ABSTRACT
Non-Alcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease in children. Epigenetic alterations, such as through DNA methylation (DNAm), may link adverse childhood exposures and fatty liver and provide non-invasive methods for identifying children at high risk for NAFLD and associated metabolic dysfunction. We investigated the association between differential DNAm and liver fat content (LFC) and liver injury in pre-adolescent children. Leveraging data from the Newborn Epigenetics Study (NEST), we enrolled 90 mother-child dyads and used linear regression to identify CpG sites and differentially methylated regions (DMRs) in peripheral blood associated with LFC and alanine aminotransferase (ALT) levels in 7–12yo children. DNAm was measured using Infinium HumanMethylationEPIC BeadChips (Illumina). LFC and fibrosis were quantified by magnetic resonance imaging proton density fat fraction and elastography. Median LFC was 1.4% (range, 0.3–13.4%) and MRE was 2.5 kPa (range, 1.5–3.6kPa). Three children had LFC ≥ 5%, while six (7.6%) met our definition of NAFLD (LFC ≥ 3.7%). All children with NAFLD were obese and five were Black. LFC was associated with 88 DMRs and 106 CpGs (FDR<5%). The top two CpGs, cg25474373 and cg07264203, mapped to or near RFTN2 and PRICKLE2 genes. These two CpG sites were also significantly associated with a NAFLD diagnosis. As higher LFC associates with an adverse cardiometabolic profile already in childhood, altered DNAm may identify these children early in disease course for targeted intervention. Larger, longitudinal studies are needed to validate these findings and determine mechanistic relevance.
Introduction
Non-Alcoholic fatty liver disease (NAFLD) is now the most prevalent chronic liver disease among children and adults in the United States [Citation1,Citation2]. Driven largely by the epidemics of obesity and insulin resistance, it is expected that NAFLD will soon become the leading cause of liver transplantation among children in the Western world and that rates of chronic progressive NAFLD will expand into children of increasingly younger ages [Citation2–4]. Given these predictions and the risks associated with more severe disease over time, the rising prevalence of NAFLD among the paediatric population is particularly alarming [Citation5,Citation6].
Mounting evidence suggests that risk for fatty liver starts very early in life and predisposes children to progressive NAFLD and liver injury as well as to increased morbidity and mortality [Citation7,Citation8]. Similar to adults, NAFLD in children develops as a result of a complex interplay between metabolic derangements in the form of increased total body fat and insulin resistance, genetic and epigenetic variation, and environmental exposures[Citation9]. Many of these influences can begin in utero, such as through adverse exposures to pre-pregnancy maternal obesity [Citation10–12], diabetes [Citation12–14], and low and high birthweight [Citation15]. All of these have been linked to increased risk for obesity and poor metabolic health, as well as the increased likelihood for progression of steatosis to non-alcoholic steatohepatitis (NASH) and advanced fibrosis [Citation16,Citation17].
Despite increased knowledge about the risks for paediatric NAFLD and NASH and its natural history, biomarkers able to identify children early in the disease course remain undefined. Standard of care techniques used to identify these children early have proven insufficient in identifying or halting disease progression[Citation18]. Thus, alternative mechanisms to identify disease risk and progression earlier are critically needed. Epigenetics may be an important tool to fill these gaps. Epigenetic mechanisms alter gene expression and subsequent phenotypes without altering the DNA sequence and can be inherited across generations[Citation19]. DNA methylation represents one of the most studied and stable epigenetic mechanisms. In humans, several studies have shown that exposure to maternal body mass index (BMI) [Citation20,Citation21] and gestational diabetes [Citation22] associate with reproducible differences in DNA methylation at birth. Despite reduced effect sizes over time, differences in DNA methylation can persist through childhood and may be an important mechanism establishing and promoting increased risk for ongoing disease throughout life[Citation23]. Despite accumulating evidence to support DNA methylation influences on childhood metabolic dysfunction, studies of DNA methylation in NAFLD have largely been performed in adults using liver biopsy tissue [Citation24–27] or peripheral blood [Citation28,Citation29], and few have assessed the association between peripheral blood differences in DNA methylation with other metabolic disease, hepatic steatosis and/or NAFLD in children[Citation30].
Although epigenetic marks are generally cell- and tissue-specific, there are data to suggest that peripheral blood DNA methylation profiles can reflect differences in DNA methylation in internal organs including the liver. [Citation31] A recent study using untargeted approaches revealed that peripheral blood DNA methylation patterns reflect cell-type changes in the liver and thus could provide a biomarker for NAFLD fibrosis[Citation32]. In addition, specific changes in peripheral blood-derived DNA hypermethylation at one CpG (cg06690548) was recently shown to be associated with lower risk of hepatic steatosis[Citation33]. More recently, investigators identified a whole blood DNA methylation profile associated with hepatic fat and NAFLD risk and found that methylation levels in blood were moderately correlated with those measured in liver tissue[Citation28]. Thus, while some DNA methylation may be tissue specific, changes in those tissues are likely reflected in blood, particularly the liver through which more than 75% of the body’s blood flows. The ability to identify non-invasive markers as a proxy of disease for more inaccessible tissues would be critical to improved mechanistic understanding, early identification and even interventions particularly in children where risk should be mitigated.
To address the gap in knowledge, we examined the epigenome-wide association between DNA methylation at >850,000 CpGs and liver fat content (LFC), fibrosis, and cardiometabolic abnormalities in pre-teenage children initially recruited to the longitudinal birth cohort, the Newborn Epigenetics Study (NEST). We assessed the association between differentially methylated regions (DMRs) and single CpG sites in peripheral blood to identify epigenetics marks that could signal a possible mechanistic risk for NAFLD and serve as future potential biomarkers of cardiometabolic health.
Materials and methods
Study population
We enrolled children currently followed in NEST, a multi-ethnic, birth cohort study designed to understand the role of environmental influences on epigenetic responses and health outcomes in later life. Between 2005 and 2011, NEST recruited pregnant women from prenatal clinics serving Duke University Obstetrics and Durham Regional Hospital Obstetrics facilities in North Carolina, respectively. A detailed description of the study participants, enrollment criteria, and data collected has been previously reported [Citation34,Citation35]. Briefly, women were consecutive visitors to the two Durham County obstetrics clinics and considered eligible for enrolment in NEST if aged 18 years and older, pregnant, and spoke English or Spanish. Women were excluded if planning to move in the next three years or to have their newborns adopted. NEST and the present study have been approved by the Duke University Institutional Review Board. Written informed consent was obtained for all participants and children aged 12 or older provided additional assent for participation.
