ABSTRACT
Background: Circular RNA (circRNA) plays a critical role in tumour progression. Circ-CCT3, a particularly abundant circRNA, was proposed to be involved in tumorigenesis. However, the role of circ-CCT3 in hepatocellular carcinoma remains elusive.Methods: Here, circ-CCT3 (a circRNA derived from exons 3, 4 and 5 of the CCT3 gene, hsa_circ_0004680) was identified by circRNA microarray and validated by qRT-PCR. RNA immunoprecipitation (RIP) was performed to confirm the binding between ALKBH5 along with METTL3 and circ-CCT3. Methylated RNA Immunoprecipitation (MeRIP) was used to detect the N6-methyladenosine (m 2A) levels of circ-CCT3. CircRNAs in vivo precipitation, luciferase reporter assay, biotin-coupled microRNA capture, and fluorescence in situ hybridization were conducted to assess the interaction between circ-CCT3 and miR-378a-3p. The functions of circ-CCT3 in HCC were evaluated both in vitro and in vivo.Results: We demonstrated that circ-CCT3 was highly expressed in HCC which indicated the poor prognosis. Circ-CCT3 expression served as an independent risk factor for overall survival in patients with HCC. Knocking-down of circ-CCT3 inhibited the proliferation, invasion and migration of HCC cells, and angiogenesis of HUVEC. Mechanistically, ALKBH5 and METTL3 could bind and regulate m A-modification of circ-CCT3. Further, circ-CCT3 upregulated the expression of FLT-1 by sponging miR-378a-3p.Conclusions: Circ-CCT3 was significantly up-regulated in HCC and promoted liver cancer development via miR-378a-3p-FLT1 axis. It was also found that circ-CCT3 was under m A-modification mediated by ALKBH5 and METTL3. Our study highlights circ-CCT3 as a potential therapeutic target of HCC treatment, which provides a novel understanding on mechanisms of circRNAs in HCC progression.
Background
Hepatocellular carcinoma (HCC), which currently accounts for the majority of liver cancer, is the 6th most common cancer worldwide [Citation1,Citation2]. The high degree of malignancy of HCC and limited effective treatments contribute to the alarming mortality, representing the 3th leading cause of all cancer-related deaths worldwide yearly (incidence >800,000 per year) [Citation2–4]. Therefore, identifying new targets and biomarkers for the treatment of HCC is vital important. CircRNAs are covalently closed RNA rings processed by back-splicing of precursor mRNA, which lack 5′-3′ ends and poly (A) tails. They can be divided into several forms, including introns, exons, introns – exon, or fusion transcripts of parental genes in terms of genomic sources [Citation5–7]. CircRNAs were first discovered in viruses as early as the 1970s [Citation8] and were originally thought to be non-functional by-products of aberrant RNA splicing and thus have not incited enough scientific attention. However, recently, accumulating studies have suggested that circRNAs have multi-functions, including microRNA sponges, transcriptional regulators, scaffolds for proteins, templates for protein synthesis and competitors of linear splicing [Citation9,Citation10]. Moreover, dysregulated expressions of circRNAs have been reported to be correlated with cancer biology [Citation11–16]. Previous studies have suggested that the great majority of circRNAs can function as microRNAs (miRNAs) sponges [Citation17–20] or bind to proteins [Citation21,Citation22] to regulate the related target genes expression. In this study, we performed circRNA sequencing to elucidate the dysregulated circRNAs in HCC tissues compared with adjacent noncancerous liver tissue.Circ-CCT3 is originated from exons 3–5 of chaperonin containing TCP1 subunit 3 (CCT3) by back-splicing [Citation23]. Current studies focusing on the mechanism of functions of circ-CCT3 in HCC remains unclear, which is urgently needed to be deeply investigated.
Materials and Methods
Details on materials and methods are described in the Supplementary data.
