ABSTRACT
The circular RNAs (circRNAs) involved in competitive endogenous RNA (ceRNA) mechanism are critical modulators affecting pathogenesis of thyroid carcinoma (TC). The study’s goal was to investigate the effects of circ 0003747 on the biological progression of papillary thyroid cancer (PTC). Normal thyroid cells Nthy-ori3–1 and TC derived cell lines were used in our study. Sanger sequencing and RNase R treatment were utilized for validating the circular structure of circ_0003747. In our work, circ_0003747 was found to be highly expressed in TC cells. Circ_0003747 knockdown reduced TC cell viability, proliferation, migration, and invasion while increasing cell apoptosis. Circ_0003747 targeted and negatively regulated miR-338-3p expression. Besides, miR-338-3p interacted with PLCD3 to repress its expression. Overexpression of miR-338-3p inhibited TC cell progression, and PLCD3 reversed these effects. Furthermore, PLCD3 overexpression reversed the effects of circ_0003747 knockdown on TC cells. Additionally, the knockdown of circ_0003747 remarkably suppressed tumour size and growth, restrained PLCD3 expression and promoted miR-338-3p expression in nude mice. In conclusion, circ_0003747 facilitated the biological progression of TC by modulating the miR-338-3p/PLCD3 axis, and it may be a new target for TC treatment.
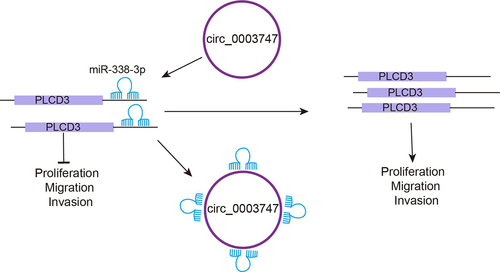
Abbreviations: TC: Thyroid carcinoma; PTC: Papillary thyroid carcinoma; CircRNAs: Circular RNAs; MiRNA: MicroRNA; EMT: Epithelial-mesenchymal transition; HCC: Hepatocellular carcinoma; PLCD3: Phospholipase C Delta 3; CeRNA: Competitive endogenous RNA
Introduction
Thyroid carcinoma (TC) accounts for about 1% of the systemic malignant tumour, which is characterized by enlarged thyroid nodules, dysphagia, enophthalmos, and other clinical symptoms [Citation1]. Due to advances in radiotherapy and chemotherapy, most TCs have low malignancy and a good prognosis [Citation2]. The common clinical treatment methods for TC are total thyroidectomy or subtotal thyroidectomy, but they are prone to recurrence and metastasis [Citation3]. The advanced or aggressive TC is resistant to radioactive iodine absorption or surgical resection and can lead to recurrence and even death [Citation2]. Therefore, it is critical in clinic to investigate the related factors that contribute to the development of TC.
As endogenous non-coding RNAs, circular RNAs (circRNAs) can interlink by covalent bonds to form a closed loop structure and regulate downstream gene expression by binding to targeted miRNAs [Citation4,Citation5]. As reported in the previous study, circular RNA N4BP2L2 (circN4BP2L2) could promote colorectal cancer process via miR-340-5p/CXCR4 pathway [Citation6]. Circular RNA TAB2 (circTAB2) inhibits lung cancer progression via the miR-3142/GLIS2 axis [Citation7]. Circular RNA DOCK1 (circDOCK1) was involved in thyroid carcinogenesis through inhibiting miR-124 expression and activating signal transduction of Janus Kinase/Signal Transducer and Activator of Transcription/Adenosine monophosphate protein kinase (JAK/STAT/AMPK) in TC cells [Citation8]. Circ_0000144 expression was notably enhanced in TC cells and exerted a cancer-promoting effect on TC via miR-217/AKT3 signal axis [Citation9]. The level of circ_0003747 was strikingly elevated in the Papillary Thyroid Carcinoma (PTC) [Citation10]. However, the molecular regulatory mechanism of circ_0003747 on TC process has not been reported. Thus, the study of ceRNA in the treatment of thyroid cancer is helpful to better understand the molecular mechanism of thyroid cancer. However, the molecular regulatory mechanism of circ_0003747 on the TC process has not been reported.