From these participants, we conducted a cross-sectional study called the Study of Early-life, Epigenetics and NAFLD (SEEN). We recruited children for SEEN based on maternal pre-pregnancy body mass index (BMI). At enrollment in NEST, questionnaires and medical abstractions gathered information regarding maternal pre-pregnancy anthropometric measurements, including current height (metres) and usual pre-pregnancy weight (kilograms). BMI was calculated from these measurements and mothers were classified as obese if BMI was 30 kg/m2 or greater, according to the World Health Organization classifications (https://www.who.int/dietphysicalactivity/childhood_what/en/). For this study, we oversampled children born to women with pre-pregnancy obesity. All participants had baseline data collected at enrolment in NEST and mother-child dyads were screened for the presence of underlying chronic liver disease. Children were excluded if they had a history of known underlying liver disease or alternative reason for fatty liver disease such as exposure to total parenteral nutrition within the prior three months, short bowel syndrome, biliopancreatic diversion or bypass, active malignancy, or contraindication to MRI. Between August 2016 and April 2019, a total of 181 eligible mother-child dyads with pre-pregnancy BMI and parity data and children between the ages of 7 and 12 from NEST were re-contacted. Of these, 60 declined to participate or were unsure of participation and 31 failed to show for appointments. A total of 90 mother-child dyads were consented for SEEN. All consented mother-child dyads completed enrollment activities. We compared the 90 participants to the overall NEST cohort (n = 2681) and they were similar with respect to maternal age at delivery (p = 0.30), maternal education (p = 0.32), sex of the child (p = 0.30), and child birthweight (p = 0.43). The 90 mother-child dyads were more likely to be African American (p < 0.05). We examined race/ethnicity as a potential confounder.
Data collection
Upon enrollment in SEEN, each parent completed a questionnaire that provided information about their own and their child’s demographics, lifestyle characteristics including employment/education, exercise and dietary habits, smoking history, comorbidities including NAFLD and other chronic liver disease, family history and medication use. As part of the parent NEST study, demographics, medical history, questionnaires, lifestyle, diet, and health characteristics were obtained for all enrolled mothers. At delivery, parturition data were obtained from medical records.
Assessment of child body size and cardiometabolic health at age 7-12 years
Children underwent a physical exam which included vital signs, weight and height measurements, and waist and hip circumference measurements. Using these values, sex and age-specific BMI percentiles were computed based on the Centers for Disease Control and Prevention protocols (https://www.cdc.gov/healthyweight/bmi/calculator.html)[Citation36]. A diagnosis of hypertension or elevated blood pressure were assessed by blood pressure percentiles calculated using the American Academy of Paediatrics (AAP) MDCalc for paediatric hypertension based on the AAP Paediatric Hypertension Guidelines according to recommended guidelines from the National Education Program Working Group on High Blood Pressure in Children and Adolescents[Citation37]. Fasting venous blood samples were collected to measure liver function and measures of metabolic health including alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels, total bilirubin, total cholesterol, triglycerides (TG), high density lipoprotein (HDL) and low density lipoprotein (LDL) levels, and glycosylated haemoglobin (HbA1c%).
Liver fat content and fibrosis assessments
We measured LFC using a 3.0 Tesla MRI system (TIMTrio, Siemens Healthineers, Erlangen, Germany). MRI was performed in children after a fasting period of more than six hours without sedation and each exam lasted approximately 25 minutes. MRI was performed using a non-contrast, breath-hold, multi-echo spoilt gradient recalled echo sequence that uses a low flip angle to reduce T1 bias, with correction for T2*decay, and uses spectral modelling to address fat-water and fat-fat signal interference effects, and proton-density fat fraction (PDFF) maps were generated [Citation38,Citation39]. Three equal-sized regions of interest measuring at least 2 cm2 each were placed in the anterior, mid, and posterior portions of the right hepatic lobe, avoiding visible vessels and artefacts on the PDFF maps. LFC was determined as the mean PDFF value of the regions of interest.
Abdominal adiposity was measured using a conventional, T1-weighted 3D gradient recalled echo sequence. Cross-sectional area of intra-abdominal fat, subcutaneous fat, and skeletal muscle wwa measured using manually-drawn regions of interest. From these, we calculated subcutaneous to visceral fat and visceral to subcutaneous fat ratios. Subcutaneous Additionally, T1 mapping of the liver was performed using an FDA-approved multi-flip angle gradient echo sequence. T1 mapping has been suggested as a surrogate for hepatic fibrosis, but not extensively validated.
MR elastography (MRE) was used to measure liver stiffness using the Resoundant system (Resoundant, Inc., Rochester, MN). The acquired images are used to generate quantitative maps of tissue stiffness referred to as elastograms and to provide an overall estimate of parenchymal stiffness with units of kiloPascals (kPa) [Citation40,Citation41]. Regions of interest covering at least 10 cm2 in total were drawn on one or more of the stiffness maps, and the liver stiffness recorded as the mean pixel-wise stiffness.
In addition to MRE, we assessed liver stiffness in children via transient elastography (TE) (FibroScan®, Echosens, Paris). The feasibility of TE in children has been shown in several studies including those as young as 5 years of age [Citation42,Citation43]. The same trained personnel performed TE on all eligible, consented children after at least a six hour fast to obtain liver stiffness measurement (LSM). The M probe was used initially for all children and the examination was considered successful when ten valid measurements and interquartile range (IQR) was less than 30% of the median LSM value. Participants with unsuccessful examinations were excluded from the analysis of LSM.
Liver fat fraction of ≥ 5% has been defined as NAFLD in adults[Citation44]. Given that thresholds for NAFLD in children less than 10 years of age have not yet been validated, our goal was to assess associations with LFC across the spectrum of disease. We first defined NAFLD as a liver fat fraction above 3.7% as 90% of children had PDFF values less than 3.1% in our study population. This decision was based on previously published work suggesting 3.5% may represent an unhealthy amount of liver fat in adolescent children and that liver fat accumulation above 2.0% is already associated with an increased risk for cardiometabolic disease [Citation30,Citation45,Citation46]. Second, we assessed the association between top tertile LFC, defined as greater than or equal to 1.7%, and baseline child and mother characteristics as well as with altered DNA methylation.