Results
Overexpression of Circ-CCT3 correlated with poor prognosis of HCC
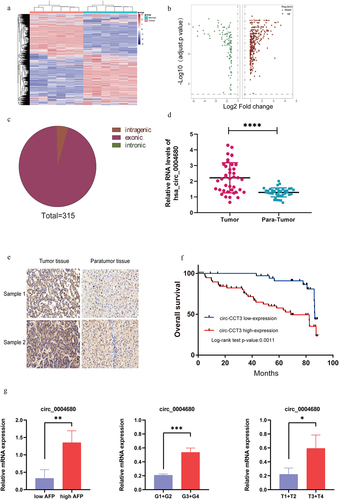
To examine the expression of circRNAs in HCC, we analysed six pairs of human HCC clinical samples and adjacent normal tissues by circRNA microarray assays. The expression analysis showed that 426 circRNAs were expressed differentially in HCC compared with matched adjacent normal liver tissues (). Among them, 315 circRNAs were upregulated (), while 113 circRNAs were downregulated. Further, we analysed the genome sources of the 315 upregulated circRNAs and found that they can be separated into three categories which were intronic, exonic, and intragenic of their parental genes. Most of the circRNAs were derived from exon clipping and cyclization (). Interestingly, we found that two circular RNAs (hsa_circ_0014717 and hsa_circ_0004680) highly expressed in HCC. To confirm our findings, the level of the two circRNAs was quantified in 40 paired HCC and adjacent normal liver tissues by qRT-PCR. We found that it was hsa_circ_0004680 but not hsa_circ_0014717 which overexpressed in HCC tissue, and we term hsa_circ_0004680 as circ-CCT3 (). To further define the expression of circRNA in HCC, another 97 paired HCC and adjacent normal liver tissues were detected with RNA in situ hybridization experiment by digoxin modified circ-CCT3 probe. The results showed that circ-CCT3 was highly expressed in HCC tissues compared with adjacent normal liver tissues (). Moreover, the Kaplan–Meier analysis indicated that the HCC patients with high circ-CCT3 expression were closely associated with poor prognosis after hepatectomy (). In addition, the expression of circ_004680 throughout the clinicopathological spectrum in HCC was investigated. The results revealed that the mRNA level of circ_004680 was correlated with pathological grade, AFP level, and TNM classification (). Collectively, these results suggested that circ-CCT3 is upregulated in HCC and may be a potential biomarker for predicting the poor prognosis of HCC.
Figure 1. Hsa_circ_0004680 is up-regulated and correlated with poor clinical outcomes in HCC patients. (a) The difference of expression in six paired of HCC samples. (b) High and low expression of circle RNAs in HCC tissue. (c) 315 circle RNAs were high expressed in HCC. (d) circ_0004680 was high expression in HCC. (e) HCC tissue array was stained by RNA in situ hybridization. (f) High expression of circ-CCT3 showed poor prognosis in HCC patients, p = 0.0011. (g)Relationships between clinicopathology (including pathological grade, AFP level, and TNM stage) and circ_004680 in HCC tissues. The results revealed that the mRNA level of circ_004680 was correlated with pathological grade, AFP level and TNM classification.
Characterization of Circ-CCT3 in HCC
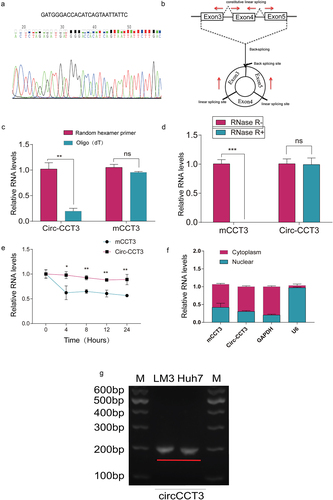
According to circBase, circ-CCT3 is derived from exons 3–5 of CCT3 gene. To identify the circular structure and characteristics of circ-CCT3, we first designed a pair of divergent primers targeting at circ-CCT3 and a pair of convergent primers for CCT3. Then, we use Sanger sequencing to examine the specific back splicing site of circ-CCT3(). Further, the reverse transcription experiments were performed with the random hexamer and oligo(dT)18 primers. The results showed that compared with random hexamer primers, the relative expression of circ-CCT3 was significantly downregulated when using the oligo (dT)18 primers, while CCT3 mRNA (mCCT3) was not (). Circ-CCT3 was proved to have no poly-A tail. Additionally, circ-CCT3 was resistant to RNase R, a highly processive 3’ to 5’exoribonuclease that digests linear RNAs, implying that circ-CCT3 was circular (). Moreover, we used actinomycin D to inhibit transcription. Then, we measured the half-life of circ-CCT3 and CCT3 mRNA in HCC-LM3 cells. As expected, circ-CCT3 was more stable than mCCT3 (). In addition, nucleocytoplasmic fractionation assays showed that cir-CCT3 is mainly distributed in the cytoplasm (). Besides, we performed the electrophoresis experiments to determine the size of circ-CCT3 (). Collectively, these findings indicated that circ-CCT3 is a circular and stable transcript and distributed both in cytoplasm and nuclear.