MicroRNAs (miRNAs) are small non-coding RNAs that play a vital role in the post-transcriptional regulatory process [Citation11]. It was demonstrated that dysregulation of miRNAs regulated tumour cell progression [Citation12]. The expression changes of various miRNAs might be the main mechanism of thyroid carcinogenesis [Citation13]. The expression of miR-338-3p was lower in gastric cancer tissues, and miR-338-3p restrained the migration and invasion of gastric cancer cells and suppressed epithelial–mesenchymal transition progress [Citation14]. The miR-338-3p was down-regulated in hepatocellular carcinoma (HCC) tissues, and miR-338-3p could repress tumour growth and sensitize HCC cells to sorafenib via inhibiting HIF-1α [Citation15].. As anovel tumour suppressor, miR-338-3p restrained cell proliferation and migration, whereas accelerated apoptosis of TC by targeting AKT3[Citation16].
Phospholipase C (PLC) is a key enzyme in the phosphoinositol pathway and is involved in signal transduction in eukaryotes. As a member of the PLC family, phospholipase C Delta 3 (PLCD3) plays a crucial role in multiple biological processes, such as survival of cardiomyocytes and trophoblast cells, maintenance of normal heart function, and promotion of neurite expansion [Citation17,Citation18]. PLCD3 also serves as an oncogene, which has been shown to promote tumorigenesis and progression of TC [Citation19]. In this work, a CircInteractome database analysis found that circ_0003747 had a target binding site with miR-338-3p. StarBase predicted that miR-338-3p had binding site with PLCD3. Our initial findings showed that circ_0003747 targeted miR-338-3p, which was negatively correlated with the target gene PLCD3, suggesting that circ_0003747, as a ceRNA, regulated the expression of downstream target gene PLCD3 by directly binding to miR-338-3p.
Herein, the function of circ_0003747 in TC was investigated. The aim of this study was to the investigate the function of circ_0003747 in TC and unveil the relationship between circ_0003747 and miR-338-3p and their roles in TC. Moreover, the potential regulatory mechanism involving the miR-338-3p/PLCD3 axis was analysed as well. Our study provided important evidence for circ_0003747 as a promising target for TC treatment.
Materials and methods
Cell culture and treatment
The normal thyroid cells (Nthy-ori3–1) and TC derived cell lines (TPC-1, IHH4, B-CPAP, and CAL-62) were provided by the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in the DMEM/F12 medium containing 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco, USA) at 37°C in a 5% CO2 incubator. When the cells covered about 80% of the bottom area of the culture flask, the cells were sub cultured, and the cells at logarithmic growth phase were used in the further experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted according to TRIZOL reagent (Thermo Fisher Scientific, USA). The RNA levels were quantified using a Nanodrop Spectrophotometer (IMPLEN GmbH, Germany). Reverse transcription was conducted according to instructions of reverse transcription kit (Takara, China). The relative RNA expression levels were detected by qRT-PCR assay using the SYBR Green Master PCR mix (Applied Biosystems, USA) as per the manufacturer’s protocol following previous study [Citation20]. GAPDH and U6 were used as the internal reference. The primers used in the present work are in .
Table 1. Primers used for qRT-PCR in this study.
Western blot assay
The Western blot assay was performed following the protocol outlined in [Citation20]. Total proteins were extracted according to the instructions of RIPA reagent (Beyotime, China); then, the protein concentrations were measured. Equal amounts of proteins were taken to SDS-PAGE for separation, and then electrically transferred to the PVDF membranes (Invitrogen, USA). The samples were blocked in 5% non-fat milk for 60 min. Subsequently, for the detection of PLCD3 protein, the anti-PLCD3 (1/2000, Abcam, USA) primary antibody was added and incubated overnight at 4°C. Then, the goat horseradish peroxidase-conjugated anti-rabbit IgG was added. The immunoreactive bands were visualized by the electrochemical luminescence detection system following the instructions. Image J was utilized to assess grey values of target bands with GAPDH as an internal control.
Circ_0003747 identification
Sanger sequencing and RNase R treatment were utilized for validating the circular structure of circ_0003747 [Citation21]. The total RNA was treated with the presence or absence of RNase R at 37°C for 30 min. The relative levels of RNA were assessed using qRT-PCR assay, and the data were analysed with 2−∆∆Ct method.
Cell transfection
For transfection [Citation22], TC cells (1.2 × 106 cells/well) were seeded in 6-well plate. The shRNAs targeting circ_0003747 (sh-circ_0003747) and over-expression of PLCD3 (pcDNA3.1 PLCD3) were designed by GenePharma (Shanghai, China). The miR-338-3p mimics (50 nM), inhibitor (50 nM), or vectors (2 µg/mL) provided by RiboBio (Guangzhou, China) were transfected into cells with Lipofectamine 2000 (Invitrogen, USA).