DNA methylation
DNA from peripheral blood from children enrolled in this study was extracted using Puregene Reagents (QIAamp DNA Mini Kit, Qiagen, Valencia, CA, USA). Details have been described previously[Citation47]. DNA quality was assessed using a Nanodrop 1000 Spectrophotometer (Thermo Scientific; Wilmington, DE). Bisulphite conversion was performed using 500ng of DNA via the EZ DNA Methylation Kit per manufacturer’s instructions (Zymo Research; Irvine, CA). Samples were plated randomly onto 96 well plates and processed with the Illumina Infinium MethylationEPIC BeadChip Kit per the manufacturer’s recommendations (llumina Inc., San Diego, CA). The EPIC BeadChips interrogate over 850,000 methylation sites across the genome.
DMR identification
Data processing and quality control assessment of the methyl-array data was carried out using the minfi package from the R statistical programming environment [Citation48,Citation49]. A MethylSet object containing only methylated and unmethylated signals was created using the preprocessRaw function, and beta-value densities were inspected to assess sample quality. Normalization of the methylation data was carried out using stratified quantile normalization processing method, which normalizes the methylated and unmethylated intensities separately. The normalization step also addressed outliers and removed poorly performing samples of both methylated and unmethylated channels when small intensities were close to zero. Additional potential methylation beta values outliers were trimmed using values beyond the lower and the upper outer fences. These are defined by values < 25th percentile minus 3 times interquartile range (IQR) and values > 75th percentile plus 3 times IQR. We also excluded probes with a detection p-value > 0.01 in more than 20% of the samples and probes that overlapped with sex chromosomes[Citation48]. A total of 866,091 probes exist on the Illumina Methylation 850 K array of which 845,797 probes were left after removing control probes, probes with a detection p-value > 0.01 in more than 20% of the samples, and those mapping to single nucleotide polymorphisms (SNPs) and sex chromosomes.
CpG identification
To identify individual CpG sites data processing and quality control was performed using quantile normalization to eliminate systematic differences across the arrays. Probes that had a detection p-value > 0.05 in more than 10% of the cohort were filtered out prior to analysis. Probes were annotated with their genomic coordinates in the hg38 version of the human genome and the nearest gene to the probe was listed using the gene models provided by Ensembl (version 93). Chromatin state was reported for each probe based on the 18-state HMM for mononuclear cells from the Roadmap Epigenomics Project[Citation50]. Principal components analysis was performed on filtered beta values to test for confounding factors and assess for the presence of outliers in the R statistical programming environment.
DNA methylation analysis
We assessed DNA methylation across the genome using two methods: DMRs and specific CpG methylation. Multivariable linear regression analyses were used to estimate the associations between DNA methylation in peripheral blood and liver health-related outcomes including LFC, top tertile LFC, top tertile ALT, and a diagnosis of NAFLD. Models were adjusted for maternal pre-pregnancy obesity, maternal age at delivery, maternal cigarette smoking, child sex, and cell proportions. The Houseman cell type count were generated using ExpirmentalHub package and combined with the covariate data table. Regression models for specific CpG site methylation were fit using the limma package in R. The DMRs analysis was performed using DMRcate [Citation51] R package. Significant DMRs were defined based on the following criteria: 1) region with p-value <0.05 after correcting for multiple comparisons using the Benjamini and Hochberg approach [Citation52]; 2) Epigenome-wide association (EWAS) effect estimates for the individual CpGs in a DMR have the same direction; 3) more than two CpGs in the DMR; and 4) minimum distance separating DMRs of 10 lambda (conseclambda).
Next, we examined our CpG results to determine if there was any overlap with 22 CpG sites measured in peripheral blood previously associated with liver fat accumulation in adults[Citation28]. In addition, we examined our DMR results for LFC to determine any overlap with differential methylation associated with BMI as measured in peripheral blood of children aged 2–18 years[Citation53].
Statistical analysis
Mother and child characteristics were summarized by the mean and standard deviation for symmetrical distributions and median and interquartile range (IQR) for asymmetric distributions.
Differences in normally distributed data were analysed using Student’s t test or analysis of variance, while non-normally distributed data were analysed using the Mann-Whitney U test. Chi-square or Fisher’s exact test, as appropriate, were used to compare proportions. All statistical tests were two-sided and based on a significance level of 5%. Data were analysed using SAS JMP statistical software for Windows (version 14.0; SAS Institute, Inc., Cary, NC.)
Pathway analysis and comparison to CpG associated with hepatic steatosis in adults
We used the Ingenuity Pathway Analysis (IPA, Ingenuity systems, Inc., Redwood City, CA, www.ingenuity.com) tool to examine canonical pathways, upstream regulators, diseases and functions associated with significant genes annotated to the DMRs with peripheral blood of NAFLD, ALT and top tertile LFC. The significance of canonical pathways and upstream regulators was determined by IPA’s default threshold [-log(Benjamini Hochberg p-value) > 1.3]. We also conducted aggregate comparison analysis to find pathways common in PDFF, NAFLD and ALT.
Results
Participant characteristics
Of the 90 children enrolled in the study, good quality MRI-PDFF values were obtained in 79 (87.8%). Baseline characteristics of the 79 children and mothers are summarized in . These are shown by LFC dichotomized by the top-tertile PDFF. Breakdown of baseline characteristics by child BMI (obese/overweight (OW) versus non-obese/OW) is shown in Supplementary . Children enrolled in the study were a median age of 9.2 years (IQR 8.1–9.8) and 44.3% identified as male at birth. A majority (64.5%) of children were African American (AA) and significantly more AA children were considered obese or OW and were born to mothers with pre-pregnancy obesity (66.1% vs. 33.9%, p < 0.0001) (Supplemental Table 1). LFC did not differ significantly by race or ethnicity in this cohort. Children with higher LFC were significantly more likely to be older, have higher BMI and larger waist circumference, and fall into the obese BMI category. Several parameters indicative of the metabolic syndrome in adults (e.g., increased systolic blood pressure, serum triglyceride concentrations, and serum TG/HDL ratio [Citation54–56]) tended to be higher in the top-tertile LFC than in the lower two tertiles of LFC, but none of these achieved statistical significance. In contrast, ALT values were significantly higher in children in the top-tertile LFC.
Table 1. Child characteristics by top tertile liver fat content (PDFF).