Figure 2. Characteristics of hsa_circ_0004680.(a) Sanger sequencing of RT-PCR products performed by circ-CCT3 primer. (b) circ_0004680 derived from exon 3,4 and 5 of gene CCT3. (c) circ-CCT3 can not be digested by RNase R, while it’s parental gene CCT3 was seriously digested.***p < 0.001. (d) circ-CCT3 can be amplified by random hexamer primer. (e) circ-CCT3 was restrained by Actinomycin D. (f) PCR results showed circ-CCT3 was mainly located in cytoplasm. (g) the electrophoresis experiments result of circ-CCT3 in LM3 and Huh7 cell lines.
Knock-down Circ-CCT3 inhibits HCC growth and metastasis in vivo and in vitro
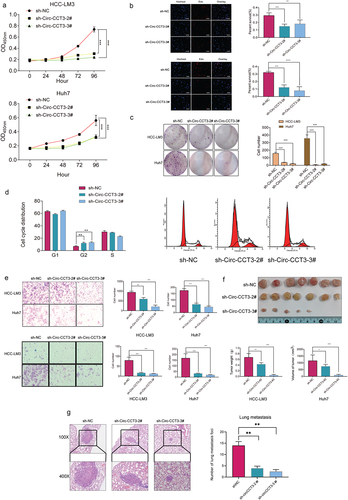
We performed a series of experiments both in vivo and in vitro to explore the biological functions of circ-CCT3 in HCC. According to the expression of circ-CCT3 in various HCC cell lines, we infected lentivirus of circ-CCT3 into HCC-LM3 and Huh7 cell lines with high expression of circ-CCT3 and constructed two stable circ-CCT3 knock-down cell lines in both kinds of HCC cell lines verified by RT-PCR. The Cell Counting Kit-8 (CCK-8) assay, EDU assay, and colony formation assays indicated that circ-CCT3 deficiency markedly impaired the proliferation ability of HCC cells (). In addition, we conducted flow cytometry to monitor cell cycle and found that HCC cells were mostly arrested at cell cycle G2 phase when the expression of circ-CCT3 was decreased, indicating that the knock-down of circ-CCT3 inhibited the cell cycle of tumour cells (). Further, cell migration and invasion were dramatically impaired by silencing of circ-CCT3 (). To verify the biological role of circ-CCT3 in vivo, we constructed tumour xenograft models by subcutaneously injecting HCC cells with stable knock-down (sh-circ-CCT3-#2, #3) into nude mice (ten mice in each group). The silencing of circ-CCT3 dramatically inhibited tumour growth (). Subsequently, to find out the potential metastatic ability of circ-CCT3 in vivo, we injected the above two stable cell lines into the lateral tail vein of nude mice. Ten weeks later, haematoxylin and eosin (H&E) staining of dissected lungs further validated that knock-down of circ-CCT3 could suppress lung metastasis().