MTT assay
Cells (1 × 104 cells/well) were seeded in a 96-well plate and incubated for 24 h at room temperature. Then, MTT (20 μL) was mixed into each well at 24 h, 48 h, and 72 h points, and incubated for 4 h at 37°C. Next, 150 μL dimethyl sulphoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA) was also added into each well at 37°C with 5% of CO2 for 3 h. The absorbance was measured at 490 nm [Citation23] using a microwell reader (Analytik Jena AG, Germany) at room temperature.
Colony formation assay
Colony formation assay was conducted refer to previous study [Citation24]. TC cells were transfected for 48 h and incubated into a six-well plate at a density of 1 × 103 cells per well. Then, the cells were cultured for 14 d at 37°C with 5% CO2. The cells were fixed with paraformaldehyde for 30 min and stained with 0.1% crystal violet (Sigma, USA). Subsequently, excessive crystal violet was wiped off, and the cells were rinsed with PBS buffer. The cells were photographed, and visible colonies were counted using Image J software.
Wound healing assay
Referring to the study in [Citation25], TC cells (2 × 105 cells/mL) were incubated into a 24-well plate and cultured at 37°C in a 5% CO2 incubator. After incubating to confluence, the cells were scratched by a sterile pipette tip. Subsequently, the plates were washed with fresh medium twice and incubated. Cells migrated from the wound edge, and the distance between the two sides of the wound was monitored at 0 h and 24 h under a microscope. The degree of cell migration was quantified by the ratio of measured gap distance.
Transwell assay
The invasion ability of the TC cells was assessed by transwell assay using a chamber (Corning Costar, Inc., USA) (8 µm pore size) with the addition of 10 μL Matrigel (BD Biosciences, USA) at 1:3 dilution. After transfection, TC cells were seeded at 5 × 105/well on a polycarbonate membrane with a fibronectin coating inserted into the upper transwell chamber. Then, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min. Subsequently, the number of invasive cells was counted under a microscope in five different fields [Citation26].
Apoptosis detection
Cell apoptosis of TC cells was evaluated by flow cytometry using Annexin V-FITC/PI staining kit (Invitrogen, USA) following a published protocol [Citation26]. The TC cells were trypsinized, rinsed by cold PBS buffer and stained with Annexin V-FITC/PI solution. Then, a FACScan flow cytometer (Beckman Coulter, USA) was applied, and the cell apoptosis rate was quantitatively assessed.
Dual-luciferase reporter gene detection
The targeted binding site between circ_0003747 and miR-338-3p was predicted through the online website CircInteractome (https://circinteractome.irp.nia.nih.gov/). Meanwhile, the StarBase online website (http://starbase.sysu.edu.cn/index.php) was utilized for predicting the binding site between miR-338-3p and PLCD3. The wild-type and mutant-type full-length sequences involving the predicted binding sequence of circ_0003747 (circ_0003747-WT/circ_0003747-MUT) and PLCD3 3’-UTR (PLCD3-WT/PLCD3-MUT) were cloned into the Xhol I-Not I restriction sites of pmirGLO firefly luciferase plasmid. All constructs were sequenced to verify the integrity. Lipofectamine 3000 (Invitrogen, USA) was used for Circ_0003747-WT/circ_0003747-MUT and miR-338-3p mimics (GenePharma, China) co-transfection. Luciferase assay was then carried out using a dual-luciferase reporter assay kit according to the manufacturer’s protocol (Promega) 48 h after transfection [Citation25]. Data were normalized to Renilla Iuciferase activity and the relative activities of Luciferase were calculated. Besides, the correlation between miR-338-3p and PLCD3 was validated using the same method.
Mice xenograft assay
BALB/c female nude mice (4 weeks) were provided by the Animal Center of the Chinese Academy of Science and housed under controlled laboratory conditions. The mice were randomly divided into sh-circ_0003747 group and sh-NC group, with 6 mice in each group. TPC-1 cells (5 × 106) were treated with sh-NC or sh-circ_0003747 before being injected subcutaneously into the dorsal sides of the mice, referring to a previous study [Citation25]. The length and width of the tumour were measured every 3 d, and the tumour volume was calculated using (length × width2)/2. After euthanizing on the 28th d, the tumour tissues were imaged and prepared for further experimental use. Our protocol was approved by the Ethics Committee of Xiangya Hospital, Central South University.
