV in its etiopathogenesis. Virchows Arch. 2015 Oct;467(4):405–415.
- Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011 Aug;2(2):123–150.
- Fisser MC, Rommer A, Steinleitner K, et al. Induction of the proapoptotic tumor suppressor gene Cell Adhesion Molecule 1 by chemotherapeutic agents is repressed in therapy resistant acute myeloid leukemia. Mol Carcinog. 2015 Dec;54(12):1815–1819.
- Mao X, Seidlitz E, Truant R, et al. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004 Jul 22;23(33):5632–5642.
- Chen K, Wang G, Peng L, et al. CADM1/TSLC1 inactivation by promoter hypermethylation is a frequent event in colorectal carcinogenesis and correlates with late stages of the disease. Int J Cancer. 2011 Jan 15;128(2):266–273.
- Overmeer RM, Henken FE, Snijders PJ, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008 Aug;215(4):388–397.
- Vallath S, Sage EK, Kolluri KK, et al. CADM1 inhibits squamous cell carcinoma progression by reducing STAT3 activity. Sci Rep. 2016 Apr 1;6(1):24006.
- Steenbergen RD, Kramer D, Braakhuis BJ, et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004 Feb 18;96(4):294–305.
- van Kempen PM, Noorlag R, Braunius WW, et al. Differences in methylation profiles between HPV-positive and HPV-negative oropharynx squamous cell carcinoma: a systematic review. Epigenetics. 2014 Feb;9(2):194–203.
- van Kempen PM, van Bockel L, Braunius WW, et al. HPV-positive oropharyngeal squamous cell carcinoma is associated with TIMP3 and CADM1 promoter hypermethylation. Cancer Med. 2014 Oct;3(5):1185–1196.
- Woo HJ, Kim SJ, Song KJ, et al. Hypermethylation of the tumor-suppressor cell adhesion molecule 1 in human papillomavirus-transformed cervical carcinoma cells. Int J Oncol. 2015;46(6):2656–2662.
- Guerrero-Preston R, Michailidi C, Marchionni L, et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics. 2014 Jul;9(7):1031–1046.
- Hayashi M, Guerrero-Preston R, Sidransky D, et al. Paired box 5 methylation detection by droplet digital PCR for ultra-sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer Prev Res (Phila). 2015 Nov;8(11):1017–1026.
- Wu D, Zhang J, Fan P, et al. Methylation in the promoter regions of WT1, NKX6-1 and DBC1 genes in cervical cancer tissues of Uygur women in Xinjiang. Genet Mol Biol. 2018 Jan-Mar;41(1):9–17.
- Scharnhorst V, van der EAJ, Jochemsen AG. WT1 proteins: functions in growth and differentiation. Gene. 2001;273(2):141–161. 2001/08//.
- Ribeiro IP, Caramelo F, Marques F, et al. WT1, MSH6, GATA5 and PAX5 as epigenetic oral squamous cell carcinoma biomarkers - a short report. Cell Oncol (Dordr). 2016 Dec;39(6):573–582.
- Oji Y, Yamamoto H, Nomura M, et al. Overexpression of the Wilms’ tumor gene WT1 in colorectal adenocarcinoma. Cancer Sci. 2003 Aug;94(8):712–717.
- Lotan R. Retinoids in cancer chemoprevention. FASEB J. 1996 Jul;10(9):1031–1039.
- McGregor F, Muntoni A, Fleming J, et al. Molecular Changes Associated with Oral Dysplasia Progression and Acquisition of Immortality: Potential for Its Reversal by 5-Azacytidine. Cancer Res . 2002;62(16): 4757–4766.
- Maruya S, Issa JP, Weber RS, et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin Cancer Res. 2004 Jun;10(11):3825–3830.
- Youssef EM, Lotan D, Issa JP, et al. Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis. Clin Cancer Res. 2004 Mar;10(5):1733–1742.
- Zhang X, Yang X, Wang J, et al. Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int J Clin Exp Pathol. 2015;8(9):11452–11457.