Figure 3. Knock-down of circ-CCT3 suppresses the proliferation of HCC cell lines both in vitro and in vivo. (a) CCK8 assay indicated that knock-down circ-CCT3 inhibited the proliferation of HCC-LM3 and Huh7 cells. (b) EDU assay showed that knock-down of circ-CCT3 inhibited the proliferation of HCC-LM3 and Huh7 cells. (c) Colony formation assay showed knock-down of circ-CCT3 restrained the growth of HCC-LM3 and Huh7 cells. (d) Cell cycle analysis showed knock-down of circ-CCT3 arrested HCC-LM3 and Huh7 cells G2 phase. (e) Transwell assay indicated that knock-down of circ-CCT3 inhibited the invasion and metastasis of HCC-LM3 and Huh7 cells in vitro. (f) Knock-down of circ-CCT3 restrained the growth of xenograft mice tumour.(g) Knock-down of circ-CCT3 inhibited lung metastasis in nude mice..
Circ-CCT3 may function as a sponge for miR-378-3p
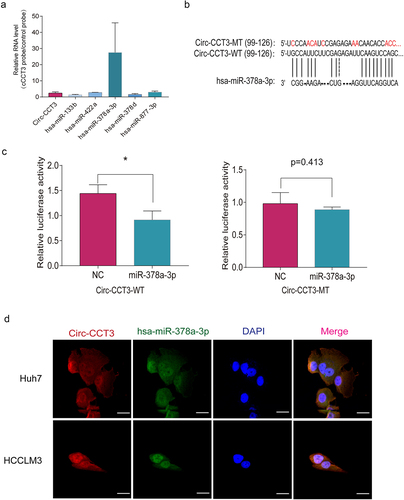
As previously reported, circRNAs primarily regulated gene expression by sponging to miRNA. We aimed to find out the possible miRNA targets for circ-CCT3, which are correlated with HCC. Through miRNA target prediction software, five potential microRNAs were predicted. In order to find out the functional miRNA which may interact with circ-CCT3 in HCC cells, we use probes specifically against circ-CCT3 to RIP and analysed the 5 candidate miRNAs in the eluted complex. We found a specific enrichment of circ-CCT3 and miR-378a-3p as compared to the controls, while the other miRNAs had very low enrichment (), indicating that miR-378a-3p is the circ-CCT3-associated miRNA in HCC cells. To further confirm the interaction between circ-CCT3 and miR-378a-3p, we predicted the binding sequence of circ-CCT3 and miR-378a-3p with BiBiserv prediction tool (https://bibiserv.cebitec.uni-bielefeld.de/sessionTimeout.jsf) (). Then, we mutated the target sites of circ-CCT3 for miR-378a-3p and constructed them into MCS of pmirGLO luciferase reporter, named circ-CCT3-WT and circ-CCT3-MT. To confirm the direct interaction between miR-378a-3p and circ-CCT3, we performed dual-luciferase reporter assays, where Huh7 cells was transfected with a reporter plasmid containing circ-CCT3-WT or circ-CCT3-MT, respectively. We found that the luciferase activity of WT group was moderately weakened, while that of Mut group had no impact (). Furthermore, the double FISH assay was performed and showed the co-localization of circ-CCT3 and miR-378a-3p (). Furthermore, the rescue experiments were carried out. As shown in Fig S1, the results revealed that the knock-down of miR-378a-3p could reverse the activity of circ-CCT3 silenced tumour cells through CCK-8 and cloning formation assays. Together, our findings supported that circ-CCT3 functioned as a sponge of miR-378a-3p.
Figure 4. Downregulation of circ-CCT3 plays a tumour suppression role in HCC via the miR-378a-3p-FLT-1pathway. (a) the 11 potential target genes of miR-378-3p. (b) Luciferase reporter assays were performed with both WT and Mut plasmids. (c, d) Expression of FLT1 following circ-CCT3 overexpression or knock-down was evaluated by RT-Qpcr and western blotting in HCC cell lines. (e) Rescue experiments were performed to confirm the regulation of FLT-1 was under the control of circ-CCT3 and miR-378a-3p. (f) Tube formation assay was performed to evaluate the angiogenesis after knock-down of circ-CCT3 in HUVECs. (g) Transwell assay was conducted to evaluate the metastasis of HCC cells after overexpressing circ-CCT3.