Immunohistochemistry assay
The tumour tissues were fixed, embedded in paraffin, and then cut into sections of 4 μm. Specific rabbit anti-PLCD3 antibody (1/100, Santa Cruz Biotechnology, USA) was added and incubated at 4°C overnight. Then, the tissues were incubated with goat anti-rabbit secondary antibody. The complexes were detected by HRP–streptavidin conjugate and visualized using diaminobenzidine (DAB) [Citation25].
Statistical analysis
Each experimental group repeated three times and obtained three independent data. Statistical data was analysed by GraphPad Prism 8, and the values were shown as means ± standards deviation (SD). For the normally distributed data, the Student’s t-test was adopted for pairwise comparison, and multi-group comparison was conducted using one-way ANOVA. P value <0.05 indicates statistically significant. * Indicates p < 0.05; **indicates p < 0.01 and *** indicates p < 0.001.
Results
Relative expressions of circ_0003747, miR-338-3p, and PLCD3 in TC cells
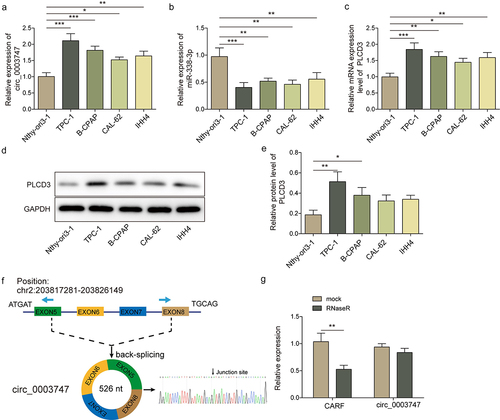
As shown in the transcription levels of circ_0003747 and PLCD3 in TC cells were higher than those in Nthy-ori3–1 cells. In contrast, miR-338-3p level was reduced in TC cells than that in Nthy-ori3–1 cells (). Moreover, PLCD3 protein level was up-regulated in TC cells (, e). Circ_0003747 was located on exon 5–8 of CARF gene of chromosome 2, with a total length of 526 nt, and its molecular characteristic structure was determined by Sanger sequencing (). After treatment with RNase R, the relative expression level of CARF was dramatically reduced, and circ_0003747 level was basically unchanged, indicating that circ_0003747 was a stable circular structure ().
Figure 1. The expressions of circ_0003747 and PLCD3 are increased, miR-338-3p expression is reduced in TC cells.
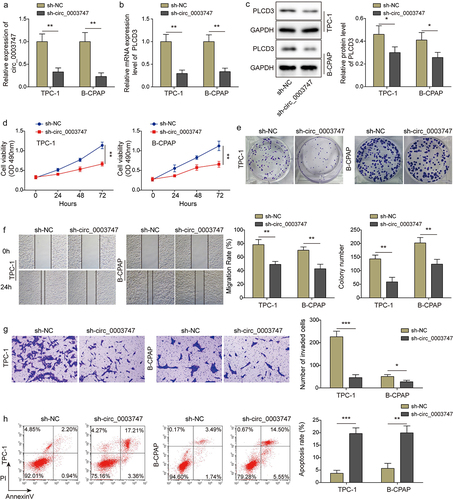
Knockdown of circ_0003747 represses TC cell proliferation and migration but promotes apoptosis
To investigate the effect of circ_0003747 on TC progression, sh-circ_0003747 was transfected into TPC-1 and B-CPAP cells. As shown in ) the levels of circ_0003747 and PLCD3 were reduced in TC cells after sh-circ_0003747 transfection. Besides, PLCD3 protein expression was strikingly inhibited in transfected TC cells (). Knockdown of circ_0003747 inhibited viability and proliferation of TC cells (). In addition, the downregulation of circ_0003747 significantly depleted the migration and invasion of TC cells (). Additionally, the apoptosis rate was notably elevated in TC cells after knockdown of circ_0003747 (). The above results illuminated that circ_0003747 could positively regulate PLCD3 expression and circ_0003747 knockdown restrained progression of TC cells.
Figure 2. Circ_0003747 knockdown suppresses TC cells progression.