Imaging characteristics of children
shows the imaging characteristics for children enrolled in SEEN by LFC (i.e., top tertile PDFF). This is shown by obese/OW status in Supplementary Table 2. Liver fibrosis was assessed by liver stiffness measurement (LSM) via TE in 85 (94%) and MRE in 81 (90%). Median LFC was 1.4% (range, 0.3–13.4%), LSM was 4.3 kPa (range, 2.3–14.5 kPa), and MRE was 2.5 kPa (range, 1.5–3.6kPa). Three children had LFC ≥ 5%, while six (7.6%) met our definition of NAFLD. All children meeting our definition of NAFLD were considered obese by BMI category (z > 95th percentile) and interestingly, five of the six were AA. Five children had MRE values ≥ 3 kPa and the majority were considered obese by BMI. MRE was significantly higher in those with lower LFC and whether this is of clinical significance remains uncertain. Subcutaneous to visceral fat ratio and subcutaneous fat area were significantly higher in children with top-tertile LFC as was visceral fat area. As expected, obese and overweight children also had significantly greater subcutaneous and visceral fat area than normal weight children (Supplementary Table 2). Visceral and subcutaneous adipose tissue have been shown to confer different metabolic risks in children. In adults, visceral adipose tissue (VAT) is more strongly associated with cardiometabolic risk than subcutaneous adipose tissue (SAT) [Citation57,Citation58]. In children, however, the relationship is less clear with some data suggesting there may be a threshold above which VAT and/or SAT confers risk while other data found SAT to be more strongly associated with insulin resistance and obesity.
Table 2. Imaging characteristics of children by top tertile liver fat content (PDFF).
Differentially methylated regions and liver fat content, NAFLD and ALT in pre-teenage children
First, we assessed associations between top tertile LFC, a diagnosis of NAFLD, and top tertile ALT and DMRs to determine potential mechanistic pathways as well as non-invasive risk markers for early signs of liver fat and injury. At an FDR adjusted p-value of <0.005, we identified 88 DMRs associated with top-tertile LFC, 71 with a diagnosis of NAFLD, and 123 with top tertile ALT values. The top ten significant DMRs associated with LFC are shown in . The most significant of these was for a DMR of 13 CpG sites in the maestro heat like repeat family member 6 (MROH6) gene. This gene shows cell type specificity to hepatocytes but low overall tissue specificity and is known to affect expression of transforming growth factor beta-1 (TGFß-1) according to the human protein atlas. KEGG pathway annotations reveals it is most associated with primary bile acid biosynthesis and nitrogen metabolism in humans and a SNP in the gene was found to be associated with IL-18 levels in patients with acute coronary syndrome[Citation59]. The full list of significant DMRs by outcome are listed in Supplementary Tables 3–5. We then assessed overlap of significant DMRs among the three analyses of liver fat and injury. Of the 88, 71, and 123 significant DMRs, we did not identify any overlap between LFC and NAFLD while six DMRs overlapped between NAFLD and ALT. These DMRs mapped to six genes, namely, AURKC, BHMT, FAM47E-STBD1, KIAA0040, NBPF19 and ZNF264 (Supplementary Table 6).
Table 3. Significant DMR associated with top tertile liver fat content (by MRI-PDFF).
CpG site methylation and liver fat content, NAFLD and ALT
Next, we analysed the association of individual CpG sites with LFC, ALT and a diagnosis of NAFLD in pre-teenage children. 106 CpG sites were significantly associated with LFC and 122 with ALT at an adjusted p-value <0.005 (Supplementary Tables 7–9). Only two CpG sites were significantly associated with a diagnosis of NAFLD at the adjusted p-value <0.005. Interestingly, the genes associated with these top two CpG sites on Chromosomes 2 and 3, cg25474373 and cg07264203, correspond to RFTN2 and PRICKLE2, both of which were also the top two CpG sites found to be differentially methylated in the LFC analysis. RFTN2, Raftlin Family Member 2, is a protein coding gene that mediates clathrin-dependent internalization of TLR4 in some cells upon bacterial lipopolysaccharide stimulation and may regulate B-cell antigen receptor-mediated signalling[Citation60]. More recently, lower levels of raftlin in maternal serum and placental tissue were found in women who gave birth to foetus with intrauterine growth retardation (IUGR) [Citation61] which is interesting as IUGR has been associated with metabolic dysfunction including insulin resistance and NAFLD later in life[Citation62]. The exact function of PRICKLE2, Prickle Planar Polarity Protein 2 gene, is not well described but the gene encodes a postsynaptic protein involved in neuronal architecture and function. Its function has been related to the Wnt Signalling pathway. Altered DNA methylation of this gene in sperm has been associated with reduced fecundity[Citation63]. Altered methylation of this CpG or gene has not previously been associated with the liver or fatty liver in the published literature.
Overlap with DNA methylation in adult NAFLD
Next, we assessed the overlap between 22 CpGs differentially methylated in adult NAFLD [Citation28] with the significantly differentially methylated CpGs in the peripheral blood of children with liver fat accumulation (top tertile PDFF). Two CpGs, cg18120259 corresponding to the LOC100132354 gene, and cg19016694 corresponding to the TBCD gene, overlapped between the two analyses. In both analyses, hypomethylation of cg10916694 was associated with higher LFC while hypermethylation of cg18120259 was associated with higher LFC in our analysis and mixed in the adult NAFLD analysis. Many others overlapped in terms of gene name but not by specific CpG site (i.e., SKI, PHGDH, SLC7A11, RPS6KA2, SLC43A1, CPT1A, SLC9A3R1, ZFR2, LINC00649, ABCG1).
Overlap with DNA methylation and paediatric body mass index
DNA methylation has also been associated with adiposity in adults. To determine whether changes in DNA methylation may underlie earlier associations between exposures and adiposity, a large meta-analysis of children in five continents was recently conducted[Citation53]. These investigators identified a relationship between DNA methylation and BMI in different age groups. We compared our differential methylation results for LFC with those associated with BMI to determine a potential overlap. Of the 13 genes that met genome-wide significance in their analysis, we identified an overlap three SNORD genes and SNHG1 (SNORD29, SNORD30 and SNORD31, SNHG1).
Pathway and upstream regulator analyses
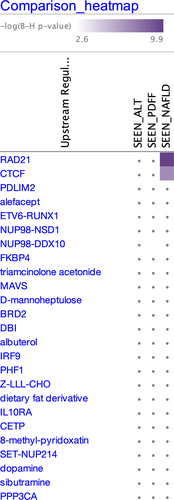
In order to assess how DMRs associate with one another and significant biological processes and pathways, we conducted an Ingenuity Pathway Analysis of the three outcomes. For the canonical pathway analyses, Type I Diabetes Mellitus signalling, Type II Diabetes Mellitus signalling, Antioxidant Action of Vitamins and MSP-RON signalling pathways were significant across all three outcomes while those most involved with DMRs from the hepatic fat content (PDFF) analysis also included Endocannabinoid Cancer, TGF-β signalling, and Interferon signalling (). Glycine Betain Degradation was the most significant canonical pathway involved with NAFLD. For the analysis of upstream transcriptional regulators, IPA identified two significant genes, CTCF and RAD21, for the NAFLD DMRs while there were no significant upstream regulators identified for ALT or PDFF ().