Downregulation of circ-CCT3 inhibits the growth, metastasis, and angiogenesis of HCC through the miR-378a-3p-FLT-1 pathway
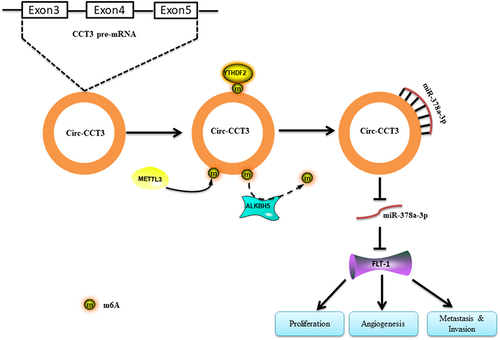
In order to understand whether circ-CCT3 exerts its oncogenic effect through miR-378a-3p, we predicted the potential target genes of miR-378-3p with miRanda genes prediction tools, and selected 11 potential genes with a score of more than 85. Next, we overexpressed miR-378a-3p mimics and measured the expression of targets by qRT-PCR. Interestingly, FLT-1 was the most downregulated (). To substantiate the requirement of FLT-1 for miR-378a-3p, luciferase reporter assays were performed with a wild-type (WT) and a mutant (Mut) plasmids. For Mut reporters, the 3’UTR sequence of FLT-1 that may be binded with miR-378a-3p were replaced, while WT reporter contained the complete sequence. As expected, the luciferase reporter activity was reduced in the WT group, but it did not remarkably change in the Mut group (). Next, we conducted qRT-qPCR and Western blot to evaluate the impact of circ-CCT3 on FLT-1. Among which, FLT-1 was significantly up-regulated following circ-CCT3 overexpression, and it was moderately down-regulated when circ-CCT3 was silenced (). Moreover, the attenuation of FLT-1 induced by miR-378a-3p could be retrieved by circ-CCT3, suggesting that the FLT-1 was under the control of circ-CCT3-miR-378a-3p axis ().
Figure 5. Circ-CCT3 serves as sponge for miR-378a-3p. (a) RNA immunoprecipitation assay showed circ-CCT3 interacted with miR-378a-3p.(b) Examples of the potential bindings between circ-CCT3 conserved sequence and miR-378a-3p. (c) Dual-Luciferase assay showed miR-378a-3p combined with wild type circ-CCT3 .(d) Dual-Luciferase assay showed miR-378a-3p combined with mutant type circ-CCT3. (e) Co-localization between miR-378-3p and circ-CCT3 observed by RNA in situ hybridization in Huh7 and HCC-LM3 cells. Nuclei were stained with DAPI.
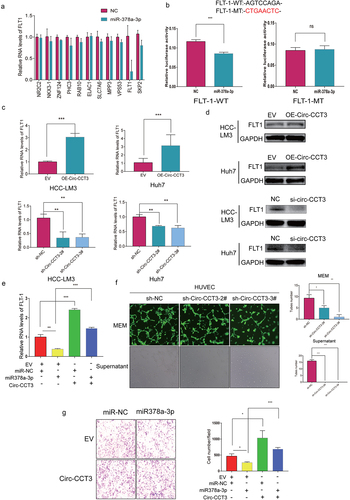
FLT1, a tyrosine-protein kinase, that acts as a cell-surface receptor for PGF, VEGFA, and VEGFB, and plays a critical role in the regulation of cell survival, angiogenesis, cell invasion, and migration [Citation24,Citation25]. As our experiments have showed that circ-CCT3 may sponge miR-378a-3p to upregulate FLT-1, we infected lentivirus of knock-down circ-CCT3 into Human Umbilical Vein Endothelial Cells (HUVECs). Tube formation assay was performed to evaluate the angiogenesis, and tube numbers highly decreased after knock-down of circ-CCT3 in HUVECs (). Meanwhile, the supernatant derived from Huh7 stable lentivirus of knock-down circ-CCT3 inhibited the tube formations compared with normal controls (). Additionally, transwell assays showed that miR-378a-3p could suppress the metastasis of HCC and that the inhibition could be blocked by circ-CCT3 overexpression (). Collectively, these observations demonstrated that circ-CCT3 promotes the HCC growth and metastasis mainly by regulating the miR378a-3p-FLT1 axis.