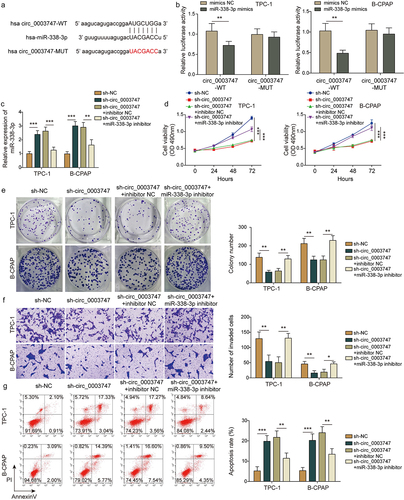
Circ_0003747 serves as a miR-338-3p sponge in TC cells
CircInteractome database analysis found that circ_0003747 had a target-binding site with miR-338-3p (). miR-338-3p mimics restrained luciferase activities in circ_0003747-WT cells, but there were no significant changes in circ_0003747-MUT cells (). The expression of miR-338-3p in TC cells was elevated after circ_0003747 was knocked down and enormously declined after miR-338-3p inhibitor reintroduction (). The cell viability, proliferation ability, migration, and invasion activity were drastically restrained in sh-circ_0003747 transfected TC cells, but these effects were counteracted due to the miR-338-3p inhibitor reintroduction (). Additionally, sh-circ_0003747 transfection promoted apoptosis of TC cells, while miR-338-3p inhibitor weakened this effect (). Taken together, circ_0003747 acted as a sponge of miR-338-3p and regulated the progression of TC cells.
Figure 3. Circ_0003747 negatively regulates miR-338-3p expression in TC cells.
miR-338-3p targets PLCD3 and inhibits TC process
StarBase predicted that miR-338-3p had binding site with PLCD3 (), and the potential regulatory mechanism of miR-338-3p and PLCD3 in TC was explored. The luciferase activities were remarkably suppressed after miR-338-3p mimics transfection in PLCD3-WT cells, but there were no significant changes in PLCD3-MUT cells (). Meanwhile, miR-338-3p mimic transfection could repressed the mRNA expression level of PLCD3 in TC cells (). Furthermore, the overexpression of PLCD3 reversed the down-regulation effect of miR-338-3p on PLCD3 gene expression (). Cell viability, proliferation ability, migration, and invasion activity were markedly restrained in TC cells with miR-338-3p mimic transfection, but the effect was abrogated due to PLCD3 overexpression ( and Fig. S2). In addition, miR-338-3p mimic transfection promoted apoptosis of TC cells, while simultaneous upregulation of PLCD3 attenuated this effect (). Collectively, it was illustrated that miR-338-3p targeted PLCD3 to negatively regulate its expression and repressed TC cell progression.
Figure 4. MiR-338-3p enrichment inhibits TC cell progression through targeting PLCD3.
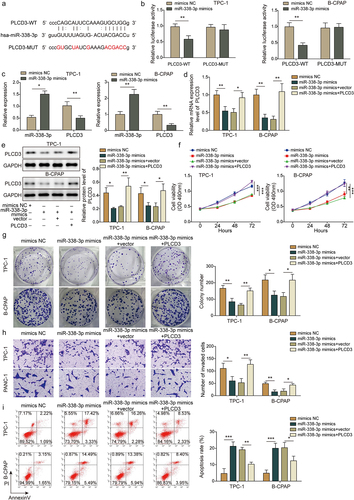
Overexpression of PLCD3 eliminates the effects of circ_0003747 knockdown on TC cells
To further address whether circ_0003747 involved in TC cell malignant phenotypes via regulating PLCD3 expression, TC cells were co-transfected with sh-circ_0003747 and PLCD3. PLCD3 overexpression attenuated the inhibition effect of sh-circ_0003747 on its gene expression (). In addition, upregulation of PLCD3 eliminated the inhibition effects of cell viability, proliferation ability, migration, and invasion activity by sh-circ_0003747 transfections in TC cells (). Moreover, the promoted apoptosis of TC cells by sh-circ_0003747 transfections were weakened after PLCD3 reintroduction (). These findings indicated that circ_0003747 affected TC cell progression by regulating PLCD3 expression.
Figure 5. Overexpression of PLCD3 reverses the effects of circ_0003747 knockdown.