Discussion
To our knowledge, this is one of the first epigenome-wide association studies of liver fat accumulation and liver injury in a multi-ethnic group of pre-teenage children. The aim was to determine these associations in order to identify markers of future progressive liver disease and poor metabolic health as well as genes and pathways involved. In this cohort of pre-teenage children, we identified 88 DMRs and 106 CpGs significantly associated with LFC accumulation measured as a both higher versus lower LFC and continuous values. Many of the identified DMRs and CpGs were in or near genes regulating biological pathways relevant to the development of liver steatosis and associated metabolic risk factors. These include the DMR of the MROH6 promoter which affects expression of TGFB-1 and genetic variants have been previously identified as playing an important role in determining IL-18 levels in a large study of patients with acute coronary syndrome[Citation59]. IL-18 promotes atherosclerotic plaque formation in humans. Another was the DMR of the WAP, follistatin/Kazal, Immunoglobulin, Kunitz and Netrin Domain Containing 2 (WFIKKN2) gene containing 20 CpGs on chromosome 17. This gene controls the action of many types of proteases and a recently published study assessing the plasma proteome in obesity [Citation64] found that this gene has a potentially causal role in the development of obesity likely via regulating some members of the transforming growth factor-beta (TGFB) family. Thus, altered methylation of this gene may associate with an increased risk for liver fat accumulation through promotion of obesity. In our analysis of CpG site methylation with continuous LFC, the two most significant genes overlapped between the LFC and NAFLD analyses, RFTN2 and PRICKLE2. These did not overlap with the significant DMRs, however. Ingenuity Pathway Analysis for differentially methylated DMRs and CpGs did identify many common pathways associated with altered DNA methylation and increased LFC including TGFβ signalling, IL-12 and IL-22 signalling and production of macrophages, BEX signalling pathway, and FXR/RXR activation.
In our assessment of DNA methylation and our diagnosis of NAFLD in children, the most significant DMR was downstream of the gene ZNRD1 (Zinc ribbon domain-containing 1), also known as RNA polymerase I subunit N. This gene is known to play a role in regulation of cell proliferation and in the development of several kinds of cancers including hepatocellular carcinoma, but has not yet been associated with NAFLD or obesity in the published literature[Citation65]. The second most significant DMR in the NAFLD analysis was in the promoter region of the GLTPD2, glycolipid transfer protein domain containing 2, gene. Glycolipid transfer proteins accelerate the intermembrane transfer of glycolipids and can regulate sphingolipid homoeostasis, inflammation and autophagy. Very little is published about this specific GLTP, but a recent large scale study of genetic regulation of the lipidome and its association with cardiovascular disease found a variant of this gene is associated with atherosclerosis[Citation66]. Six genes were identified in common between the NAFLD and ALT DMRs analyses including AURKC, BHMT, FAM47E-STBD1, KIAA0040, NBPF19 and ZNF264. Of these, BHMT (betaine-homocysteine S-methyltransferase), is the best documented for its role in methylation as it is an enzyme that catalyzes the conversion of homocysteine into methionine by replacing a hydrogen on homocysteine with a methyl group from betaine. BHMT is important in metabolic health as it provides the body with the essential amino acid methionine which is key to maintaining normal patterns of DNA methylation, one carbon metabolism, and choline metabolism which is important to lipoprotein formation[Citation67]. There is ample published literature in animal models to suggest that alterations in these amino acids (i.e., choline, methionine, betaine) in utero and early life alter an offspring’s phenotype and risk for obesity and diabetes as well as hepatic steatosis [Citation68,Citation69]. In addition, groundbreaking work by Robert Waterland, Randy Jirtle and colleagues revealed the epigenetic mechanisms likely play a significant role in the regulation of transgenerational nutritional effects on obesity[Citation70]. For example, in one study using agouti mice, they showed that a methyl-supplemented diet (folic acid, vitamin B12, betaine and choline) during development prevented the effect of maternal obesity on offspring obesity[Citation71]. This affect did not appear to be related to methylation of the agouti allele itself, but through establishment of epigenetic mechanisms at other weight related genomic loci that interact with methyl supplementation, thus supporting the role for epigenetic variation contributing to the interindividual effects of nutrition on offspring health. Using IPA, we found that significant DMRs in the NAFLD analysis were identified in the Glycine Betain Degradation pathway.
To identify potential mechanistically important upstream regulators to these genes, we conducted an upstream regulators analysis in IPA. We identified to significant genes, CTCF and RAD21, in the NAFLD analysis. CCCTC-binding factor (CTCF) is a highly conserved zinc finger protein and serves as transcriptional activator, repressor or insulator for thousands of genes in the mammalian genome. CTCF is known to be involved in the H19/IGF2 locus where it plays an important role in the imprinting control region (ICR) of the gene[Citation72]. There is precedent for the H19 gene in obesity as it is a long noncoding RNA and plays a role in the negative regulation of body weight and cell proliferation and its expression is controlled via imprinting. Only a few studies have been published about CTCF and fatty liver. One published in 2021 found that liver-specific deletion of CTCF lead to fatty liver in mice via augmented PPARγDNA-binding activity[Citation73]. RAD21 (RAD21 cohesin complex component) is a protein coding gene involved in the repair of DNA double-strand breaks and chromatid cohesion during mitosis. It serves a diversity of cellular functions and also interacts with an element of CTCF. Thus far, there are no data published for a role of RAD21 in fatty liver, but one recent study identified RAD21 as part of a gene signature able to identify patients with hepatocellular carcinoma[Citation74].
