ABSTRACT
The detection of methylated templates in cell-free DNA (cfDNA) is increasingly recognized as a valuable, non-invasive tool for diagnosis, monitoring and prognostication in a range of medical contexts. The importance of controlling pre-analytical conditions in laboratory workflows prior to cfDNA quantification is well-established. Significant variations in the recovery of DNA following processes such as cfDNA extraction and sodium bisulphite modification may confound downstream analysis, particularly when accurate quantification of templates is required. Given the wealth of potential applications for this emerging molecular technology, attention has turned to the requirement to recognize and minimize pre-analytical variables prior to cfDNA methylation analysis. We recently described the development of an approach using an exogenous DNA construct to evaluate the recovery efficiency of cfDNA following the extraction and bisulphite modification steps (CEREBIS). Here, we report our experience in the practical application of this technique in 107 consecutive patient plasma samples submitted for quantitative cfDNA methylation analysis. The mean recovery of cfDNA (as estimated using cerebis), following extraction and bisulphite modification, was 37% ± 7%. Nine (8.4%) of the 107 samples were found to be outside of control limits, where the recovery of cerebis indicated significant differences in the efficiency of the pre-analytical processing of these samples. Recognition of these out-of-control samples precluded subsequent molecular analysis. Implementation of data-driven quality control measures, such as the one described, has the potential to improve the quality of liquid biopsy methylation analysis, interpretation and reporting.
Introduction
The analysis of methylation patterns in cell-free DNA (cfDNA) is increasingly being recognized as an invaluable, non-invasive tool for diagnosis, monitoring and prognostication across a range of medical disciplines. In oncology, the quantitative analysis of cfDNA fragments based on their methylation status at a target locus has demonstrated utility in screening, targeted diagnosis and as a predictor of treatment response in a number of cancers. [Citation1–4]
The importance of controlling pre-analytical conditions in laboratory workflows prior to cfDNA quantification is well-recognized[Citation5]. Significant variations in the recovery of DNA following processes such as cfDNA extraction from the sample and sodium bisulphite modification of cfDNA may confound downstream analysis. This issue is particularly pertinent in disease monitoring when performing quantitative analysis of a methylated cfDNA target in serial samples from the same patient. Variations in pre-analytical processes may occur despite the use of standardized protocols due to insensible losses of the DNA analyte to plasticware, pipetting errors or lot-to-lot variation in laboratory reagents and consumables. Given the wealth of potential applications for this emerging molecular technology and the importance of precision, attention has turned towards minimization of pre-analytical variables prior to the analysis of methylated cfDNA. [Citation6–8]
One approach adopted to address some of these issues has been the incorporation of ‘spike-in’ molecules to act as controls during pre-analytical laboratory processes. The quantification of spike-in controls in the analyte following these steps has been used to compare the performance of different cfDNA extraction and bisulphite modification kits and has been shown to reflect the recovery of target, endogenous cfDNA (mainly in the setting of quantifying rare mutant alleles). [Citation7,Citation9,Citation10]
We recently described the development of an approach using an exogenous DNA construct to evaluate the recovery efficiency of cfDNA following the extraction and bisulphite modification steps prior to cfDNA methylation analysis (CEREBIS)[Citation11]. This synthetic DNA construct, containing a random, non-human DNA sequence was designed to mimic the mono-nucleosomal fragment size of cfDNA and incorporated strategically placed cytosine-free regions to enable PCR amplification, with the same primer pair, both before and after bisulphite modification.
In this proof-of-concept study, a known concentration of CEREBIS was spiked into a small cohort of plasma samples prior to cfDNA extraction and bisulphite modification. CEREBIS recovery was subsequently quantified following these steps using droplet digital PCR (ddPCR), by employing a set of primers targeting non-cytosine regions of the construct. This approach enabled the estimation of cfDNA recovery in samples in this small cohort following these key steps in the laboratory workflow; providing a measure of quality control for each plasma sample with the potential to identify problems with pre-analytical processes.
Here, we report our experience and the feasibility of the practical application of this technique in a larger cohort of 107 consecutive patient plasma samples submitted for quantitative cfDNA methylation analysis.
Methods
This study is reported in accordance with the updated ‘Minimum Information for Publication of Quantitative Digital PCR Experiments (dMIQE) guidelines’ (Suppl. Table S1). [Citation12]
Human Research Ethics Committee approval was granted by the Austin Hospital (HREC/18/Austin/147). Plasma samples included in this study were obtained from a cohort of consenting adult liver transplant recipients at a large Australian liver transplantation centre. They were intended for downstream quantification of graft-specific methylated cfDNA templates as part of a clinical study to develop non-invasive biomarkers to monitor graft health post-transplantation.