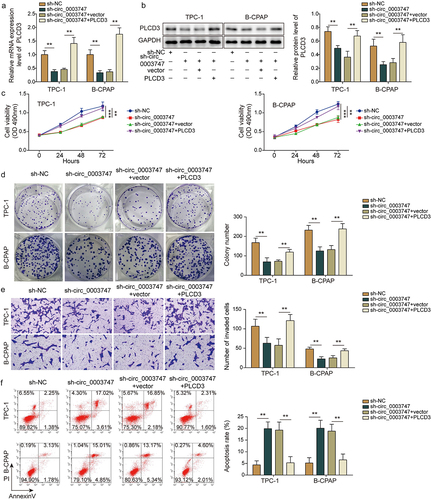
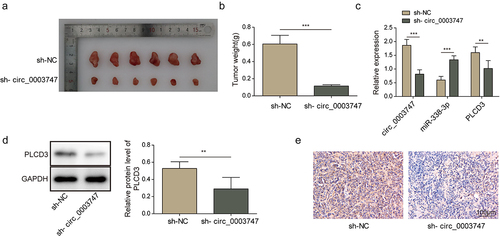
Circ_0003747 facilitates tumour growth in vivo through miR-338-3p/PLCD3 signalling axis
To validate the effects of circ_0003747 on tumorigenesis in vivo, TC cells were subcutaneously injected into the nude mice. Knockdown of circ_0003747 dramatically suppressed the tumour size and growth in vivo (). Circ_0003747 knockdown restrained the mRNA expression levels of PLCD3 and promoted miR-338-3p expression (). Furthermore, western blot and immunohistochemistry assays showed that PLCD3 protein expression was inhibited in circ_0003747 knockdown group in comparison with that in the NC group (). Based on these data, circ_0003747 plays a crucial role in TC cell progression by modulating the miR-338-3p/PLCD3 axis.
Figure 6. Circ_0003747 knockdown inhibits tumour growth in nude mice.
Discussion
TC is the most common malignancy of the endocrine system, and the prognosis and treatment at diagnosis depends on the type and stage of the tumour. The treatment strategies for TC can improve survival rate and reduce recurrence rate in patients with TC [Citation27]. However, the molecular regulatory mechanism of TC is complex, and the exploration of specific biomarkers will be beneficial to the diagnosis and treatment of TC in clinic. Therefore, it is critical to investigate the signal cascade in TC process. In our study, it was proved for the first time that circ_0003747-modulated miR-338-3p/PLCD3 axis was responsible for regulating the progress of TC cells. This work provides a new insight into the possible pathogenesis of TC.
CircRNAs are produced by back-splicing and regulate the expression of downstream genes through binding to miRNAs [Citation28]. The dysregulation of the circRNA-miRNA-mRNA regulatory network may contribute to multiple disease progression, including the development of cancer [Citation29]. As demonstrated by Chu et al., circRNA runx family transcription factor 1 (circRUNX1) regulated the progression of PTC cells through the miR-296-3p/DDHD2 axis [Citation30]. The level of circ_0003747 was strikingly elevated in Papillary Thyroid Carcinoma (PTC) [Citation10]. Therefore, we paid attention to the role of circ_0003747 in TC. In our research, it was found that circ_0003747 was highly expressed in TC cells and circ_0003747 knockdown inhibited TC cell progression.
MiRNAs play a vital role in human cancer and can be used as biomarkers for the diagnosis, prognosis, and treatment of cancer [Citation31]. miR-199a-3p impaired cell proliferation and migration in PTC-derived cell lines [Citation32]. In our research, miR-338-3p was found to be lowly expressed in TC, and the overexpression of miR-338-3p inhibited TC cell progression, implying that miR-338-3p may be a suppressor of biological behaviours at least in the TC process. A large number of evidences have shown that circRNAs can perform their biological function as ceRNAs by competitively reducing the contents of active miRNAs, which may be prognostic and predictive factors for clinical assessment of disease condition and outcome [Citation33,Citation34]. Circ_0009294 was reported to promote tumour cell proliferation, migration, and invasion via binding to miR-451a in Papillary Thyroid Carcinoma [Citation35]. As reported, in hepatocellular carcinoma cells, circ_104566 promoted cell proliferation, migration, and invasion, and knockdown of circ_104566 restrained HCC progression via miR-338-3p/FOXP1 axis [Citation36]. Consistent with the above studies, in the present work, miR-338-3p was verified as target of circ_0003747, and miR-338-3p inhibitor abolished the effects of circ_0003747 knockdown on TC cell progression.
PLCD3 plays vital roles in many biological processes, such as promoting neurite extension, cardiomyocyte survival, and normal heart function, regulating tumorigenesis and progression of TC. To investigate the function of miR-338-3p in TC, we focused on PLCD3 because PLCD3 has been proved to be an oncogene in TC cell lines [Citation19]. Our findings discovered that miR-338-3p directly targeted PLCD3 to negatively regulate its expression. To further verify whether circ_0003747 could function by miR-338-3p, we co-transfected sh-circ_0003747 and PLCD3 in TC cells. We found that interference with circ_0003747 and then overexpression of PLCD3 might reverse the role of circ_0003747 in TC cell progression. Moreover, the knockdown of circ_0003747 remarkably suppressed tumour size and growth, restrained PLCD3 expression and promoted miR-338-3p expression in nude mice. Existing reports have suggested that circRNAs served mainly as an miRNA molecular sponge and contained numbers and types of miRNA response elements at varying degrees [Citation37]. For example, a previous report by Zhang W, et al. [Citation38] demonstrated that circ_102002 might promote PTC process by sponging miR-488-3p and upregulating HAS2 expression. The same results were obtained in our study, where we reported for the first time that circ_0003747 could directly bind to miR-338-3p and increase PLCD3 expression, thus promoting TC cell progression.