METTL3 and ALKBH5 mediates the m6A modification of Circ-CCT3 in HCC
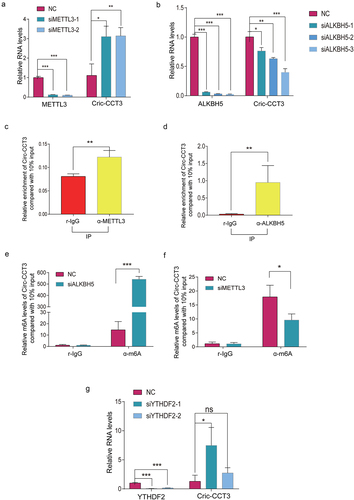
Current studies have indicated that N6-methyladenosine (m 2A) modification in circRNAs was widely spread and performed critical biological functions [Citation26,Citation27]. To illustrate whether circ-CCT3 was regulated by m6 A modification, we first use starBase (http://starbase.sysu.edu.cn/index.php) to predict RNA binding proteins associated with m6 A modification in circ-CCT3 (supplementary),which are m6A eraser ALKBH5 and FTO, m6 A writer METTL3 and METTL14, m 6A reader YTHDC1 and YTHDF1and YTHDF2. To further confirm the role of m6 A in regulation of circ-CCT3, we knocked down the above genes in Huh7 cells. Intriguingly, knock-down of METTL3 led to elevated circ-CCT3 expression and circ-CCT3 was downregulated after knocking down of ALKBH5(). However, no response was observed when silencing YTHDC1 and HNRNPA2B1. Consistent with these results, the RNA-binding protein (RBP) ALKBH5 and METTL3 were prone to binding with circ-CCT3 (). Thus, we supposed that ALKBH5 and MEETL3 may regulate circ-CCT3. To further verify that ALKBH5 and METTL3 mediate m6 A of circ-CCT3, MeRIP assay was used to analysem6 A levels of circ-CCT3. As expected, the m6 A levels of circ-CCT3 increased after knocking down of ALKBH5, the m6A levels of circ-CCT3 decreased after knocking down of METTL3 (). YTHDF2 was well known as a m6 A reader protein and frequently participated in the process of mRNA degradation [Citation28]. Consistently, knock-down of YTHDF2 caused the upregulation of circ-CCT3 expression (). After Act.D treatment, we found that silence of YTHDF2 would prolong the half-life of circ-CCT3 RNA (Fig. S2). In brief, ALKBH5 and METTL3 mediate m6 A-modification of circ-CCT3.
Figure 6. Circ-CCT3 is modified by m6A . (a) Knock-down of METTL3 increased circ-CCT3 level. (b) Knock-down of ALKBH5 decreased circ-CCT3 level. (c, d) RNA pull-down showed METTL3 and ALKBH5 combined with circ-CCT3. (e) Knock-down of ALKBH5 increased circ-CCT3 m6A level. (f) Knock-down of METTL3 decreased circ-CCT3 m6A level. (g) Knock-down of YTHDF2 increased circ-CCT3 m6A level.
Discussion
Circ-CCT3 (hsa_circ_0004680) is a covalently circular from exon 3, 4, 5 of CCT3 gene, and it plays an important role in many functions containing cell proliferation, migration, and angiogenesis in HCC, which indicated circ-CCT3 can be as a novel therapeutic target of HCC. Moreover, the overexpression of circ-CCT3 is associated with the poor prognosis of HCC. Mechanically, circ-CCT3 can sponge miR-378a-3p to regulate the expression of FLT1 and to promote HCC progression.