Because we found several meaningful alterations in DNA methylation in the blood of children with higher LFC, NAFLD and elevated ALT, we assessed whether any of the CpG sites matched hepatic-fat associated CpGs in adults. Several gene names overlapped between adult NAFLD and those found altered in LFC and ALT and NAFLD diagnosis in children. Two specific CpG sites were significant in the adult NAFLD analysis as well as the analysis of LFC in children. These included: a) cg18120259, a CpG site downstream of the LOC100132354 gene which was hypermethylated in children with top-tertile LFC vs. bottom two tertile LFC, and b) cg19016694, an intronic CpG site on the TBCD gene which was hypomethylated in top tertile LFC children and also found to be hypomethylated the blood of adult patients with increased hepatic fat[Citation28]. TBCD, tubulin folding cofactor D gene, is a protein coding gene involved in the pathway leading to correctly folded beta-tubulin. Cofactor D is one of four proteins and is thought to play a role in capturing and stabilizing beta-tubulin. LOC100132354 is a long-noncoding RNA that has been found to regulate the adjacent gene, vascular endothelial growth factor A (VEGFA). VEGF is a key factor for inducing endothelial cell proliferation and angiogenesis, particularly in tumours[Citation75]. LOC100132354 was previously identified as a potential mediator of prenatal adversity and risk factor for metabolic disease in an analysis of DNA methylation patients from the Dutch Hunger Winter study. Investigators suggested that DNA methylation alterations in this gene play a role in mediating the relationship between famine exposure during gestation and high triglyceride values in adulthood[Citation76]. In addition, a SNP near this gene has been associated with heritability of circulating VEGF levels which may play a role in increasing risk for cardiometabolic disease[Citation77]. We performed a similar comparison with a large meta-analysis of DNA methylation and BMI throughout childhood[Citation53]. Four genes associated with significant differential methylation with LFC overlapped with those findings. These included SNORD29, SNORD30 and SNORD31, SNHG1. These genes encode small Nucleolar RNA C/D Box 29, 30 and 31 and small nucleolar RNA host gene 1. All are noncoding genes and functions include guiding modifications of RNAs with C/D associated with methylation as well as regulation of expression of tumour suppressor and cell proliferation genes. The function of these genes as it relates to obesity, fatty liver, or cardiometabolic dysfunction has yet to be defined. It is also unknown if methylation changes with age or if these differences are cause or consequence of adiposity. Further investigation of the relationship between DNA methylation, adiposity, and LFC throughout the lifespan is needed.
Only one prior published study has assessed the epigenome-wide association between DNA methylation and liver fat accumulation in children. This was a large study of 10-year-old children enrolled in the Generation R Study from the Netherlands. In this study, Geurtsen and colleagues [Citation30] found a median fat fraction of 2% (95% range, 1.3, 5.1) in their majority (81%) healthy weight Caucasian children. They assessed associations of DNA methylation in umbilical cord blood in one cohort and in peripheral blood of a different cohort of 10-year-old children. Like in our analysis, they analysed DMRs as well as CpG sites using several regression models. Unlike in ours however, they did not identify any significant CpG methylation or DMRs associated with LFC. This difference may have resulted from their population which included largely Caucasian, lean, healthy children born to healthy weight mothers. By comparison, our cohort comprised a more diverse, more overweight and obese group of children, with higher percentages of liver fat and who were more likely to be exposed to obesity in utero. Such early life exposures likely impacted our ability to identify significant changes in DNA methylation and such deserves further study.
Strengths and limitations
The strengths of this study include its prospective design in an ethnically and socioeconomically diverse population and the use of an established, comprehensive, and reproducible methylation array. Sensitive imaging-based methods were used to enable non-invasive measurement of liver fat content in children without the need for contrast, sedation, or invasive liver biopsies [Citation78,Citation79]. Limitations of the study include the relatively small sample size and inability to make causal associations for alterations in DNA methylation in this investigation. Imaging results were not confirmed by liver histology but given the accuracy of MRI based imaging for liver fat in adults and the risks associated with liver biopsy in asymptomatic patients we did not feel that histologic validation was warranted. In addition, our goal is to identify non-invasive markers of liver fat accumulation and risks for future progressive NAFLD and fibrosis thus supporting the move away from liver biopsy. Currently this is only the second published study of DNA methylation and liver fat accumulation in young children and the first in an ethnically diverse population. Our cohort had a high percentage of AA children which makes it unique in that other studies of paediatric fatty liver typically include high numbers of Caucasian and/or Hispanic children. This heterogeneity may have impacted our ability to find increased LFC in AA children. We anticipate that this finding also increases the awareness of providers to consider fatty liver in children of all races and ethnicities. We acknowledge that this requires further studies in diverse populations. This is a critical need as recent studies of obese children in Indian and Malaysian found alarmingly high rates of NAFLD of over 60% by ultrasound in both populations [Citation80,Citation81]. Whether there are biologically relevant differences in methylation by race that impact LFC or metabolic dysfunction is an area that needs further study. Additionally, we were able to identify early signs of poor metabolic health in these young children. One recently published study in children aged 4–8 years from the Healthy Start Study, found that even low LFC was independently associated with estimated insulin resistance and suggested that hepatic fat may be an early indicator of metabolic dysfunction in youth. While we did not directly measure insulin resistance in our study, other markers of metabolic dysfunction such as TG and HbA1c were abnormal in our obese children with higher LFC[Citation82]. Together, these studies suggests that even a lower LFC in children should trigger providers to follow these children more closely and focus on interventions that decrease the likelihood of progressive liver injury in the future.
Conclusions
We identified significant alterations in blood DNA methylation associated with increased liver fat content and liver injury in pre-teenage children. These alterations may merely be an association but could identify important epigenetic variations resulting from developmental and/or nutritional exposures. It will therefore be critically important to follow children with ongoing metabolic risks prospectively to enable the identification of causal vs. consequential changes in DNA methylation. In addition, larger, prospective studies would aid in determining the timing and potential mechanistic associations of these changes in relation to liver and metabolic phenotypes. These advances are critical for the development of non-invasive biomarkers, prevention, and novel therapeutic targets in the fight against the rising epidemic of paediatric and adult NAFLD.
Availability of data and materials
Data from the SEEN study cohort are available from the corresponding author on reasonable request.
Supplemental Material
Download Zip (124.8 KB)Acknowledgments
We would like to thank all participants of the SEEN study for their contribution to this study. We gratefully acknowledge Carole Greiner for her laboratory expertise and contribution to the DNA methylation analyses, and David Corcoran and Jennifer Modliszewski from the Duke Center for Genomic and Computational Biology for bioinformatics assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Sahota AK, Shapiro WL, Newton KP, et al. Incidence of Nonalcoholic Fatty Liver Disease in Children: 2009-2018. Pediatrics. 2020;146(6). DOI:10.1542/peds.2020-0771
- Temple JL, Cordero P, Li J, et al. A Guide to Non-Alcoholic Fatty Liver Disease in Childhood and Adolescence. Int J Mol Sci. 2016;17(6):947.
- Schwimmer JB, Deutsch R, Kahen T, et al. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118(4):1388–1393.
- Vos MB, Abrams SH, Barlow SE, et al. NASPGHAN Clinical Practice Guideline for the Diagnosis and Treatment of Nonalcoholic Fatty Liver Disease in Children: recommendations from the Expert Committee on NAFLD (ECON) and the North American Society of Pediatric Gastroenterology, Hepatology and Nutrition (NASPGHAN). J Pediatr Gastroenterol Nutr. 2017;64(2):319–334.