150,000 copies of CEREBIS in 10 μL of DNA elution buffer were spiked into each 4 ml thawed plasma sample prior to cfDNA extraction and subsequent bisulphite modification (see Suppl. Figure S1 for detail on the CEREBIS construct). The blood samples had been previously processed using 2 step-centrifugation within three hours of collection (800 g 10 min AT, 3000 g 10 min AT) to obtain the cell-free plasma fraction prior to frozen storage at −80°C. cfDNA extraction from samples (into an end volume of 50uL of elution buffer) and bisulphite modification (into an end volume of 20uL of elution buffer) were performed using the QIAamp Circulating Nucleic Acid Kit (Qiagen) and EZ DNA Methylation-Lightning™ kit (Zymo Research) respectively, according to the manufacturers’ instructions. Each of the extraction and bisulphite batches included negative extraction controls (blanks). Eluted DNA (in AVE buffer [Qiagen]) was stored at 4°C prior to analysis. The recovery of CEREBIS was quantified in the eluted cfDNA following bisulphite modification using ddPCR (Bio-Rad QX200 ddPCR™ system [Bio-Rad, Pleasanton, CA]) as previously described (Suppl. Table S2 gives details on the reaction mixture and PCR conditions)[Citation11]. All reactions met the minimum number of partitions required for analysis as recommended by the manufacturer (>10,000). The raw data were analysed using the QuantaSoft™ Analysis Pro v1.0 (Bio-Rad) employing the default thresholding settings.
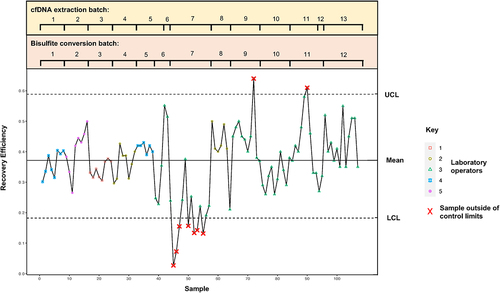
A control chart was created to visualize performance for each consecutive sample (). Data derived from the previous pilot study was used to calibrate the expected performance of these pre-analytical steps (mean DNA recovery efficiency following bisulphite modification: 38% ± 6%). Upper and lower control limits were set, based on the beta probability distribution. [Citation13] The Beta chart control limits are computed from percentiles of the cumulative distribution function (cdf) of the Beta distribution; a 90% confidence interval was employed.
Figure 1. Control chart illustrating cfDNA recovery in 107 consecutive patient samples following cfDNA extraction and bisulphite modification as measured by ddPCR using CEREBIS. The mean recovery efficiency was 37% ± 6. Upper and lower control limits (UCL, LCL) were set according to the beta probability distribution. Samples breaching control limits (n = 9) are marked with an X. cfDNA extraction and bisulphite modification batches are marked.
Results
As illustrated in , 9 (8.4%) samples were found to be outside of the control limits, where the recovery of CEREBIS indicated significant variability in efficiency of the pre-analytical processing of these samples. Following review, decreased recovery observed in samples 44 to 56 was deemed likely to be attributed to one or more of a combination of factors including (i) the use of dated extraction and bisulphite modification consumables (nearing, but not past, the recommended expiration date) for this batch of samples, (ii) a new laboratory operator and (iii) repeated freeze-thaw of the CEREBIS construct resulting in diminished performance. These variables were addressed and rectified through (i) the use of new cfDNA extraction and bisulphite modification kits, (ii) further training of the new operator and (iii) the use of a new lot of CEREBIS in single-use aliquots. As a result, the performance of the workflow was restored for the plasma samples that followed. Interestingly, samples 72 and 90 were also identified to fall outside of the control limits of the workflow. Review of run-sheets showed that the operator inadvertently doubled a reagent during the cfDNA extraction process for these samples, resulting in an ‘over-performance’ of cfDNA recovery following bisulphite modification. Further experiments are currently being conducted in our laboratory to identify potential mechanisms for this observation.
Recognition of these out-of-control samples precluded subsequent molecular analysis of the bisulphite-modified DNA as it was considered that the technical errors identified in these pre-analytical steps were likely to confound quantitative cfDNA analysis and lead to a potential misinterpretation of the true underlying biology.