In summary, circ_0003747 was overexpressed in PTC cells. The proliferation and migration of TC cells were repressed, and apoptosis was promoted by circ_0003747 knockdown. Circ_0003747 mainly sponged miR-338-3p and negatively regulated its expression. In addition, PLCD3 was identified as a target of miR-338-3p in PTC. Circ_0003747 promoted the TC process in a miR-338-3p/PLCD3-dependent manner. These findings about the signal axis of circ_0003747/miR-338-3p/PLCD3 may help to understand the PTC pathogenesis and improve clinical treatment. However, it is still unknown whether circ_0003747 can regulate other miRNAs in TC. Future study should conduct to investigate the in-depth mechanisms of circ_0003747.
Authors’ contribution
XLD: Conceptualization; Writing-original draft; Methodology; Formal analysis;
FDX: Investigation: Funding acquisition; Project administration; Writing-review & editing;
XYL: Investigation: Funding acquisition; Project administration; Writing-review & editing.
All authors have read and approved the final version of this manuscript to be published.
Availability of data and material
All data generated or analysed during this study are included in this article. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable. This article does not contain any studies with human participants performed by any of the authors.
Ethical approval
Our protocol was approved by the Ethics Committee of Xiangya Hospital, Central South University.
Supplemental Material
Download Zip (2.2 MB)Acknowledgments
We would like to express our sincere gratitude to the reviewers for their constructive comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2210339.
Additional information
Funding
References
- Topstad D, Dickinson JA. Thyroid cancer incidence in Canada: a national cancer registry analysis. CMAJ Open. 2017;5(3):E612–14.
- Xu J, Li Z, Su Q, et al. TRIM29 promotes progression of thyroid carcinoma via activating P13K/AKT signaling pathway. Oncol Rep. 2017;37(3):1555–1564.
- Jisheng H, Nan Z, Rui K, et al. Sun: total thyroidectomy as primary surgical management for thyroid disease: surgical therapy experience from 5559 thyroidectomies in a less-developed region. World J Surg Oncol. 2016;14(1):20.
- Zhang P, Ming Y, Ye Q, et al. Comprehensive circRNA expression profile during ischemic postconditioning attenuating hepatic ischemia/reperfusion injury. Sci Rep. 2019;9(1). DOI:10.1038/s41598-018-36443-8
- Liu Q, Zhang X, Hu X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Rep. 2016;6:22572.
- Yang K, Wang Y, Zhang F. et al. CircN4BP2L2 promotes colorectal cancer growth and metastasis through regulation of the miR-340-5p/CXCR4 axis. Lab Invest. 2022;102(1):38–47.
- Man W, Cui, Y, Li, J, et al. circTAB2 inhibits lung cancer proliferation, migration and invasion by sponging miR-3142 to upregulate GLIS2. Apoptosis. 2023;28:471–484.
- Cui W, Xue J. Circular RNA DOCK1 downregulates microRNA-124 to induce the growth of human thyroid cancer cell lines. Biofactors. 2020;46(4):591–599.
- Fan Y, Shi H, Hu Y, et al. Circ_0000144 facilitates the progression of thyroid cancer via the miR-217/AKT3 pathway. J Gene Med. 2020;22(12):e3269.
- Lan X, Xu J, Chen C, et al. The landscape of circular RNA expression profiles in papillary thyroid carcinoma based on RNA sequencing. Cell Physiol Biochem. 2018;47(3):1122–1132.
- Zinovyev A, Morozova N, Gorban AN, et al. Mathematical modeling of microRna–mediated mechanisms of translation repression. Adv Exp Med Biol. 2013;774:189–224.
- Wang J, Zeng H, Li H, et al. MicroRNA-101 inhibits growth, proliferation and migration and induces apoptosis of breast cancer cells by targeting sex-determining region Y-Box 2. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2017;43:717.