- Nobili V, Alisi A, Newton KP, et al. Comparison of the Phenotype and Approach to Pediatric vs Adult Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150(8):1798–1810.
- Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20.
- Feldstein AE, Charatcharoenwitthaya P, Treeprasertsuk S, et al. The natural history of non-alcoholic fatty liver disease in children: a follow-up study for up to 20 years. Gut. 2009;58(11):1538–1544.
- Holterman A-XL, Guzman G, Fantuzzi G, et al. Nonalcoholic fatty liver disease in severely obese adolescent and adult patients. Obesity (Silver Spring). 2013;21(3):591–597.
- Vittorio J, Lavine JE. Recent advances in understanding and managing pediatric nonalcoholic fatty liver disease. F1000Research. 2020;9:377.
- Ayonrinde OT, Adams LA, Mori TA, et al. Sex differences between parental pregnancy characteristics and nonalcoholic fatty liver disease in adolescents. Hepatology. 2018;67(1):108–122.
- Ayonrinde OT, Oddy WH, Adams LA, et al. Infant nutrition and maternal obesity influence the risk of non-alcoholic fatty liver disease in adolescents. J Hepatol. 2017;67(3):568–576.
- Brumbaugh DE, Tearse P, Cree-Green M, et al. Intrahepatic fat is increased in the neonatal offspring of obese women with gestational diabetes. J Pediatr. 2013;162(5):930–936 e931.
- Modi N, Murgasova D, Ruager-Martin R, et al. The influence of maternal body mass index on infant adiposity and hepatic lipid content. Pediatr Res. 2011;70(3):287–291.
- Patel KR, White FV, Deutsch GH. Hepatic steatosis is prevalent in stillborns delivered to women with diabetes mellitus. J Pediatr Gastroenterol Nutr. 2015;60(2):152–158.
- Newton KP, Feldman HS, Chambers CD, et al. Low and High Birth Weights Are Risk Factors for Nonalcoholic Fatty Liver Disease in Children. J Pediatr. 2017;187:141–146 e141.
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114(2):147–152.
- Angulo P. Obesity and nonalcoholic fatty liver disease. Nutr Rev. 2007;65(6 Pt 2):S57–63.
- Xanthakos SA, Lavine JE, Yates KP, et al. Progression of Fatty Liver Disease in Children Receiving Standard of Care Lifestyle Advice. Gastroenterology. 2020;159(5):1731–1751. e1710.
- Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398.
- Sharp GC, Salas LA, Monnereau C, et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum Mol Genet. 2017;26(20):4067–4085.
- Martin CL, Jima D, Sharp GC, et al. Maternal pre-pregnancy obesity, offspring cord blood DNA methylation, and offspring cardiometabolic health in early childhood: an epigenome-wide association study. Epigenetics. 2019;14(4):325–340.
- Girchenko P, Lahti J, Czamara D, et al. Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clin Epigenetics. 2017;9(1):49.
- Czamara D, Eraslan G, Page CM, et al. Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nat Commun. 2019;10(1):2548.
- Murphy SK, Yang H, Moylan CA, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–1087.
- Hotta K, Kitamoto A, Kitamoto T, et al. Identification of differentially methylated region (DMR) networks associated with progression of nonalcoholic fatty liver disease. Sci Rep. 2018;8(1):13567.
- Gerhard GS, Malenica I, Llaci L, et al. Differentially methylated loci in NAFLD cirrhosis are associated with key signaling pathways. Clin Epigenetics. 2018;10(1):93.
- Ahrens M, Ammerpohl O, von Schonfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302.
- Ma J, Nano J, Ding J, et al. A Peripheral Blood DNA Methylation Signature of Hepatic Fat Reveals a Potential Causal Pathway for Nonalcoholic Fatty Liver Disease. Diabetes. 2019;68(5):1073–1083.
- Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66(7):1321–1328.
- Geurtsen ML, Jaddoe VWV, Salas LA, et al. Newborn and childhood differential DNA methylation and liver fat in school-age children. Clin Epigenetics. 2019;12(1):3.
- Slieker RC, Bos SD, Goeman JJ, et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin. 2013;6(1):26.
- Johnson ND, Wu X, Still CD, et al. Differential DNA methylation and changing cell-type proportions as fibrotic stage progresses in NAFLD. Clin Epigenetics. 2021;13(1):152.
- Nano J, Ghanbari M, Wang W, et al. Epigenome-Wide Association Study Identifies Methylation Sites Associated With Liver Enzymes and Hepatic Steatosis. Gastroenterology. 2017;153(4):1096–1106. e1092.
- Hoyo C, Murtha AP, Schildkraut JM, et al. Folic acid supplementation before and during pregnancy in the Newborn Epigenetics STudy (NEST). BMC Public Health. 2011;11(1):46.
- Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012;7(7):735–746.
- Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, et al. CDC growth charts: united States. Adv Data. 2000(314):1–27.
- Flynn JT, Kaelber DC, Baker-Smith CM, et al. Clinical Practice Guideline for Screening and Management of High Blood Pressure in Children and Adolescents. Pediatrics. 2017;140(3). DOI:10.1542/peds.2017-1904
- Bashir MR, Zhong X, Nickel MD, et al. Quantification of hepatic steatosis with a multistep adaptive fitting MRI approach: prospective validation against MR spectroscopy. AJR Am J Roentgenol. 2015;204(2):297–306.
- Zhong X, Nickel MD, Kannengiesser SA, et al. Liver fat quantification using a multi-step adaptive fitting approach with multi-echo GRE imaging. Magn Reson Med. 2014;72(5):1353–1365.
- Serai SD, Towbin AJ, Podberesky DJ. Pediatric liver MR elastography. Dig Dis Sci. 2012;57(10):2713–2719.
- Goyal NP, Sawh MC, Ugalde-Nicalo P, et al. Evaluation of Quantitative Imaging Biomarkers for Early-phase Clinical Trials of Steatohepatitis in Adolescents. J Pediatr Gastroenterol Nutr. 2020;70(1):99–105.
- Zeng J, Zhang X, Sun C, et al. Feasibility study and reference values of FibroScan 502 with M probe in healthy preschool children aged 5 years. BMC Pediatr. 2019;19(1):129.
- Engelmann G, Gebhardt C, Wenning D, et al. Feasibility study and control values of transient elastography in healthy children. Eur J Pediatr. 2012;171(2):353–360.
- Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–357.
- Geurtsen ML, Santos S, Felix JF, et al. Liver Fat and Cardiometabolic Risk Factors Among School-Age Children. Hepatology. 2020;72(1):119–129.