Clinically, we found that circ-CCT3 was overexpressed in HCC tissue, and the Kaplan–Meier analysis indicated that overexpression of circ-CCT3 was closely associated with poor prognosis in HCC after hepatectomy. Taken together, circ-CCT3 served a oncogene in HCC. Functionally, in vitro assays showed that knock-down of circ-CCT3 inhibited the proliferation of HCC cells and induced G2 cell cycle arrest. The transwell assay indicated that knock-down circ-CCT3 suppressed the invasion and metastasis of HCC cells. Further, in vivo assay demonstrated that knock-down circ-CCT3 inhibited the growth of xenograft tumours in nude mice. Subsequently, a lung metastasis model suggested knock-down circ-CCT3 restrained the lung metastasis of HCC cells.
Mechanically, circRNAs have been reported to function as miRNA sponges in most tumours [Citation29]. As circ-CCT3 predominantly distributed in cytoplasm, we predicted microRNAs could bind to circ-CCT3 and purify the circ-CCT3-associated RNAs, by circRIP, using probes specifically against circ-CCT3, and analysed the 5 candidate miRNAs in the eluted complex. The results showed a specific enrichment of circ-CCT3, miR-378a-3p compared with the controls. Meanwhile, luciferase report experiment confirmed that circ-CCT3 may function as a sponge for miR-378a-3p. Interestingly, miR-378a-3p was downregulated in HCC and was a putative biomarker for hepatocellular carcinoma diagnosis and prognosis [Citation30]. To find the downstream gene of miR-378a-3p, we predicted and selected 11 target genes, RT-PCR assay showed FLT1 was downregulated with the overexpression of miR-378a-3p mimics, luciferase report assay also confirmed that miR-378a-3p interacted with FLT-1. As previously reported, the differential up-regulation of FLT1 may have an important role in angiogenesis in HCC [Citation31]. Interestingly, the tube formation assay indicated that tube numbers highly decreased after knock-down circ-CCT3 in HUVECs. When we knocked down the expression of FLT1 in vitro, the proliferation, migration, invasion, and angiogenesis abilities of liver cancer cells were suppressed (supplementary).
m6 A plays a crucial role in regulation of circle RNA [Citation32]. We predicted m6 A sites of circ-CCT3, and RNA immunoprecipitation showed that ALKBH5 and METTL3 can bind to circ-CCT3. MeRIP assay showed that the m6 A levels of circ-CCT3 increased after knocking down ALKBH5 and YTHDF2. In brief, ALKBH5 and METTL3 mediate m6 A-modification of circ-CCT3.
To sum up, knock-down of circ-CCT3 restrains the growth, migration, and angiogenesis in HCC. Mechanistically, ALKBH5 and METTL3 mediate the m6 A modification of circ-CCT3. circ-CCT3 promotes HCC progression via sponging miR-378a-3p/FLT1 axis in the downstream ().
Conclusion
In conclusion, the present study revealed a novel circ-CCT3/miR-378a-3p/FLT1 regulatory axis in HCC development and progression. We found that the expression of circ-CCT3 is remarkably upregulated in HCC and associated with poor prognosis. Functionally and mechanistically, circ-CCT3 promotes tumour growth by the sponge activity on miR-378a-3p and improve FLT1 expression, indicating its potential tumour promoter role in liver cancer development. Further, we found that the m6 A regulator (ALKBH5 and METTL3) could bind and regulate m6 A-modification of circ-CCT3. Our work therefore suggested that a role for m6 A RNA modification in controlling specific signalling pathways and highlighted circ-CCT3 as a considerable potential prognosis predictor and therapeutic target for HCC.
Abbreviations
| HCC | = | Hepatocellular carcinoma |
| CircRNA | = | Circular RNA |
| QPCR | = | quantitative polymerase chain reaction |
| HUVEC | = | Human Umbilical Vein Endothelial Cells |
| CCT3 | = | T-complex protein 1 subunit gamma |
| RIP | = | RNA-immunoprecipitation RNA |
| MeRIP | = | Methylated (m6 A) RNA ImmunoPrecipitation RNA |
| FISH | = | Fluorescence in situ hybridization |
| FLT1 | = | Vascular endothelial growth factor receptor 1 |
| RBP | = | RNA-binding protein RNA |
| m6 A | = | N6‑Methyladenine |
| ALKBH5 | = | alk B homolog 5 alKB |
Supplemental Material
Download Zip (4.4 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
10.57760/sciencedb.j00001.00512
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2204772
Additional information
Funding
References
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. DOI:10.1038/s41572-020-00240-3
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: gLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–13. DOI:10.3322/caac.21660
- Khan AA, Liu ZK, Xu X. Recent advances in immunotherapy for hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2021;20(6):511–520.