- Saiselet M, Pita JM, Augenlicht A, et al. Maenhaut C: miRNA expression and function in thyroid carcinomas: a comparative and critical analysis and a model for other cancers. Oncotarget. 2016;7(32). DOI:10.18632/oncotarget.9655
- Huang N, Wu Z, Lin L, et al. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6(17):15222–15234.
- Xu H, Zhao L, Fang Q, et al. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1α. PLoS ONE. 2014;9(12):e115565.
- Sui GQ, Fei D, Guo F, et al. MicroRNA-338-3p inhibits thyroid cancer progression through targeting AKT3. Am J Cancer Res. 2017;7(5):1177–1187.
- Nakamura Y, Kanemaru K, Kojima R, et al. Simultaneous loss of phospholipase Cδ1 and phospholipase Cδ3 causes cardiomyocyte apoptosis and cardiomyopathy. Cell Death Amp Dis. 2014;5(5):e1215.
- Nakamura Y, Hamada Y, Fujiwara T, et al. Phospholipase C-1 and -3 are essential in the trophoblast for placental development. Molcellbiol. 2005;25(24):10979.
- Lin L, Wen J, Lin B, et al. Phospholipase C Delta 3 inhibits apoptosis and promotes proliferation, migration, and invasion of thyroid cancer cells via Hippo pathway. Acta Biochim Biophys Sin (Shanghai). 2021;53(4):481–491.
- Tian W, Yang XL, Yang H, et al. Exosomal miR-338-3p suppresses non-small-cell lung cancer cells metastasis by inhibiting CHL1 through the MAPK signaling pathway. Cell Death Amp Dis. 2021;12(11):1030.
- Wang L, Li B, Yi X, et al. Circ_smad4 promotes gastric carcinogenesis by activating wnt/β‐catenin pathway. Cell Proliferation. 2021;54(3):e12981.
- Shen XM, Tang J, Huang YZ, et al. CircRNF111 contributes to adipocyte differentiation by elevating PPARγ expression via miR-27a-3p. Epigenetics. 2022;18:1–15.
- Wang ZL, Xia FD, Feng TC, et al. Otud6B-AS1 inhibits viability, migration, and invasion of thyroid carcinoma by targeting miR-183-5p and miR-21. Front Endocrinol. 2020;11:136.
- Li XS, Shen FZ, Huang LY, et al. Jin BZ: lncRNA small nucleolar RNA host gene 20 predicts poor prognosis in glioma and promotes cell proliferation by silencing P21. Onco Targets Ther. 2019;12:805–814.
- Cheng JL, Li DJ, Lv MY, et al. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Genet Ther. 2021;28(7–8):850–863.
- Wang DW, Lou XQ, Liu ZL, et al. LncRNA SNHG1 protects SH-SY5Y cells from hypoxic injury through miR-140-5p/Bcl-XL axis. Int J Neurosci. 2021;131(4):336–345.
- Plodkowski RA, Lee EJ, Huang MG. Diagnosis and treatment of patients with thyroid cancer. Am Health Drug Benefits. 2015;8(1):30–40.
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847.
- Cai H, Hu B, Ling J, et al. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10(6):1690–1702.
- Chu J, Tao L, Yao T, et al. Circular RNA circRUNX1 promotes papillary thyroid cancer progression and metastasis by sponging MiR-296-3p and regulating DDHD2 expression. Cell Death Amp Dis. 2021;12(1):112.
- Zhou J, Wei W. Analysis of microRNA expression profiling identifies microRNA-503 regulates metastatic function in hepatocellular cancer cell. J Surg Oncol. 2011;104(3):278–283.
- Minna E, Romeo P, Cecco LD, et al. MiR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget. 2014;5(9):2513–2528.
- Long F, Lin Z, Li L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20(1). DOI:10.1186/s12943-021-01318-6
- Liu L, Wu SQ, Zhu X, et al. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma. 2019;66(5):736–745.
- Zhang ZY, Xia FD, Yao L, et al. circSSU72 promotes cell proliferation, migration and invasion of papillary thyroid carcinoma cells by targeting miR-451a/S1PR2 axis. Front Cell Dev Biol. 2022;10:817028.
- Liu G, Guo W, Rao M, et al. circRNA hsa_circ_104566 sponged miR-338-3p to promote hepatocellular carcinoma progression. Cell Transplant. 2020;29(1):963689720963948.
- Qu SB, Yang XS, Li XL, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Zhang W, Liu T, Li TS, et al. Hsa_circrna_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. Cancer Genet Ther. 2021;28(3–4):279–293.