- Di Martino M, Pacifico L, Bezzi M, et al. Comparison of magnetic resonance spectroscopy, proton density fat fraction and histological analysis in the quantification of liver steatosis in children and adolescents. World J Gastroenterol. 2016;22(39):8812–8819.
- Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One. 2012;7(7):e40924.
- Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369.
- R: a language and environment for statistcal computing. [ computer program]. 2019.
- Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330.
- Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8(1):6.
- Benjamini YH, Yosef. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Society of Statistical Society, Series B. 1995;57:289–300.
- Vehmeijer FOL, Kupers LK, Sharp GC, et al. DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies. Genome Med. 2020;12(1):105.
- Urbina EM, Khoury PR, McCoy CE, et al. Triglyceride to HDL-C ratio and increased arterial stiffness in children, adolescents, and young adults. Pediatrics. 2013;131(4):e1082–1090.
- Nur Zati Iwani AK, Jalaludin MY, Wan Mohd Zin RM, et al. TG : HDL-C Ratio Is a Good Marker to Identify Children Affected by Obesity with Increased Cardiometabolic Risk and Insulin Resistance. Int J Endocrinol. 2019;2019:8586167.
- Young KA, Maturu A, Lorenzo C, et al. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance, β-cell function, and diabetes in Hispanics and African Americans. J Diabetes Complications. 2019;33(2):118–122.
- Liu J, Fox CS, Hickson DA, et al. Impact of abdominal visceral and subcutaneous adipose tissue on cardiometabolic risk factors: the Jackson Heart Study. J Clin Endocrinol Metab. 2010;95(12):5419–5426.
- Maffeis C, Manfredi R, Trombetta M, et al. Insulin sensitivity is correlated with subcutaneous but not visceral body fat in overweight and obese prepubertal children. J Clin Endocrinol Metab. 2008;93(6):2122–2128.
- Johansson Å, Eriksson N, Becker RC, et al. NLRC4 Inflammasome Is an Important Regulator of Interleukin-18 Levels in Patients With Acute Coronary Syndromes: genome-Wide Association Study in the PLATelet inhibition and patient Outcomes Trial (PLATO). Circ Cardiovasc Genet. 2015;8(3):498–506.
- Tatematsu M, Yoshida R, Morioka Y, et al. Raftlin Controls Lipopolysaccharide-Induced TLR4 Internalization and TICAM-1 Signaling in a Cell Type–Specific Manner. J Immunol. 2016;196(9):3865–3876.
- Ozer A, Tolun F, Aslan F, et al. The role of G protein-associated estrogen receptor (GPER) 1, corin, raftlin, and estrogen in etiopathogenesis of intrauterine growth retardation. J Matern Fetal Neonatal Med. 2021;34(5):755–760.
- Spradley FT, Smith JA, Alexander BT, et al. Developmental origins of nonalcoholic fatty liver disease as a risk factor for exaggerated metabolic and cardiovascular-renal disease. Am J Physiol Endocrinol Metab. 2018;315(5):E795–E814.
- Laqqan M, Tierling S, Alkhaled Y, et al. Spermatozoa from males with reduced fecundity exhibit differential DNA methylation patterns. Andrology. 2017;5(5):971–978.
- Zaghlool SB, Sharma S, Molnar M, et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat Commun. 2021;12(1):1279.
- Hu X, Wang R, Ren Z, et al. MiR-26b suppresses hepatocellular carcinoma development by negatively regulating ZNRD1 and Wnt/β-catenin signaling. Cancer Med. 2019;8(17):7359–7371.
- Tabassum R, Ramo JT, Ripatti P, et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun. 2019;10(1):4329.
- Zeisel SC. Choline, Other Methyl-Donors and Epigenetics. Nutrients. 2017;9(5):445.
- Wolff GL, Kodell RL, Moore SR, et al. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957.
- Cordero P, Gomez-Uriz AM, Campion J, et al. Dietary supplementation with methyl donors reduces fatty liver and modifies the fatty acid synthase DNA methylation profile in rats fed an obesogenic diet. Genes Nutr. 2013;8(1):105–113.
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23(15):5293–5300.
- Waterland RA, Travisano M, Tahiliani KG, et al. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond). 2008;32(9):1373–1379.
- Kim S, Yu N-K, Kaang B-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47(6):e166.
- Choi Y, Song M-J, Jung W-J, et al. Liver-Specific Deletion of Mouse CTCF Leads to Hepatic Steatosis via Augmented PPARγ Signaling. Cell Mol Gastroenterol Hepatol. 2021;12(5):1761–1787.
- Cai J, Li B, Zhu Y, et al. Prognostic Biomarker Identification Through Integrating the Gene Signatures of Hepatocellular Carcinoma Properties. EBioMedicine. 2017;19:18–30.
- Wang Y, Zhang F, Wang J, et al. lncRNA LOC100132354 promotes angiogenesis through VEGFA/VEGFR2 signaling pathway in lung adenocarcinoma. Cancer Manag Res. 2018;10:4257–4266.
- Tobi EW, Slieker RC, Luijk R, et al. DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci Adv. 2018;4(1):eaao4364.
- Choi SH, Ruggiero D, Sorice R, et al. Six Novel Loci Associated with Circulating VEGF Levels Identified by a Meta-analysis of Genome-Wide Association Studies. PLoS Genet. 2016;12(2):e1005874.
- Middleton MS, Van Natta ML, Heba ER, et al. Diagnostic accuracy of magnetic resonance imaging hepatic proton density fat fraction in pediatric nonalcoholic fatty liver disease. Hepatology. 2018;67(3):858–872.
- Schwimmer JB, Middleton MS, Behling C, et al. Magnetic resonance imaging and liver histology as biomarkers of hepatic steatosis in children with nonalcoholic fatty liver disease. Hepatology. 2015;61(6):1887–1895.
- Gupta N, Jindal G, Nadda A, et al. Prevalence and risk factors for nonalcoholic fatty liver disease in obese children in rural Punjab, India. Journal of Family and Community Medicine. 2020;27(2):103–108.
- Mohamed RZ, Jalaludin MY, Anuar Zaini A. Predictors of non-alcoholic fatty liver disease (NAFLD) among children with obesity. J Pediatr Endocrinol Metab. 2020;33(2):247–253.
- Cohen CC, Perng W, Sundaram SS, et al. Hepatic Fat in Early Childhood Is Independently Associated With Estimated Insulin Resistance: the Healthy Start Study. J Clin Endocrinol Metab. 2021;106(11):3140–3150.