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 2020;72(2):250–261.
- Wang M, Yu F, Li P. Circular RNAs: characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers (Basel). 2018;10(8). DOI:10.3390/cancers10080258
- Meng S, Zhou H, Feng Z, et al. Epigenetics in Neurodevelopment: emerging Role of Circular RNA. Front Cell Neurosci. 2019;13:327.
- Han TS, Hur K, Cho HS, et al. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers (Basel). 2020;12(9):2622.
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856.
- Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129.
- He L, Man C, Xiang S, et al. Circular RNAs’ cap-independent translation protein and its roles in carcinomas. Mol Cancer. 2021;20(1):119.
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. DOI:10.1016/j.jhep.2018.01.012
- Zhang C, Ding R, Sun Y, et al. Circular RNA in tumor metastasis. Mol Ther Nucleic Acids. 2021;23:1243–1257.
- Lin Z, Tang X, Wan J, et al. Functions and mechanisms of circular RNAs in regulating stem cell differentiation. RNA Biol. 2021;18(12):2136–2149.
- Cui C, Yang J, Li X, et al. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19(1):58.
- Chen J, Yang X, Liu R, et al. Circular RNA GLIS2 promotes colorectal cancer cell motility via activation of the NF-κB pathway. Cell Death Dis. 2020;11(9):788. DOI:10.1038/s41419-020-02989-7
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
- Yu L, Gong X, Sun L, et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE. 2016;11(7):e0158347.
- Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919–3931.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. DOI:10.1002/hep.29270
- Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. DOI:10.1186/s13045-016-0370-2
- Jiang T, Xia Y, Lv J, et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20(1):66. DOI:10.1186/s12943-021-01358-y
- Verduci L, Strano S, Yarden Y, et al. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680.
- Li W, Xu Y, Wang X, et al. circCCT3 Modulates Vascular Endothelial Growth Factor a and Wnt Signaling to Enhance Colorectal Cancer Metastasis Through Sponging miR-613. DNA Cell Biol. 2020;39(1):118–125. DOI:10.1089/dna.2019.5139
- Byrne AM, Bouchier-Hayes DJ, Harmey JH. Angiogenic and cell survival functions of vascular endothelial growth factor (VEGF). J Cell Mol Med. 2005;9(4):777–794.
- Ceci C, Atzori MG, Lacal PM, et al. Role of VEGFs/VEGFR-1 Signaling and its Inhibition in Modulating Tumor Invasion: experimental Evidence in Different Metastatic Cancer Models. Int J Mol Sci. 2020;21(4):1388.
- Zhang L, Hou C, Chen C, et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRnas. Mol Cancer. 2020;19(1):105. DOI:10.1186/s12943-020-01224-3
- Du A, Li S, Zhou Y, et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21(1):109. DOI:10.1186/s12943-022-01575-z
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. DOI:10.1038/nature12730
- Tao M, Zheng M, Xu Y, et al. CircRNAs and their regulatory roles in cancers. Mol Med. 2021;27(1):94.
- Qian F, Wang J, Wang Y, et al. MiR-378a-3p as a putative biomarker for hepatocellular carcinoma diagnosis and prognosis: computational screening with experimental validation. Clin Transl Med. 2021;11(2):e307. DOI:10.1002/ctm2.307
- Yamaguchi R, Yano H, Nakashima Y, et al. Expression and localization of vascular endothelial growth factor receptors in human hepatocellular carcinoma and non-HCC tissues. Oncol Rep. 2000;7(4):725–729. DOI:10.3892/or.7.4.725
- Huang X, Guo H, Wang L, et al. Recent advances in crosstalk between N6-methyladenosine (m6A) modification and circular RNAs in cancer. Mol Ther Nucleic Acids. 2022;27:947–955.