Discussion
Monitoring of CEREBIS recovery has two key advantages. Firstly, it can be used to identify samples breaching control limits. The identification of these samples offers the opportunity to recognize technical errors or processes that may confound the interpretation of disease states following molecular analysis. Equipped with this quality control information, an operator may elect to request recollection of the sample or repeat the cfDNA extraction and bisulphite modification. In cases where the above is not possible, molecular analysis could be performed with an appropriately cautious interpretation of results.
Secondly, CEREBIS can assist in further assuring that any observed biological effect in serial samples analysed from a single patient is likely true and unlikely to be related to the pre-analytical technical factors of cfDNA extraction and bisulphite modification. This is especially important in the context of quantitative cfDNA analysis used in disease monitoring e.g., in the oncological setting of monitoring treatment response or for monitoring graft health following transplantation. [Citation14–16]
The ability to demonstrate reproducibility is an important characteristic for a laboratory quality control measure such as CEREBIS. Importantly, the mean recovery of CEREBIS, following extraction and bisulphite modification steps, in the 107 plasma samples reported here was 37% ± 7%. This is highly comparable to the mean recovery efficiency reported in the initial pilot study (38% ± 6%). This consistency in the use of CEREBIS was achieved despite a) the inclusion of a number of new laboratory operators, b) different lot numbers of laboratory consumables and c) the incorporation of a new CEREBIS construct order from a different supplier. Overall, these observations indicate that CEREBIS will be highly applicable, as a quality control technique, in other laboratories.
The clinical applications of molecular diagnostics continue to broaden, and stringent quality control measures are required to improve the interpretation and validity of data. Implementation of data-driven quality control measures such as the one described has the potential to improve the quality of liquid biopsy methylation analysis.
Abbreviations
AT, ambient temperature; CEREBIS, Construct to Evaluate Recovery Efficiency after cell-free DNA extraction and Bisulfite modification; cfDNA, cell-free DNA; ddPCR, droplet digital PCR; DNA, deoxyribonucleic acid.
Supplemental Material
Download MS Word (25.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author on application upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2022.2091821
Additional information
Funding
References
- Song L, Jia J, Peng X, et al. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep. 2017;7(1):3032.
- Ding W, Chen G, Shi T. Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis. Epigenetics. 2019;14(1):67–80.
- Kondrashova O, Topp M, Nesic K, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9(1):3970.
- Liu MC, Oxnard GR, Klein EA, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31(6):745–759.
- Meddeb R, Pisareva E, Thierry AR. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin Chem. 2019;65(5):623–633.
- Kerachian MA, Azghandi M, Mozaffari-Jovin S, et al. Guidelines for pre-analytical conditions for assessing the methylation of circulating cell-free DNA. Clin Epigenetics. 2021;13(1):193.
- Worm Ørntoft MB, Jensen S, Hansen TB, et al. Comparative analysis of 12 different kits for bisulfite conversion of circulating cell-free DNA. Epigenetics. 2017;12(8):626–636.
- Van Paemel R, De Koker A, Caggiano C, et al. Genome-wide study of the effect of blood collection tubes on the cell-free DNA methylome. Epigenetics. 2021;16(7):797–807.
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406(26):6499–6512.
- Lampignano R, Neumann MHD, Weber S, et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin Chem. 2020;66(1):149–160.
- Goh SK, Cox DRA, Wong BKL, et al. A synthetic DNA construct to evaluate the recovery efficiency of cell-free DNA extraction and bisulfite modification. Clin Chem. 2021;67(9):1201–1209.
- The dMIQE Group, Huggett JF, Trypsteen W. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin Chem. 2020;66(8):1012–1029.
- Sant’Anna ÂMO, ten Caten CS. Beta control charts for monitoring fraction data. Expert Syst Appl. 2012;39(11):10236–10243.
- Bergheim J, Semaan A, Gevensleben H, et al. Potential of quantitative SEPT9 and SHOX2 methylation in plasmatic circulating cell-free DNA as auxiliary staging parameter in colorectal cancer: a prospective observational cohort study. Br J Cancer. 2018;118(9):1217–1228.
- Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67(11):1995–2005.
- Cox DRA, Low N, Goh SK, et al. Low levels of hepatocyte-specific methylation in cell-free DNA are a strong negative predictor for acute t cell-mediated rejection requiring treatment following liver transplantation. Liver Transpl. 2022;28(6):1024–1038.
